Transcriptome Analysis of the Development of Pedicel Abscission Zone in Tomato
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Sample Preparation
2.2. AZ Diameter Measurement
2.3. mRNA Sequencing by Illumina HiSeq
2.4. Differential Expression Analysis
2.5. Principal Component Analysis
2.6. Correlation Analysis
2.7. GO and KEGG Enrichment Analysis
2.8. Quantitative RT-PCR
3. Results
3.1. Development of Pedicel AZ in Tomato
3.2. Transcriptome Analysis of Pedicel AZ at Different Development Stages in Tomatoes
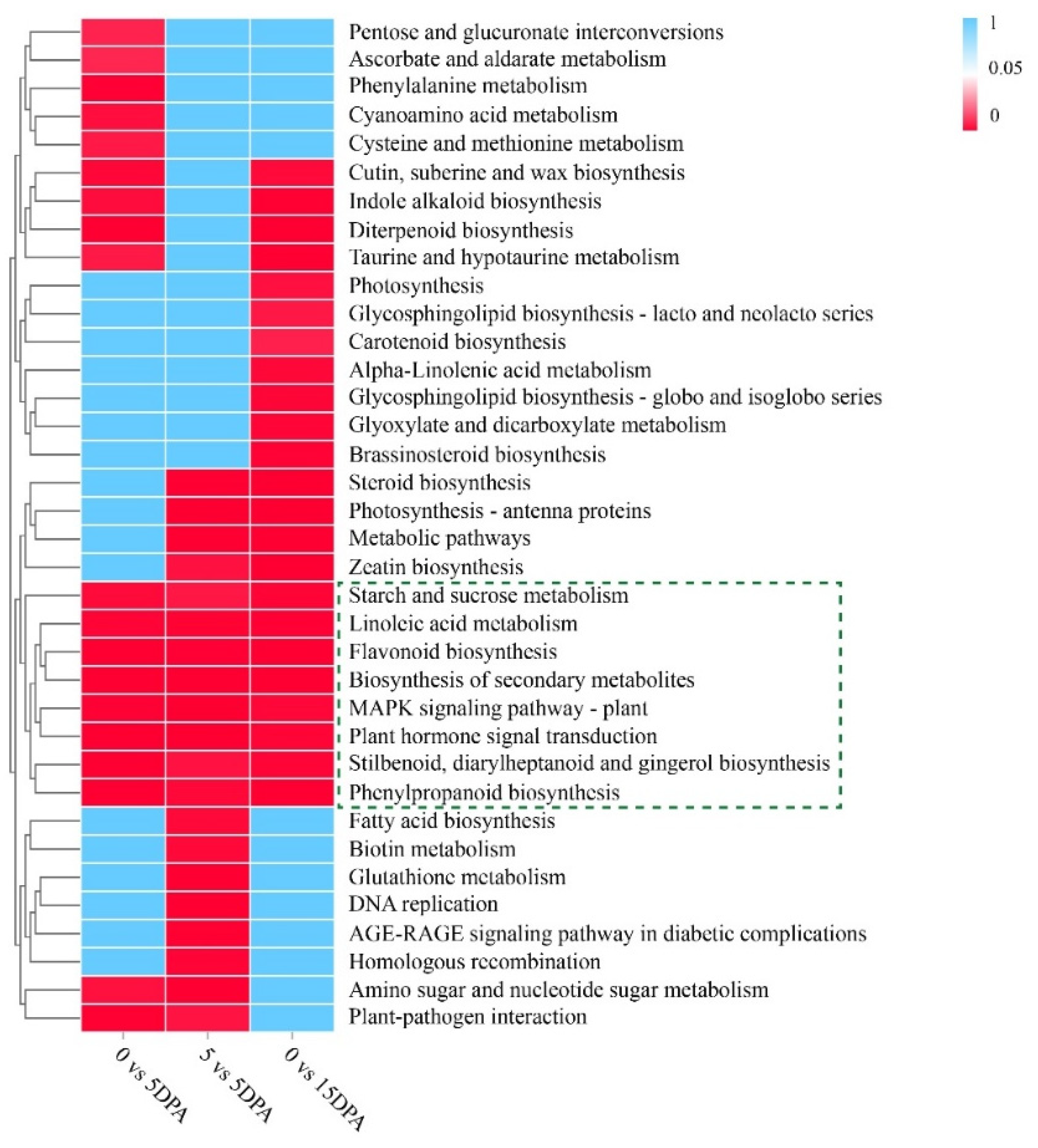
3.3. KEGG Enrichment Analysis of the DEGs
3.4. Differentially Expressed Genes That Were Selected under Three Comparisons
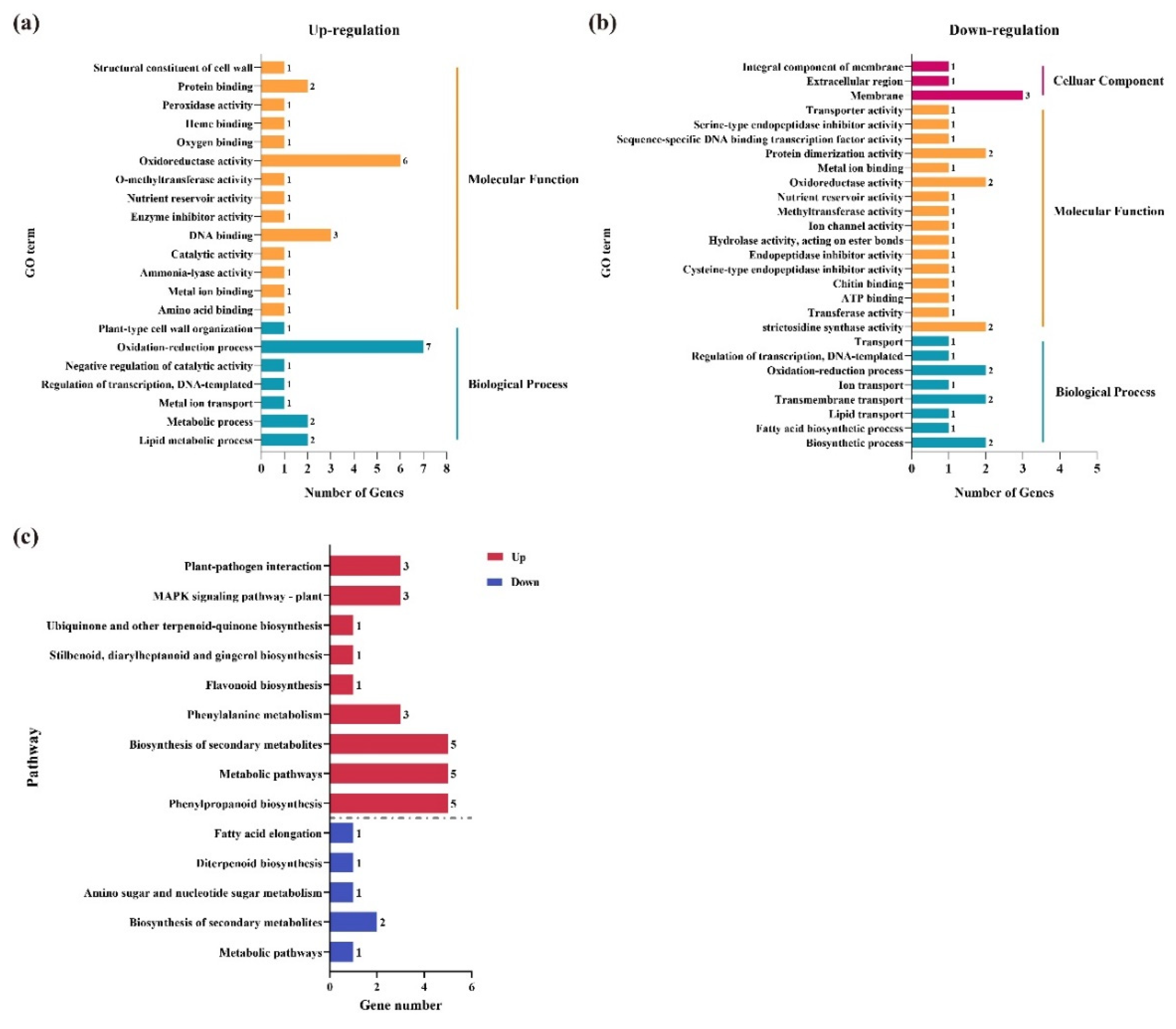
3.5. GO and KEGG Enrichment Analysis of the Key DEGs under Three Comparisons
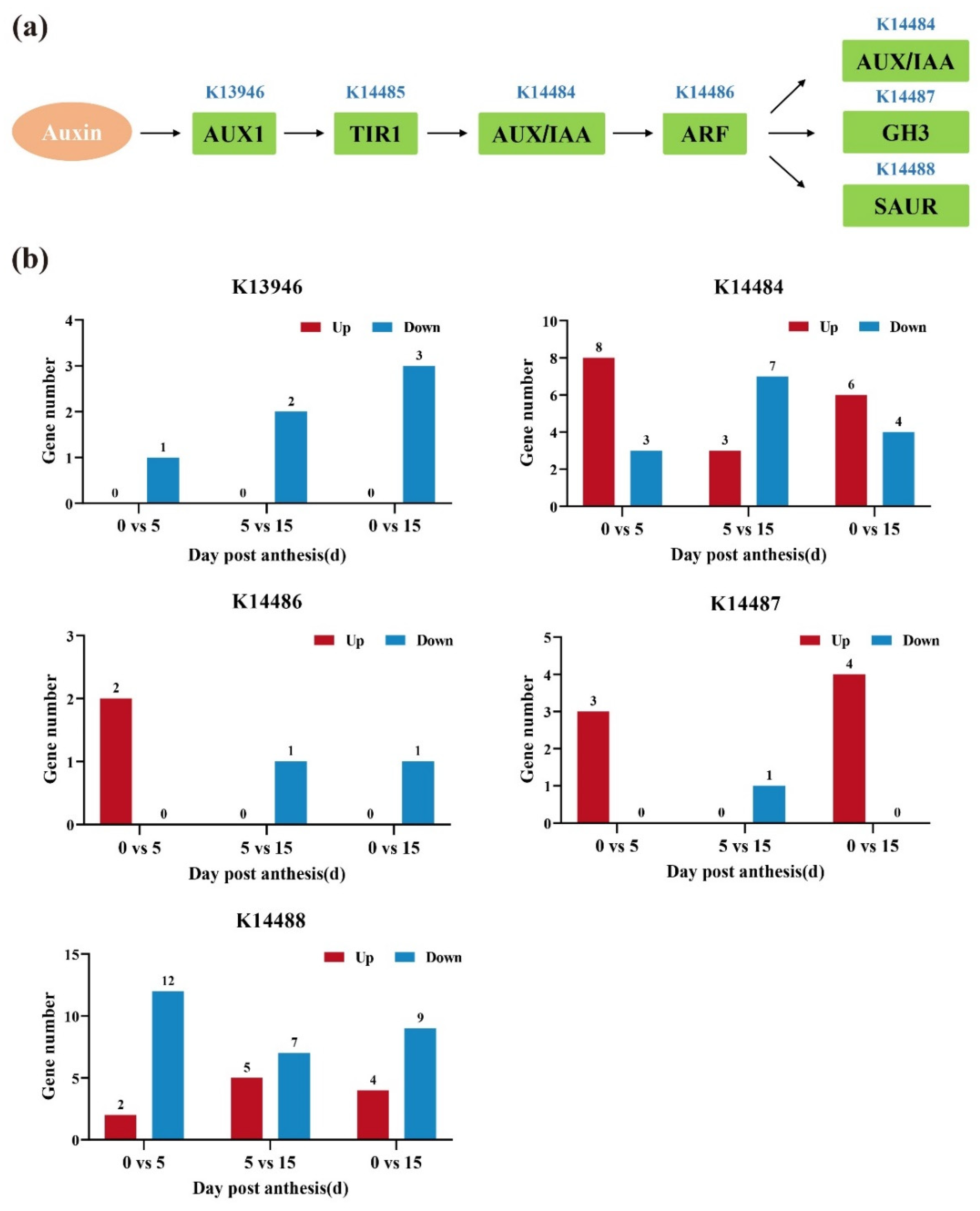
3.6. Analysis of DEGs Involved in the Auxin Signaling Pathway
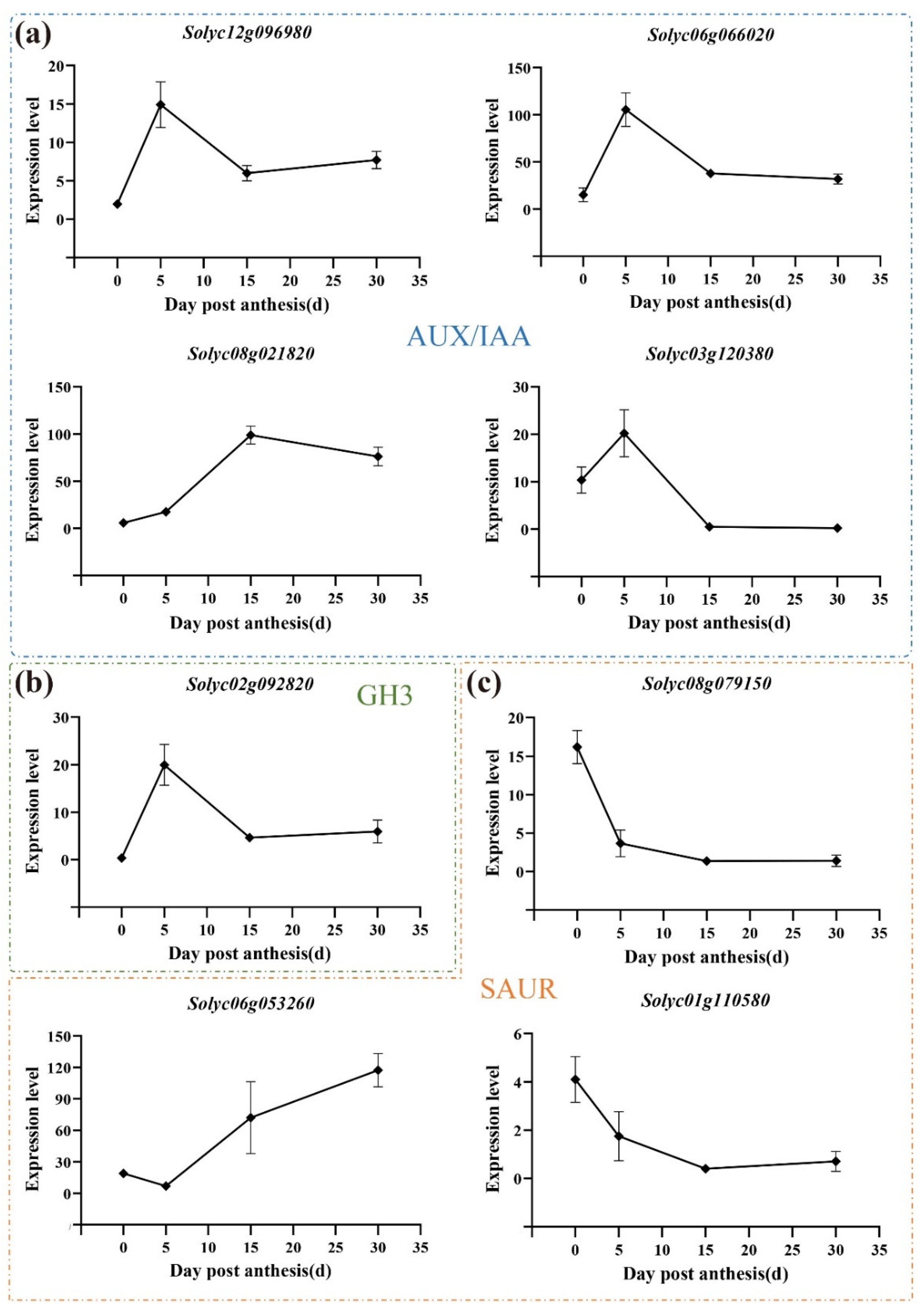
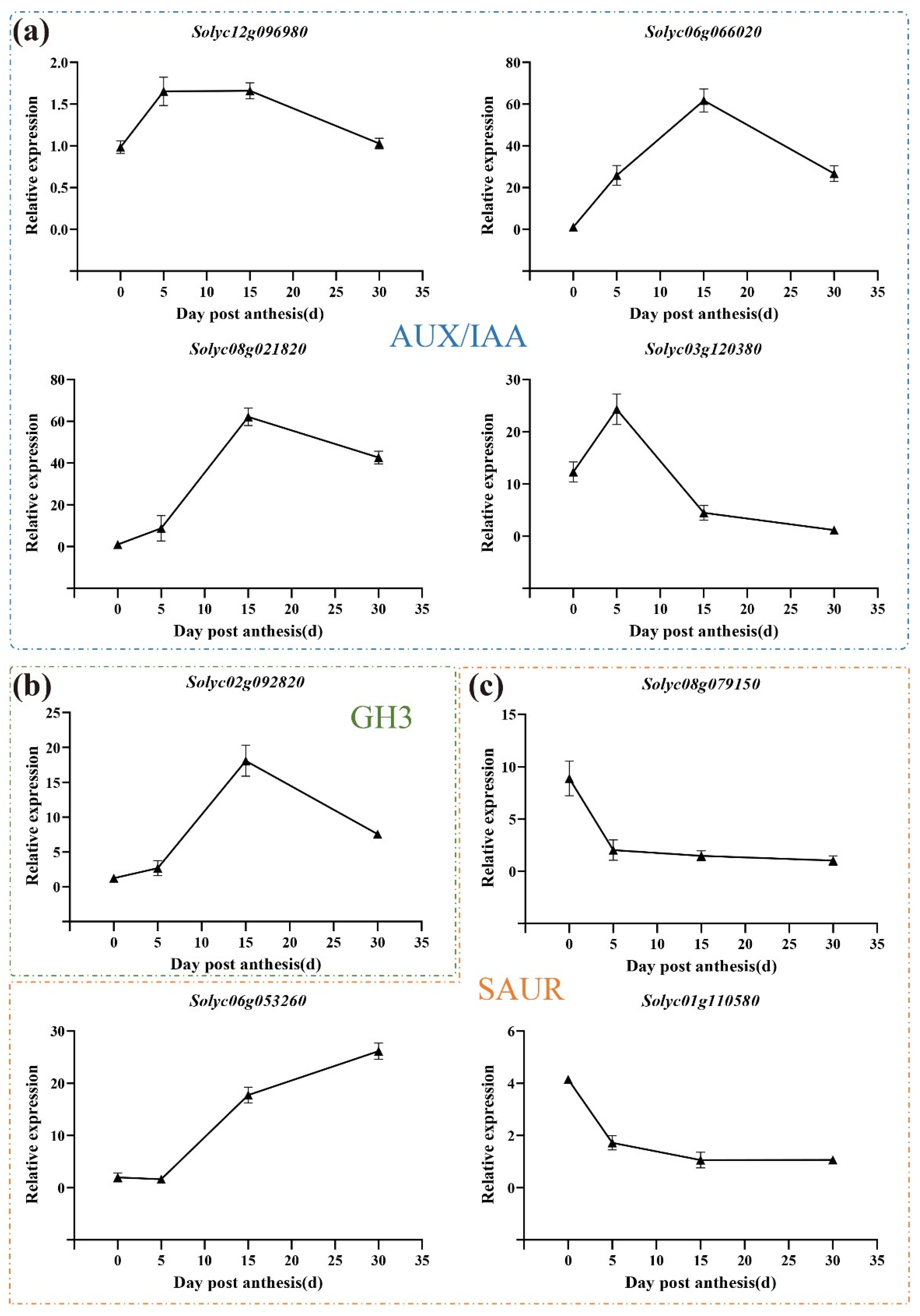
3.7. Expression Pattern of Auxin-Related Genes during All Three Development Stages
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Brown, H.S.; Addicott, F.T. The anatomy of experimental leaflet abscission in phaseolus vulgaris. Am. J. Bot. 1950, 37, 650–656. [Google Scholar] [CrossRef]
- Gawadi, A.G.; Avery, G.S., Jr. Leaf abscission and the so-called “abscission layer. Am. J. Bot. 1950, 37, 172–180. [Google Scholar] [CrossRef]
- Sexton, R.; Roberts, J.A. Cell Biology of Abscission. Annu. Rev. Plant Physiol. 1982, 33, 133–162. [Google Scholar] [CrossRef]
- Taylor, J.E.; Whitelaw, C.A. Signals in abscission. New Phytol. 2001, 151, 323–340. [Google Scholar] [CrossRef]
- Tabuchi, T. Development of the Abscission Zone in the Pedicels of Potato Plants. J. Jpn. Soc. Hortic. Sci. 1998, 67, 693–695. [Google Scholar] [CrossRef]
- Mao, L.; Begum, D.; Chuang, H.-W.; Budiman, M.A.; Szymkowiak, E.J.; Irish, E.E.; Wing, R.A. JOINTLESS is a MADS-box gene controlling tomato flower abscissionzone development. Nature 2000, 406, 910–913. [Google Scholar] [CrossRef] [PubMed]
- Pinyopich, A.; Ditta, G.S.; Savidge, B.; Liljegren, S.J.; Baumann, E.; Wisman, E.; Yanofsky, M.F. Assessing the redundancy of MADS-box genes during carpel and ovule development. Nature 2003, 424, 85–88. [Google Scholar] [CrossRef]
- Budiman, M.A.; Chang, S.B.; Lee, S.; Yang, T.J.; Zhang, H.B.; de Jong, H.; Wing, R.A. Localization of jointless-2 gene in the centromeric region of tomato chromosome 12 based on high resolution genetic and physical mapping. Theor. Appl. Genet. 2004, 108, 190–196. [Google Scholar] [CrossRef]
- McKim, S.M.; Stenvik, G.E.; Butenko, M.A.; Kristiansen, W.; Cho, S.K.; Hepworth, S.R.; Aalen, R.B.; Haughn, G.W. The BLADE-ON-PETIOLE genes are essential for abscission zone formation in Arabidopsis. Development 2008, 135, 1537–1546. [Google Scholar] [CrossRef]
- Nakano, T.; Kimbara, J.; Fujisawa, M.; Kitagawa, M.; Ihashi, N.; Maeda, H.; Kasumi, T.; Ito, Y. MACROCALYX and JOINTLESS interact in the transcriptional regulation of tomato fruit abscission zone development. Plant Physiol. 2012, 158, 439–450. [Google Scholar] [CrossRef]
- Wu, X.M.; Yu, Y.; Han, L.B.; Li, C.L.; Wang, H.Y.; Zhong, N.Q.; Yao, Y.; Xia, G.X. The tobacco BLADE-ON-PETIOLE2 gene mediates differentiation of the corolla abscission zone by controlling longitudinal cell expansion. Plant Physiol. 2012, 159, 835–850. [Google Scholar] [CrossRef] [PubMed]
- Couzigou, J.M.; Magne, K.; Mondy, S.; Cosson, V.; Clements, J.; Ratet, P. The legume NOOT-BOP-COCH-LIKE genes are conserved regulators of abscission, a major agronomical trait in cultivated crops. New Phytol. 2016, 209, 228–240. [Google Scholar] [CrossRef] [PubMed]
- Roldan, M.V.G.; Périlleux, C.; Morin, H.; Huerga-Fernandez, S.; Latrasse, D.; Benhamed, M.; Bendahmane, A. Natural and induced loss of function mutations in SlMBP21 MADS-box gene led to jointless-2 phenotype in tomato. Sci. Rep. 2017, 7, 4402. [Google Scholar] [CrossRef] [PubMed]
- Perrot-Rechenmann, C. Cellular responses to auxin: Division versus expansion. Cold Spring Harb. Perspect. Biol. 2010, 2, a001446. [Google Scholar] [CrossRef]
- Saniewski, M.; Góraj-Koniarska, J.; Gabryszewska, E.; Miyamoto, K.; Ueda, J. Auxin effectively induces the formation of the secondary abscission zone in Bryophyllum calycinum Salisb. (Crassulaceae). Acta Agrobotanica 2016, 69, 1660. [Google Scholar] [CrossRef][Green Version]
- Barbez, E.; Dünser, K.; Gaidora, A.; Lendl, T.; Busch, W. Auxin steers root cell expansion via apoplastic pH regulation in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2017, 114, E4884–E4893. [Google Scholar] [CrossRef]
- Du, M.; Spalding, E.P.; Gray, W.M. Rapid Auxin-Mediated Cell Expansion. Annu. Rev. Plant Biol. 2020, 71, 379–402. [Google Scholar] [CrossRef]
- McQueen-Mason, S.; Durachko, D.M.; Cosgrove, D.J. Two endogenous proteins that induce cell wall extension in plants. Plant Cell 1992, 4, 1425–1433. [Google Scholar]
- Nishitani, K.; Vissenberg, K. Roles of the XTH Protein Family in the Expanding Cell. In The Expanding Cell; Verbelen, J.-P., Vissenberg, K., Eds.; Springer: Berlin/Heidelberg, Germany, 2007; pp. 89–116. [Google Scholar]
- Hocq, L.; Pelloux, J.; Lefebvre, V. Connecting Homogalacturonan-Type Pectin Remodeling to Acid Growth. Trends Plant Sci. 2017, 22, 20–29. [Google Scholar] [CrossRef]
- Tao, Y.; Ferrer, J.L.; Ljung, K.; Pojer, F.; Hong, F.; Long, J.A.; Li, L.; Moreno, J.E.; Bowman, M.E.; Ivans, L.J.; et al. Rapid synthesis of auxin via a new tryptophan-dependent pathway is required for shade avoidance in plants. Cell 2008, 133, 164–176. [Google Scholar] [CrossRef]
- Sandalio, L.M.; Rodríguez-Serrano, M.; Romero-Puertas, M.C. Leaf epinasty and auxin: A biochemical and molecular overview. Plant Sci. 2016, 253, 187–193. [Google Scholar] [CrossRef] [PubMed]
- Spartz, A.K.; Lee, S.H.; Wenger, J.P.; Gonzalez, N.; Itoh, H.; Inzé, D.; Peer, W.A.; Murphy, A.S.; Overvoorde, P.J.; Gray, W.M. The SAUR19 subfamily of SMALL AUXIN UP RNA genes promote cell expansion. Plant J. 2012, 70, 978–990. [Google Scholar] [CrossRef] [PubMed]
- Spartz, A.K.; Ren, H.; Park, M.Y.; Grandt, K.N.; Lee, S.H.; Murphy, A.S.; Sussman, M.R.; Overvoorde, P.J.; Gray, W.M. SAUR Inhibition of PP2C-D Phosphatases Activates Plasma Membrane H+-ATPases to Promote Cell Expansion in Arabidopsis. Plant Cell 2014, 26, 2129–2142. [Google Scholar] [CrossRef] [PubMed]
- Lin, W.; Zhou, X.; Tang, W.; Takahashi, K.; Pan, X.; Dai, J.; Ren, H.; Zhu, X.; Pan, S.; Zheng, H.; et al. TMK-based cell-surface auxin signalling activates cell-wall acidification. Nature 2021, 599, 278–282. [Google Scholar] [CrossRef]
- Hewitt, F.; Hough, T.; O’Neill, P.; Sasse, J.; Williams, E.; Rowan, K. Effect of Brassinolide and other Growth Regulators on the Germination and Growth of Pollen Tubes of Prunus avium using a Multiple Hanging-drop Assay. Funct. Plant Biol. 1985, 12, 201–211. [Google Scholar] [CrossRef]
- Zurek, D.M.; Clouse, S.D. Molecular cloning and characterization of a brassinosteroid-regulated gene from elongating soybean (Glycine max L.) epicotyls. Plant Physiol. 1994, 104, 161–170. [Google Scholar] [CrossRef]
- Oh, M.H.; Honey, S.H.; Tax, F.E. The Control of Cell Expansion, Cell Division, and Vascular Development by Brassinosteroids: A Historical Perspective. Int. J. Mol. Sci. 2020, 21, 1743. [Google Scholar] [CrossRef]
- Schaller, G.E.; Street, I.H.; Kieber, J.J. Cytokinin and the cell cycle. Curr. Opin. Plant Biol. 2014, 21, 7–15. [Google Scholar] [CrossRef]
- Fu, X.; Harberd, N.P. Auxin promotes Arabidopsis root growth by modulating gibberellin response. Nature 2003, 421, 740–743. [Google Scholar] [CrossRef]
- Zhan, Y.; Qu, Y.; Zhu, L.; Shen, C.; Feng, X.; Yu, C. Transcriptome analysis of tomato (Solanum lycopersicum L.) shoots reveals a crosstalk between auxin and strigolactone. PLoS ONE 2018, 13, e0201124. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.; Guo, H.; Zhang, Y.; Song, Y.; Pi, E.; Yu, C.; Zhang, L.; Dong, M.; Zheng, B.; Wang, H.; et al. Identification of potential genes that contributed to the variation in the taxoid contents between two Taxus species (Taxus media and Taxus mairei). Tree Physiol. 2017, 37, 1659–1671. [Google Scholar] [CrossRef] [PubMed]
- Anders, S.; Huber, W. Differential expression analysis for sequence count data. Genome Biol. 2010, 11, R106. [Google Scholar] [CrossRef] [PubMed]
- Anders, S.; Huber, W. Differential Expression of RNA-Seq Data at the Gene Level—The DESeq Package; European Molecular Biology Laboratory (EMBL): Heidelberg, Germany, 2012. [Google Scholar]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Winch, S.; Pritchard, J. Acid-induced wall loosening is confined to the accelerating region of the root growing zone. J. Exp. Bot. 1999, 50, 1481–1487. [Google Scholar] [CrossRef]
- Gendreau, E.; Traas, J.; Desnos, T.; Grandjean, O.; Caboche, M.; Höfte, H. Cellular basis of hypocotyl growth in Arabidopsis thaliana. Plant Physiol. 1997, 114, 295–305. [Google Scholar] [CrossRef]
- Chapman, E.J.; Greenham, K.; Castillejo, C.; Sartor, R.; Bialy, A.; Sun, T.P.; Estelle, M. Hypocotyl transcriptome reveals auxin regulation of growth-promoting genes through GA-dependent and -independent pathways. PLoS ONE 2012, 7, e36210. [Google Scholar] [CrossRef] [PubMed]
- Dharmasiri, N.; Dharmasiri, S.; Weijers, D.; Lechner, E.; Yamada, M.; Hobbie, L.; Ehrismann, J.S.; Jürgens, G.; Estelle, M. Plant development is regulated by a family of auxin receptor F box proteins. Dev. Cell 2005, 9, 109–119. [Google Scholar] [CrossRef]
- Majda, M.; Robert, S. The Role of Auxin in Cell Wall Expansion. Int. J. Mol. Sci. 2018, 19, 951. [Google Scholar] [CrossRef]
- Liscum, E.; Reed, J.W. Genetics of Aux/IAA and ARF action in plant growth and development. Plant Mol. Biol. 2002, 49, 387–400. [Google Scholar] [CrossRef]


| Gene ID | Fpkm-F | Fpkm-5 | Fpkm-15 | Significant | Description |
|---|---|---|---|---|---|
| Solyc12g006750 | 0 | 3.10 | 173.10 | Up | Glutathione S-transferase zeta 1 |
| Solyc04g080540 | 0.44 | 27.95 | 129.26 | Up | DNA polymerase epsilon catalytic subunit A, putative |
| Solyc08g079270 | 0.77 | 45.09 | 130.87 | Up | R2R3MYB transcription factor 42 |
| Solyc09g074890 | 1.61 | 72.17 | 239.28 | Up | Rapid alkalinization factor |
| Solyc02g090120 | 2.89 | 117.58 | 240.35 | Up | Low quality: Inositol 1,4,5-trisphosphate receptor-interacting protein-like 2 |
| Solyc07g008240 | 0.62 | 16.34 | 41.20 | Up | Non-symbiotic hemoglobin 1 |
| Solyc11g012690 | 1.03 | 22.45 | 57.97 | Up | Heavy metal transport/detoxification superfamily protein |
| Solyc10g005010 | 1.03 | 20.42 | 49.83 | Up | NAC domain-containing protein, putative |
| Solyc07g055490 | 1.39 | 24.04 | 76.64 | Up | Cytochrome P450 |
| Solyc06g072550 | 0.59 | 9.60 | 109.96 | Up | CASP-like protein |
| Solyc07g042460 | 0.32 | 5.22 | 109.96 | Up | Respiratory burst oxidase, putative |
| Solyc11g069900 | 3.00 | 48.67 | 158.53 | Up | Alpha/beta hydrolase superfamily protein |
| Solyc02g094230 | 1.09 | 17.46 | 58.70 | Up | Low quality: Dynein heavy chain 1, axonemal |
| Solyc11g012705 | 0.68 | 10.00 | 28.37 | Up | Pollen Ole e 1 allergen and extensin family protein |
| Solyc07g053840 | 3.94 | 56.85 | 183.40 | Up | Leucine-rich repeat receptor-like protein kinase family |
| Solyc05g056170 | 8.12 | 109.98 | 223.71 | Up | Phenylalanine ammonia-lyase 2 |
| Solyc02g093270 | 8.59 | 115.79 | 313.50 | Up | Caffeoyl-CoA O-methyltransferase |
| Solyc04g076220 | 0.20 | 2.40 | 14.38 | Up | AT hook motif DNA-binding family protein |
| Solyc09g065890 | 0.30 | 3.43 | 7.79 | Up | Alpha/beta hydrolase superfamily protein |
| Solyc01g091700 | 0.13 | 1.44 | 4.22 | Up | F-box family protein |
| Solyc04g071085 | 3.09 | 33.10 | 351.47 | Up | Hydroxyproline-rich glycoprotein |
| Solyc12g042460 | 0.39 | 4.15 | 15.96 | Up | 4-coumarate—CoA ligase-like |
| Solyc01g111880 | 0.86 | 8.16 | 19.77 | Up | MAP kinase kinase kinase 11 |
| Solyc09g015350 | 0.35 | 3.19 | 8.37 | Up | Trichome birefringence-like 34 |
| Solyc12g005820 | 0.97 | 7.68 | 21.12 | Up | Basic helix-loop-helix (bHLH) DNA-binding superfamily protein |
| Solyc11g072920 | 0.14 | 1.09 | 2.26 | Up | Peroxidase |
| Solyc07g065330 | 1.74 | 12.82 | 26.00 | Up | Germin-like protein |
| Solyc01g099620 | 0.24 | 1.68 | 5.43 | Up | Respiratory burst oxidase, putative |
| Solyc06g084130 | 0.70 | 4.52 | 9.43 | Up | Bax inhibitor |
| Solyc02g084570 | 20.62 | 131.85 | 303.65 | Up | Cytochrome P450 family protein |
| Solyc07g054780 | 22.19 | 123.00 | 440.86 | Up | Low quality: Wound-responsive family protein |
| Solyc12g006560 | 0.88 | 4.83 | 51.75 | Up | Early nodulin-93 |
| Solyc12g088800 | 0.34 | 1.88 | 5.92 | Up | Lipase |
| Solyc10g084070 | 13.35 | 71.89 | 181.87 | Up | Low quality: Endoglucanase |
| Solyc12g044950 | 12.54 | 67.54 | 851.34 | Up | Lipid desaturase |
| Solyc08g081690 | 1.37 | 7.19 | 25.45 | Up | NADPH oxidase |
| Solyc06g075660 | 1.79 | 9.13 | 19.08 | Up | MYB family protein |
| Solyc12g099190 | 15.84 | 79.65 | 173.51 | Up | Low quality: Invertase inhibitor |
| Gene ID | Fpkm-F | Fpkm-5 | Fpkm-15 | Significant | Description |
|---|---|---|---|---|---|
| Solyc05g015880 | 87.66 | 6.25 | 0.63 | Down | Regulator of nonsense transcript protein |
| Solyc09g091550 | 98.55 | 7.78 | 1.99 | Down | S-adenosyl-L-methionine: salicylic acid carboxyl methyltransferase |
| Solyc03g034390 | 1647.24 | 141.12 | 13.80 | Down | Lipid transfer protein |
| Solyc09g006010 | 218.07 | 19.50 | 2.62 | Down | Pathogenesis-related protein 1 |
| Solyc06g007370 | 71.51 | 6.80 | 2.54 | Down | Low quality: 6,7-dimethyl-8-ribityllumazine synthase |
| Solyc06g051680 | 33.61 | 3.20 | 0 | Down | Protein early flowering 4 |
| Solyc01g099150 | 33.94 | 3.35 | 0.65 | Down | Desiccation-related protein PCC13-62 |
| Solyc09g072750 | 505.81 | 50.45 | 12.27 | Down | Low quality: Carbohydrate-binding X8 domain superfamily protein |
| Solyc03g114560 | 65.92 | 7.058 | 0.28 | Down | Strictosidine synthase-like protein, putative |
| Solyc09g092715 | 186.43 | 20.23 | 3.48 | Down | RGG repeats nuclear RNA-binding protein B |
| Solyc03g005910 | 46.05 | 5.19 | 0.52 | Down | Lipase, GDSL |
| Solyc01g088300 | 117.22 | 13.72 | 1.38 | Down | Germin-like protein 1 |
| Solyc03g020040 | 718.69 | 84.41 | 21.22 | Down | Pin-II type proteinase inhibitor 69 |
| Solyc09g005260 | 63.83 | 7.70 | 1.82 | Down | Vacuolar cation/proton exchanger 3 |
| Solyc00g071180 | 21.55 | 2.77 | 0 | Down | Multicystatin |
| Solyc12g039070 | 73.40 | 10.58 | 0.87 | Down | Strictosidine synthase |
| Solyc07g056670 | 190.42 | 28.74 | 10.91 | Down | Gibberellin 2-oxidase 2 |
| Solyc02g078850 | 60.27 | 9.39 | 1.20 | Down | Glycine-rich protein |
| Solyc07g007240 | 202.25 | 32.30 | 7.11 | Down | Metallocarboxypeptidase inhibitor |
| Solyc03g113560 | 32.06 | 5.24 | 0.21 | Down | Basic helix-loop-helix (bHLH) DNA-binding superfamily protein |
| Solyc09g092760 | 223.22 | 36.78 | 6.60 | Down | RNA-binding (RRM/RBD/RNP motifs) family protein |
| Solyc09g014940 | 39.74 | 7.34 | 1.38 | Down | Wound-induced protein 1 |
| Solyc08g082210 | 68.83 | 12.83 | 2.75 | Down | AP2/EREBP transcription factor |
| Solyc03g113250 | 123.55 | 23.47 | 4.69 | Down | Nitrate transporter 1:2 |
| Solyc03g097820 | 27.69 | 5.27 | 1.82 | Down | bHLH transcription factor 022 |
| Solyc12g005400 | 36.63 | 7.07 | 2.64 | Down | Cyclic nucleotide-gated ion channel, putative |
| Solyc08g067030 | 2070.64 | 400.99 | 21.90 | Down | Transmembrane protein, putative (protein of unknown function, DUF642) |
| Solyc12g055810 | 10.89 | 2.13 | 0.46 | Down | P-loop containing nucleoside triphosphate hydrolase superfamily protein |
| Solyc10g009150 | 209.96 | 42.47 | 0 | Down | Organ-specific protein S2 |
| Solyc05g009270 | 15.68 | 3.22 | 0.96 | Down | 3-ketoacyl-CoA synthase |
| Solyc10g074440 | 138.10 | 29.38 | 4.57 | Down | Chitinase |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dong, X.; Wang, Y.; Yan, Y.; Wang, H. Transcriptome Analysis of the Development of Pedicel Abscission Zone in Tomato. Horticulturae 2022, 8, 865. https://doi.org/10.3390/horticulturae8100865
Dong X, Wang Y, Yan Y, Wang H. Transcriptome Analysis of the Development of Pedicel Abscission Zone in Tomato. Horticulturae. 2022; 8(10):865. https://doi.org/10.3390/horticulturae8100865
Chicago/Turabian StyleDong, Xiufen, Yue Wang, Yanyan Yan, and Huasen Wang. 2022. "Transcriptome Analysis of the Development of Pedicel Abscission Zone in Tomato" Horticulturae 8, no. 10: 865. https://doi.org/10.3390/horticulturae8100865
APA StyleDong, X., Wang, Y., Yan, Y., & Wang, H. (2022). Transcriptome Analysis of the Development of Pedicel Abscission Zone in Tomato. Horticulturae, 8(10), 865. https://doi.org/10.3390/horticulturae8100865






