An Integrated Analysis of Transcriptome and Metabolism Reveals an Inhibitory Effect of Low Light on Anthocyanin Biosynthesis in Purple cai-tai (Brassicarapa L. var. purpurea)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials and Growth Condition
2.2. Low Light Treatment
2.3. RNA Extraction
2.4. Library Preparation and Illumina Hiseq4000 Sequencing
2.5. Read Mapping and Differential Expression Analysis
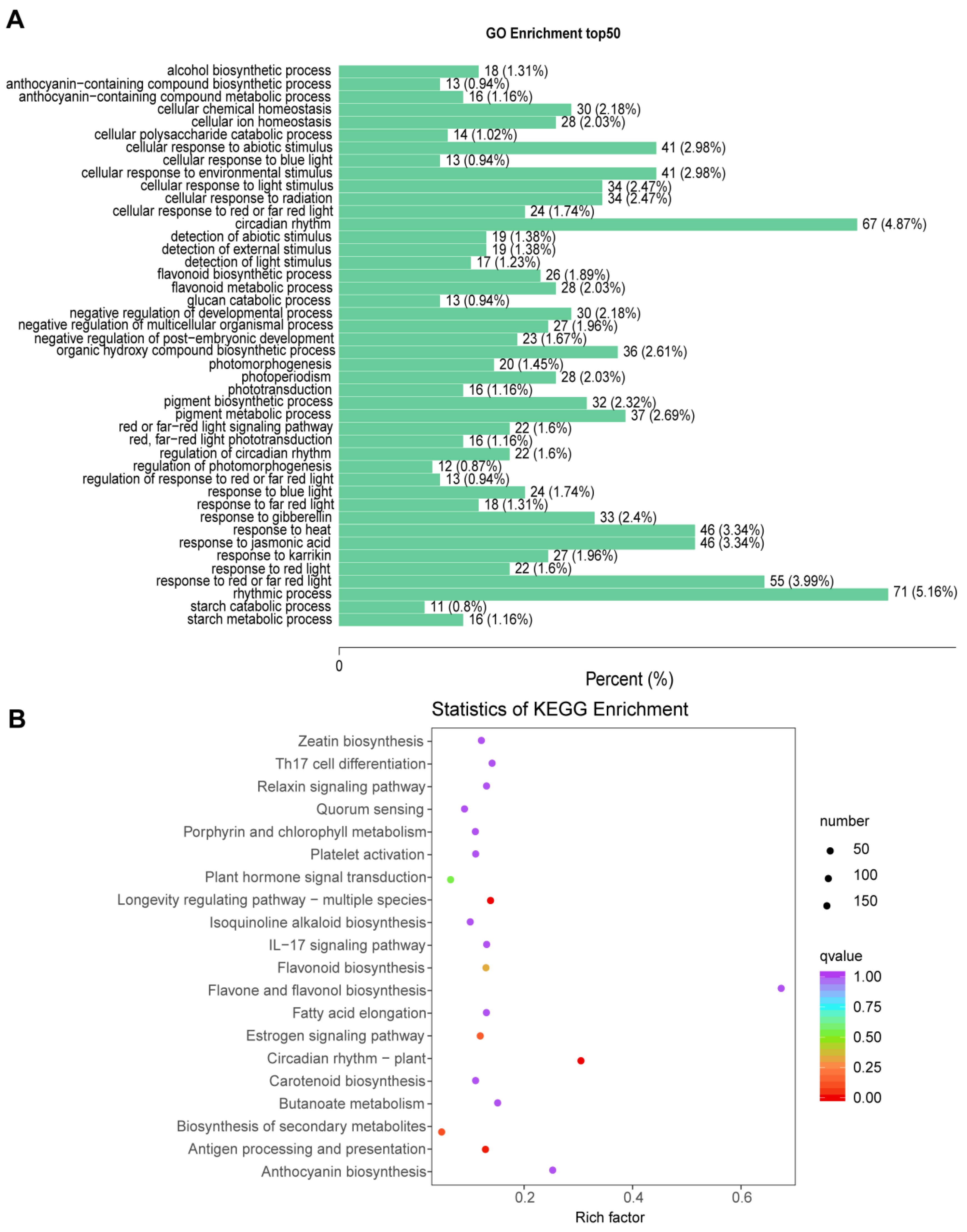
2.6. GO and KEGG Analysis
2.7. Non-Targeted Metabolism Analysis
2.8. q-RT-PCR Analysis
2.9. Statistical Analysis
3. Results
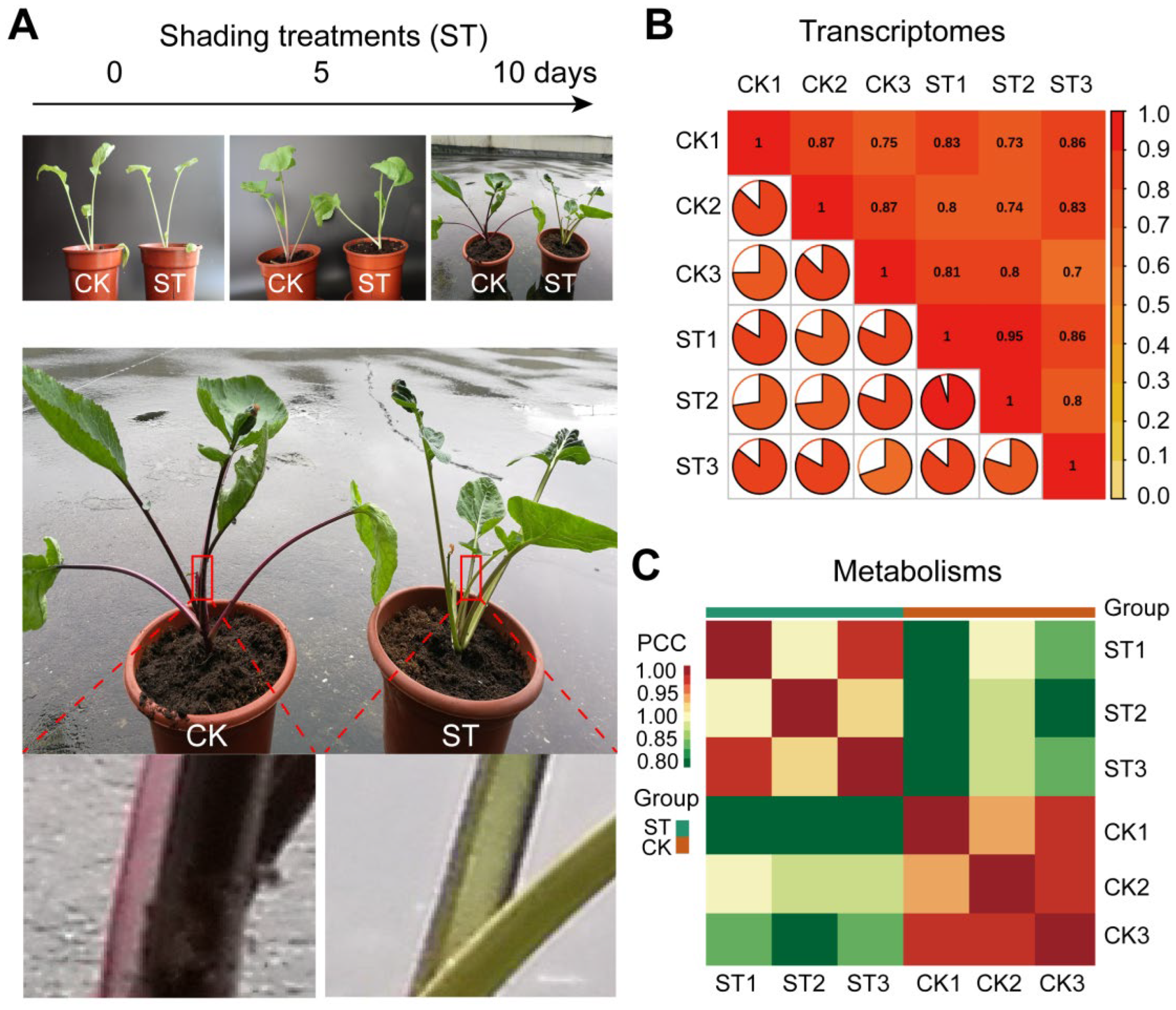
3.1. Anthocyanin Is Inhibited by Low Light in Petiole of XH1 cai-tai Seedlings
3.2. Scaffolds Assemble and Quality Control of Transcriptomes
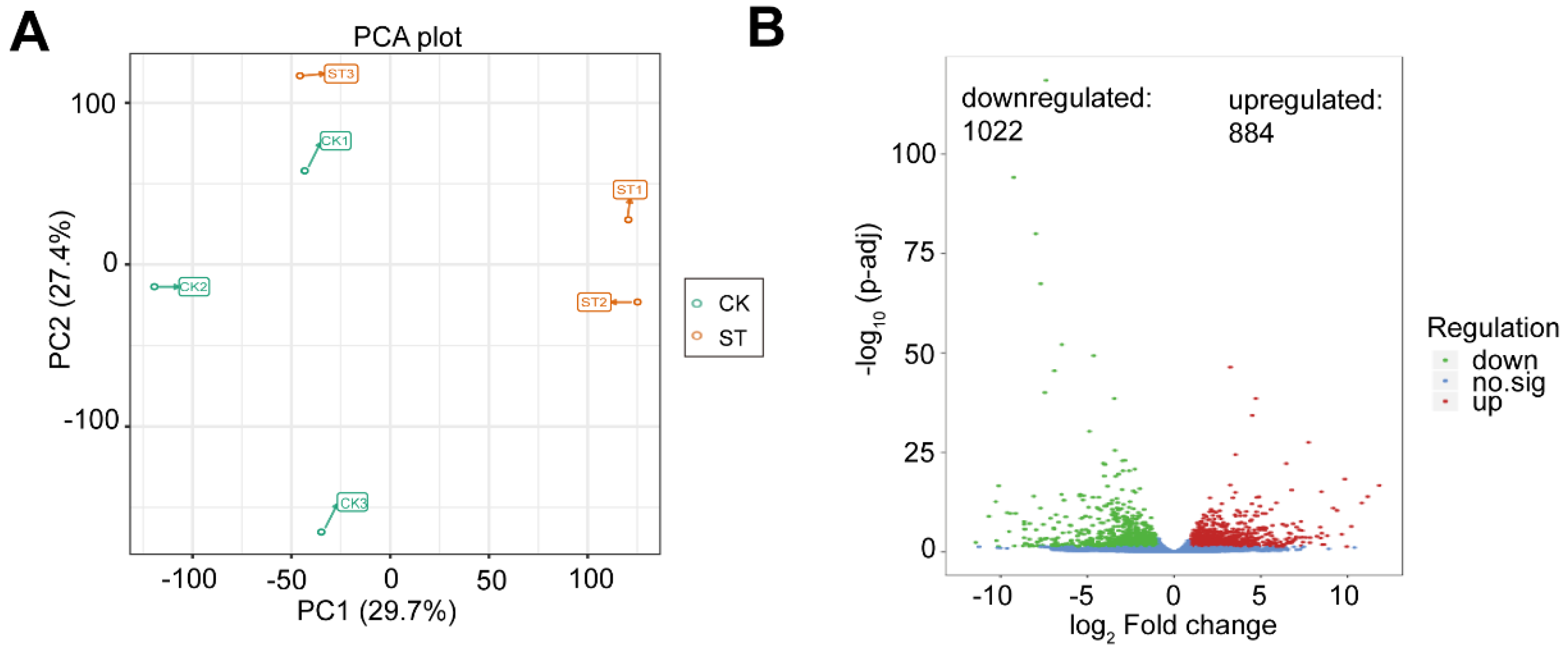
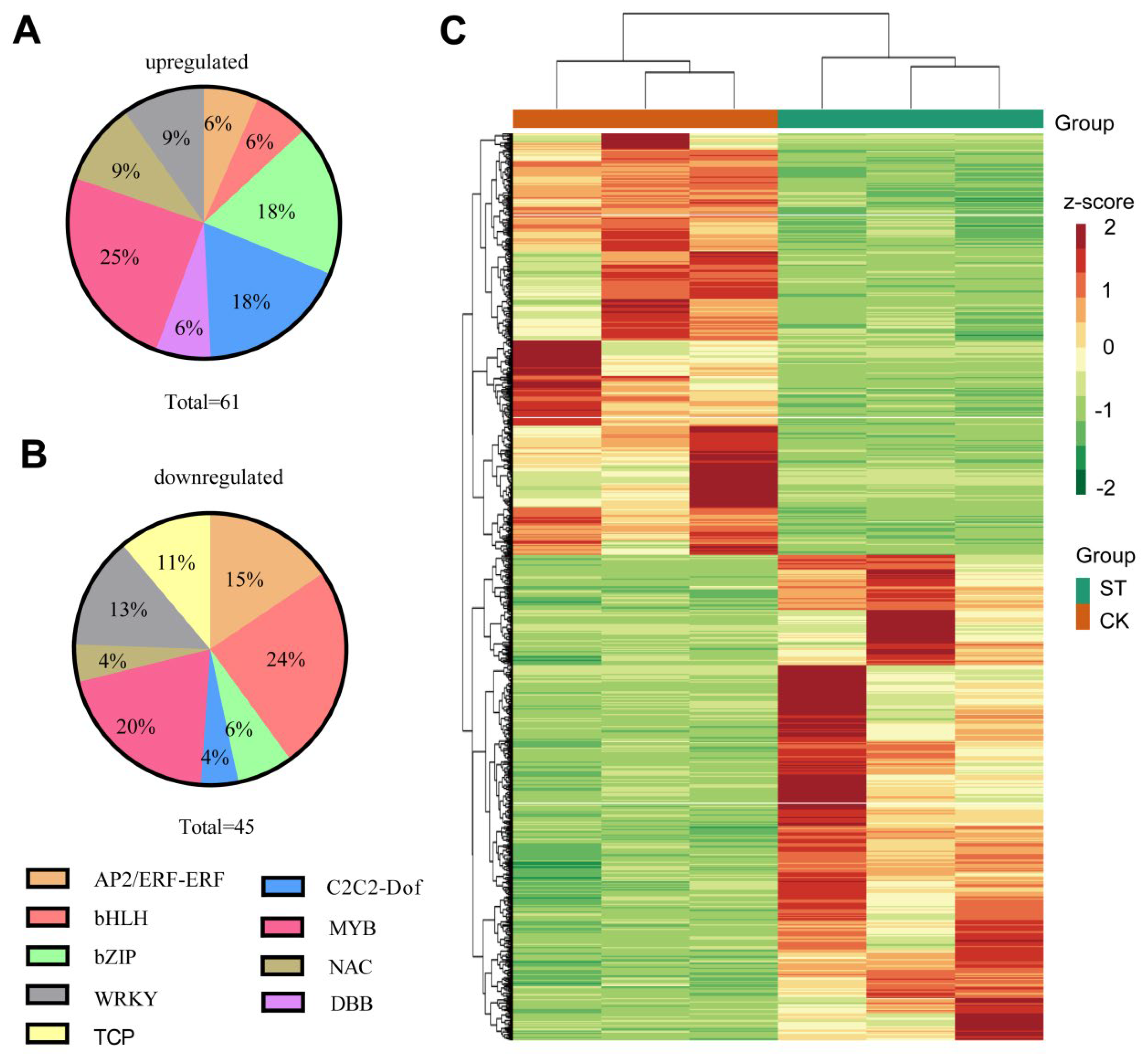
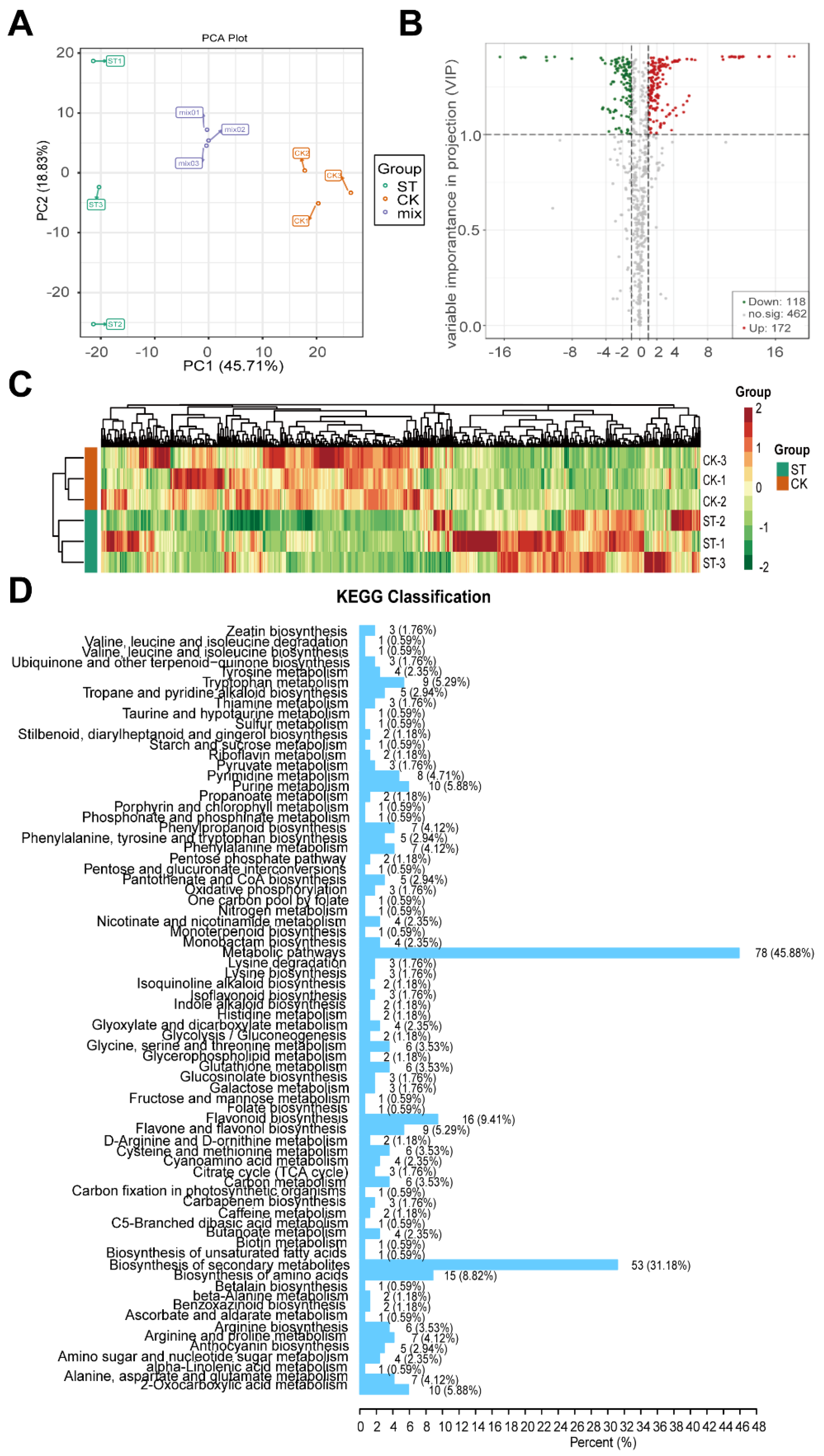
3.3. Global Gene Expression Induced by Low Light Treatments
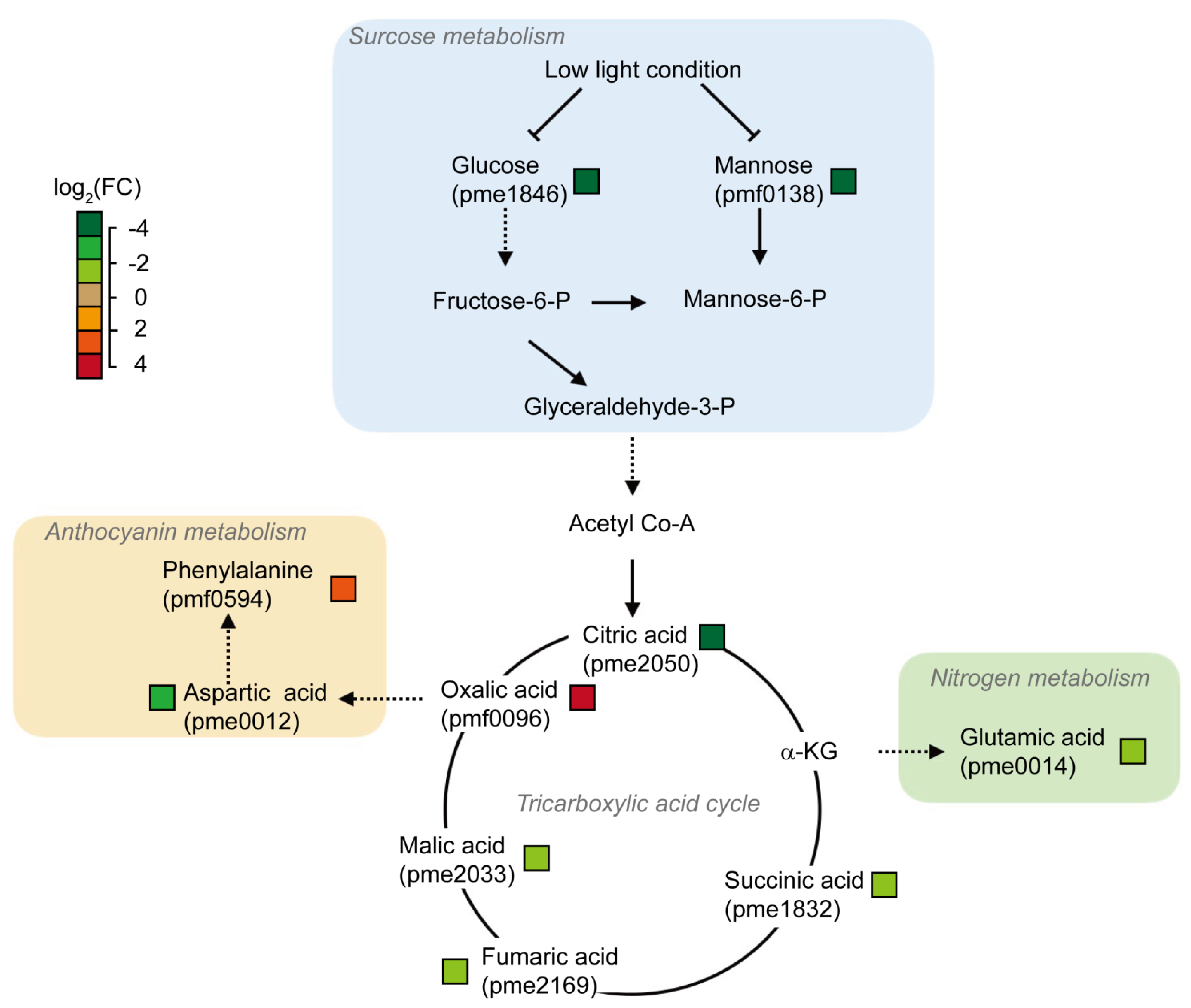
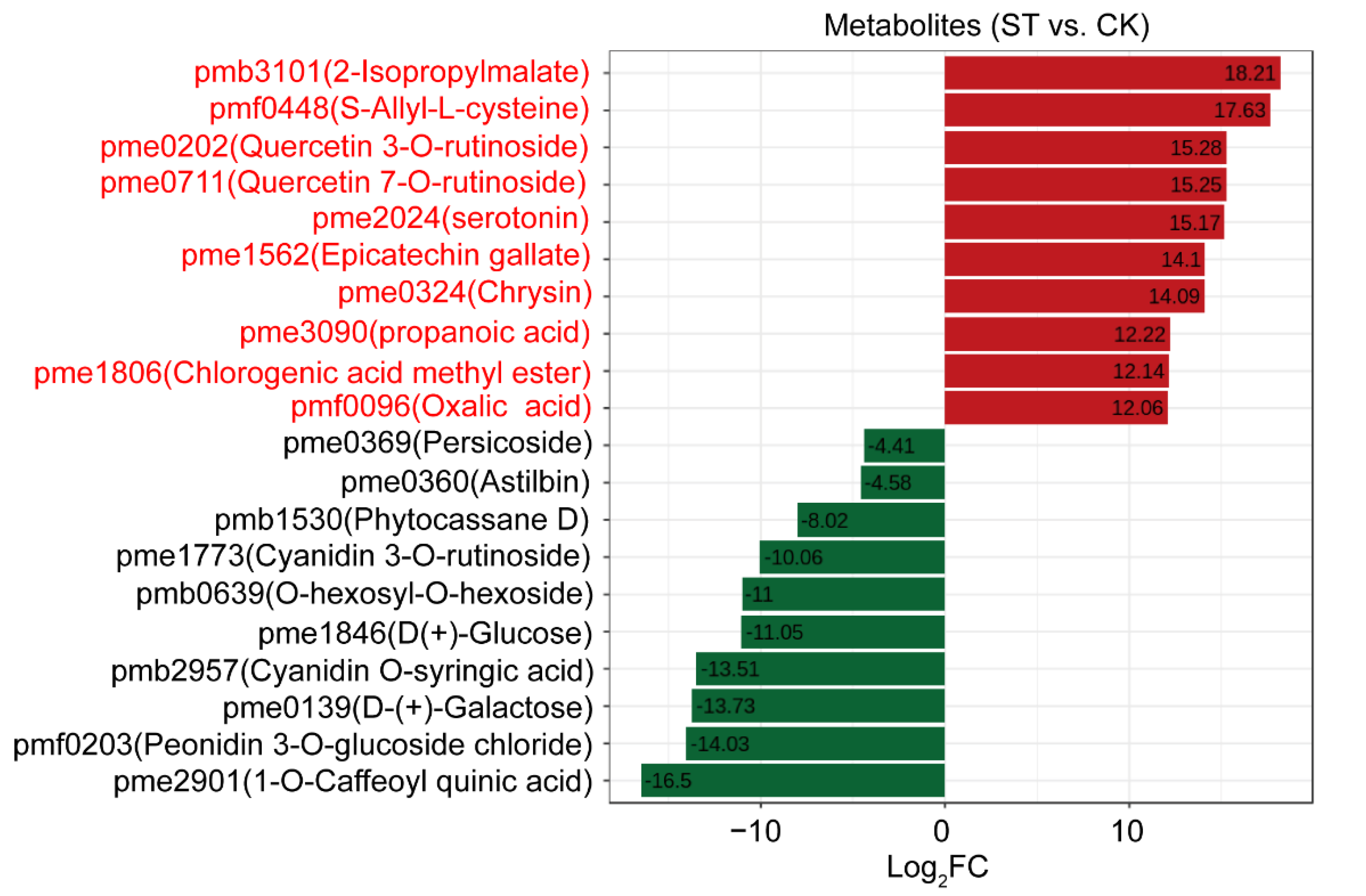
3.4. Extremely Differentially Abundant Metabolites Due to Low Light Effects
4. Discussion
- (1)
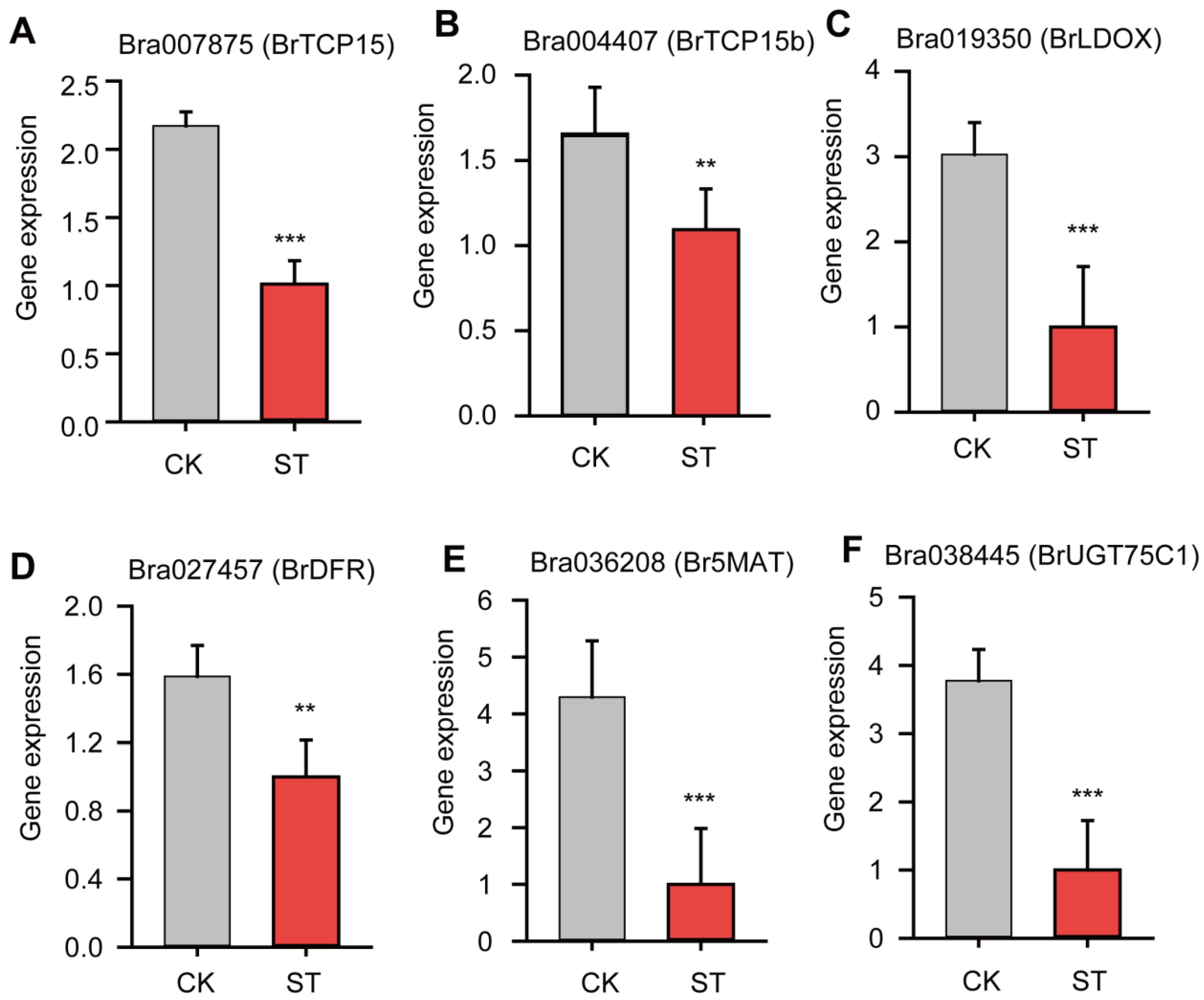
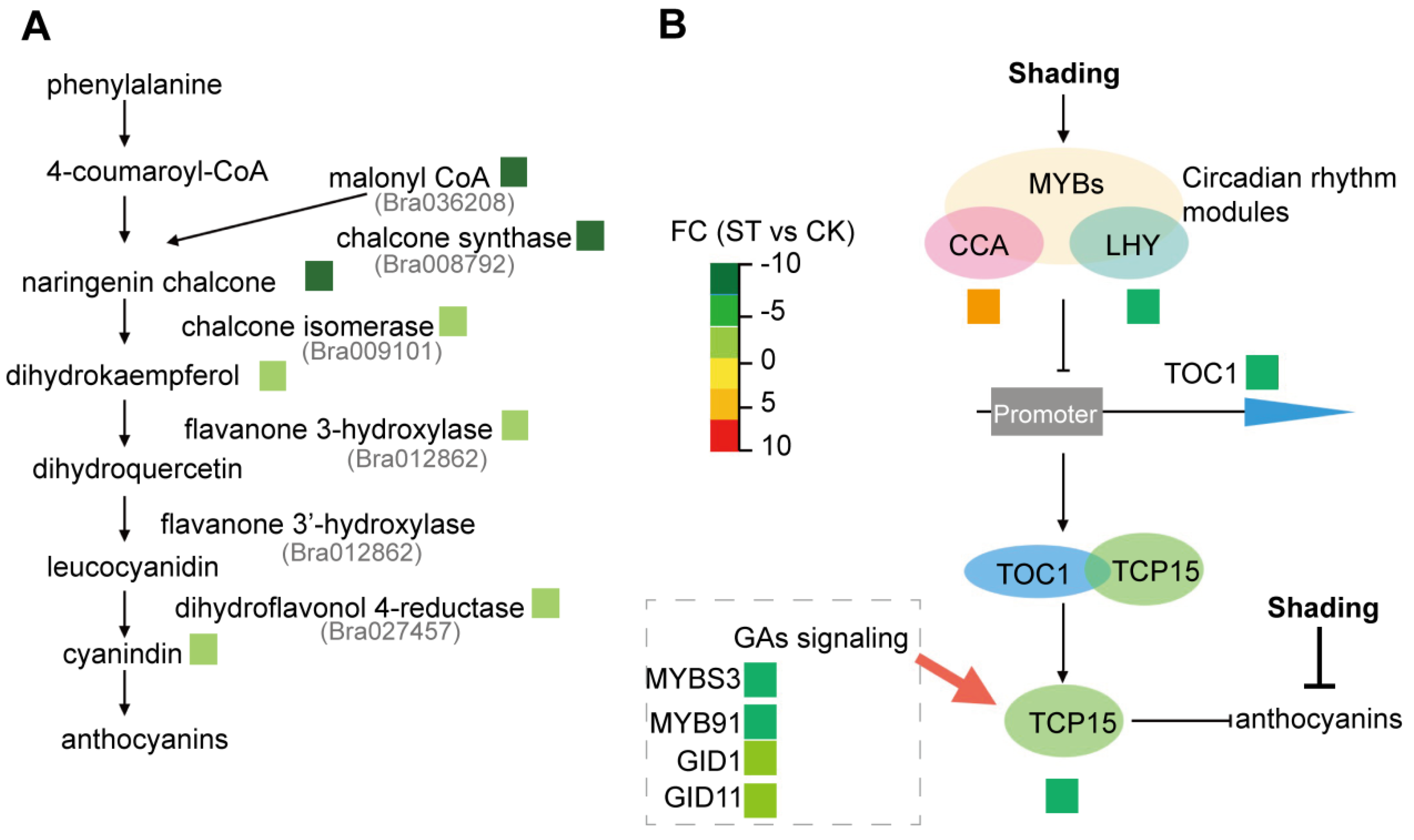
- TCP15 was reported to be related to circadian rhythm pathway [42]. Consistently, circadian rhythm pathway was the most significantly enriched in the list of DEGs according to both GO and KEGG analysis (Figure 3A,B). In Arabidopsis, there is much known about the molecular basis of the circadian clock [44,45]. The key loop includes two partially overlapped MYB domain transcription factors, CIRCADIAN CLOCK ASSOCAITED1 (CCA1) and LATE ELONGATED HYPOCOTYL (LHY). These three proteins form a negative arm that binds to TIMING OF CAB EXPRESSION1 (TOC1) promoter [44]. TOC1 could interact with CCA1 HIKING EXPENITION (CHE, i.e., BrTCP15 reported in our study), hence leads to decreased BrTCP15 gene expression [12].
- (2)
- The decreased BrTCP15 gene expression could be also related to downregulated expression of some genes involved in the gibberellin (GA) signaling pathway, such as Bra002790, Bra005177, Bra005899 and Bra009207 (Figure 9B and Table S9). TCP15 participates in the GA-signaling pathway, responded to abiotic stresses and hormone signals, for example, in Fraxinus mandshurica Rupr [45]. Therefore, low light probably inhibits the GA signaling process and hence suppresses BrTCP15 gene expression. The anthocyanin degradation under low light condition is mainly ascribed to inhibitory effects on gene expression involved in the anthocyanin biosynthetic pathway, rather than regulatory effects by BrTCP15.
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Melim, C.; Lauro, M.R.; Pires, I.M.; Oliveira, P.J.; Cabral, C. The Role of Glucosinolates from Cruciferous Vegetables (Brassicaceae) in Gastrointestinal Cancers: From Prevention to Therapeutics. Pharmaceutics 2022, 14, 190. [Google Scholar] [CrossRef]
- Rochfort, S.J.; Imsic, M.; Jones, R.; Trenerry, V.C.; Tomkins, B. Characterization of Flavonol Conjugates in Immature Leaves of Pak Choi [Brassica rapa L. Ssp. chinensis L. (Hanelt.)] by HPLC-DAD and LC-MS/MS. J. Agric. Food Chem. 2006, 54, 4855–4860. [Google Scholar] [CrossRef]
- Guo, N.; Cheng, F.; Wu, J.; Liu, B.; Zheng, S.; Liang, J.; Wang, X. Anthocyanin Biosynthetic Genes in Brassica rapa. BMC Genom. 2014, 15, 426. [Google Scholar] [CrossRef] [Green Version]
- Bell, G.E.; Danneberger, T.K.; McMahon, M.J. Spectral Irradiance Available for Turfgrass Growth in Sun and Shade. Crop Sci. 2000, 40, 189–195. [Google Scholar] [CrossRef]
- Chen, H.; Li, Q.-P.; Zeng, Y.-L.; Deng, F.; Ren, W.-J. Effect of Different Shading Materials on Grain Yield and Quality of Rice. Sci. Rep. 2019, 9, 9992. [Google Scholar] [CrossRef]
- Wang, L.; Deng, F.; Ren, W.-J.; Yang, W.-Y. Effects of Shading on Starch Pasting Characteristics of Indica Hybrid Rice (Oryza sativa L.). PLoS ONE 2013, 8, e68220. [Google Scholar] [CrossRef] [Green Version]
- Möller, M.; Assouline, S. Effects of a Shading Screen on Microclimate and Crop Water Requirements. Irrig. Sci. 2007, 25, 171–181. [Google Scholar] [CrossRef]
- Mu, H.; Jiang, D.; Wollenweber, B.; Dai, T.; Jing, Q.; Cao, W. Long-Term Low Radiation Decreases Leaf Photosynthesis, Photochemical Efficiency and Grain Yield in Winter Wheat. J. Agron. Crop Sci. 2010, 196, 38–47. [Google Scholar] [CrossRef]
- Fukuoka, N.; Suzuki, T.; Minamide, K.; Hamada, T. Effect of Shading on Anthocyanin and Non-Flavonoid Polyphenol Biosynthesis of Gynura Bicolor Leaves in Midsummer. HortScience 2014, 49, 1148–1153. [Google Scholar] [CrossRef] [Green Version]
- Zhu, H.; Xiaofeng, L.; Zhai, W.; Liu, Y.; Gao, Q.; Liu, J.; Ren, L.; Chen, H.; Zhu, Y. Effects of Low Light on Photosynthetic Properties, Antioxidant Enzyme Activity, and Anthocyanin Accumulation in Purple Pak-Choi (Brassica campestris Ssp. Chinensis Makino). PLoS ONE 2017, 12, e0179305. [Google Scholar] [CrossRef]
- Ramsay, N.A.; Glover, B.J. MYB-BHLH-WD40 Protein Complex and the Evolution of Cellular Diversity. Trends Plant Sci. 2005, 10, 63–70. [Google Scholar] [CrossRef]
- Zhang, F.; Gonzalez, A.; Zhao, M.; Payne, C.T.; Lloyd, A. A Network of Redundant BHLH Proteins Functions in All TTG1-Dependent Pathways of Arabidopsis. Development 2003, 130, 4859–4869. [Google Scholar] [CrossRef] [Green Version]
- Gonzalez, A.; Zhao, M.; Leavitt, J.M.; Lloyd, A.M. Regulation of the Anthocyanin Biosynthetic Pathway by the TTG1/BHLH/Myb Transcriptional Complex in Arabidopsis Seedlings. Plant J. 2008, 53, 814–827. [Google Scholar] [CrossRef]
- Li, J.; Essemine, J.; Shang, C.; Zhang, H.; Zhu, X.; Yu, J.; Chen, G.; Qu, M.; Sun, D. Combined Proteomics and Metabolism Analysis Unravels Prominent Roles of Antioxidant System in the Prevention of Alfalfa (Medicago sativa L.) against Salt Stress. Int. J. Mol. Sci. 2020, 21, 909. [Google Scholar] [CrossRef] [Green Version]
- Qu, M.; Chen, G.; Bunce, J.; Zhu, X.; Sicher, R. Systematic Biology Analysis on Photosynthetic Carbon Metabolism of Maize Leaf Following Sudden Heat Shock under Elevated CO2. Sci. Rep. 2018, 8, 7849. [Google Scholar] [CrossRef]
- Wang, N.; Shu, X.; Zhang, F.; Zhuang, W.; Wang, T.; Wang, Z. Comparative Transcriptome Analysis Identifies Key Regulatory Genes Involved in Anthocyanin Metabolism During Flower Development in Lycoris radiata. Front. Plant Sci. 2021, 12, 761862. [Google Scholar] [CrossRef]
- Wagner, G.P.; Kin, K.; Lynch, V.J. Measurement of MRNA Abundance Using RNA-Seq Data: RPKM Measure Is Inconsistent among Samples. Theory Biosci. 2012, 131, 281–285. [Google Scholar] [CrossRef]
- Li, B.; Dewey, C.N. RSEM: Accurate Transcript Quantification from RNA-Seq Data with or without a Reference Genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [Green Version]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. EdgeR: A Bioconductor Package for Differential Expression Analysis of Digital Gene Expression Data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [Green Version]
- Xie, C.; Mao, X.; Huang, J.; Ding, Y.; Wu, J.; Dong, S.; Kong, L.; Gao, G.; Li, C.-Y.; Wei, L. KOBAS 2.0: A Web Server for Annotation and Identification of Enriched Pathways and Diseases. Nucleic Acids Res. 2011, 39, W316–W322. [Google Scholar] [CrossRef] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Jaakola, L.; Zoratti, L.; Giongo, L.; Karppinen, K.; Uleberg, E.; Martinussen, I.; Häggman, H. Influence of Light and Temperature Conditions on Anthocyanin Accumulation in Vaccinium Spp. Berries. In Proceedings of the Acta Horticulturae; International Society for Horticultural Science (ISHS), Leuven, Belgium, 25 November 2017; pp. 321–326. [Google Scholar]
- Wang, X.; Wang, H.; Wang, J.; Sun, R.; Wu, J.; Liu, S.; Bai, Y.; Mun, J.-H.; Bancroft, I.; Cheng, F.; et al. The Genome of the Mesopolyploid Crop Species Brassica rapa. Nat. Genet. 2011, 43, 1035–1039. [Google Scholar] [CrossRef] [Green Version]
- Cai, C.; Wang, X.; Liu, B.; Wu, J.; Liang, J.; Cui, Y.; Cheng, F.; Wang, X. Brassica rapa Genome 2.0: A Reference Upgrade through Sequence Re-Assembly and Gene Re-Annotation. Mol. Plant 2017, 10, 649–651. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, J.; Pang, W.; Chen, B.; Zhang, C.; Piao, Z. Transcriptome Analysis of Brassica rapa Near-Isogenic Lines Carrying Clubroot-Resistant and -Susceptible Alleles in Response to Plasmodiophora brassicae during Early Infection. Front. Plant Sci. 2015, 6, 1183. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Munekage, Y.; Hojo, M.; Meurer, J.; Endo, T.; Tasaka, M.; Shikanai, T. PGR5 Is Involved in Cyclic Electron Flow around Photosystem I and Is Essential for Photoprotection in Arabidopsis. Cell 2002, 110, 361–371. [Google Scholar] [CrossRef] [Green Version]
- Johnson, G.N. Physiology of PSI Cyclic Electron Transport in Higher Plants. Biochim. Biophys. Acta 2011, 1807, 384–389. [Google Scholar] [CrossRef]
- Wolf, B.-C.; Isaacson, T.; Tiwari, V.; Dangoor, I.; Mufkadi, S.; Danon, A. Redox Regulation of PGRL1 at the Onset of Low Light Intensity. Plant J. 2020, 103, 715–725. [Google Scholar] [CrossRef]
- Liu, Y.; Pan, T.; Tang, Y.; Zhuang, Y.; Liu, Z.; Li, P.; Li, H.; Huang, W.; Tu, S.; Ren, G.; et al. Proteomic Analysis of Rice Subjected to Low Light Stress and Overexpression of OsGAPB Increases the Stress Tolerance. Rice 2020, 13, 30. [Google Scholar] [CrossRef]
- Huang, D.; Yuan, Y.; Tang, Z.; Huang, Y.; Kang, C.; Deng, X.; Xu, Q. Retrotransposon Promoter of Ruby1 Controls Both Light- and Cold-Induced Accumulation of Anthocyanins in Blood Orange. Plant. Cell Environ. 2019, 42, 3092–3104. [Google Scholar] [CrossRef]
- Dong, T.; Han, R.; Yu, J.; Zhu, M.; Zhang, Y.; Gong, Y.; Li, Z. Anthocyanins Accumulation and Molecular Analysis of Correlated Genes by Metabolome and Transcriptome in Green and Purple Asparaguses (Asparagus officinalis, L.). Food Chem. 2019, 271, 18–28. [Google Scholar] [CrossRef]
- Dubos, C.; Le Gourrierec, J.; Baudry, A.; Huep, G.; Lanet, E.; Debeaujon, I.; Routaboul, J.-M.; Alboresi, A.; Weisshaar, B.; Lepiniec, L. MYBL2 Is a New Regulator of Flavonoid Biosynthesis in Arabidopsis thaliana. Plant J. 2008, 55, 940–953. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.-F.; Fitzsimmons, K.; Khandelwal, A.; Kranz, R.G. CPC, a Single-Repeat R3 MYB, Is a Negative Regulator of Anthocyanin Biosynthesis in Arabidopsis. Mol. Plant 2009, 2, 790–802. [Google Scholar] [CrossRef]
- Broun, P. Transcriptional Control of Flavonoid Biosynthesis: A Complex Network of Conserved Regulators Involved in Multiple Aspects of Differentiation in Arabidopsis. Curr. Opin. Plant Biol. 2005, 8, 272–279. [Google Scholar] [CrossRef] [PubMed]
- Zhu, M.; Assmann, S.M. Metabolic Signatures in Response to Abscisic Acid (ABA) Treatment in Brassica napus Guard Cells Revealed by Metabolomics. Sci. Rep. 2017, 7, 12875. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- de Leonardis, A.M.; Fragasso, M.; Beleggia, R.; Ficco, D.B.M.; de Vita, P.; Mastrangelo, A.M. Effects of Heat Stress on Metabolite Accumulation and Composition, and Nutritional Properties of Durum Wheat Grain. Int. J. Mol. Sci. 2015, 16, 30382–30404. [Google Scholar] [CrossRef] [Green Version]
- Naing, A.H.; Kim, C.K. Abiotic Stress-Induced Anthocyanins in Plants: Their Role in Tolerance to Abiotic Stresses. Physiol. Plant. 2021, 172, 1711–1723. [Google Scholar] [CrossRef] [PubMed]
- Stracke, R.; Ishihara, H.; Huep, G.; Barsch, A.; Mehrtens, F.; Niehaus, K.; Weisshaar, B. Differential Regulation of Closely Related R2R3-MYB Transcription Factors Controls Flavonol Accumulation in Different Parts of the Arabidopsis thaliana Seedling. Plant J. 2007, 50, 660–677. [Google Scholar] [CrossRef] [Green Version]
- Zhou, B.; Leng, J.; Ma, Y.; Fan, P.; Li, Y.; Yan, H.; Xu, Q. BrmiR828 Targets BrPAP1, BrMYB82, and BrTAS4 Involved in the Light Induced Anthocyanin Biosynthetic Pathway in Brassica rapa. Int. J. Mol. Sci. 2020, 21, 4326. [Google Scholar] [CrossRef]
- Liang, N.; Zhan, Y.; Yu, L.; Wang, Z.; Zeng, F. Characteristics and Expression Analysis of FmTCP15 under Abiotic Stresses and Hormones and Interact with DELLA Protein in Fraxinus mandshurica Rupr. Forests 2019, 10, 343. [Google Scholar] [CrossRef] [Green Version]
- Viola, I.L.; Camoirano, A.; Gonzalez, D.H. Redox-Dependent Modulation of Anthocyanin Biosynthesis by the TCP Transcription Factor TCP15 during Exposure to High Light Intensity Conditions in Arabidopsis. Plant Physiol. 2016, 170, 74–85. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.A.; Kim, H.-S.; Choi, S.-H.; Jang, J.-Y.; Jeong, M.-J.; Lee, S.I. The Importance of the Circadian Clock in Regulating Plant Metabolism. Int. J. Mol. Sci. 2017, 18, 2680. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McClung, C.R. Comes a Time. Curr. Opin. Plant Biol. 2008, 11, 514–520. [Google Scholar] [CrossRef] [PubMed]
- Harmer, S.L. The Circadian System in Higher Plants. Annu. Rev. Plant Biol. 2009, 60, 357–377. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pruneda-Paz, J.L.; Breton, G.; Para, A.; Kay, S.A. A Functional Genomics Approach Reveals CHE as a Component of the Arabidopsis Circadian Clock. Science 2009, 323, 1481–1485. [Google Scholar] [CrossRef] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guo, J.; Wu, T.; Fu, M.; Li, G.; Luo, W.; Kang, Y.; Wang, T. An Integrated Analysis of Transcriptome and Metabolism Reveals an Inhibitory Effect of Low Light on Anthocyanin Biosynthesis in Purple cai-tai (Brassicarapa L. var. purpurea). Horticulturae 2022, 8, 566. https://doi.org/10.3390/horticulturae8070566
Guo J, Wu T, Fu M, Li G, Luo W, Kang Y, Wang T. An Integrated Analysis of Transcriptome and Metabolism Reveals an Inhibitory Effect of Low Light on Anthocyanin Biosynthesis in Purple cai-tai (Brassicarapa L. var. purpurea). Horticulturae. 2022; 8(7):566. https://doi.org/10.3390/horticulturae8070566
Chicago/Turabian StyleGuo, Juxian, Tingquan Wu, Mei Fu, Guihua Li, Wenlong Luo, Yunyan Kang, and Tingqin Wang. 2022. "An Integrated Analysis of Transcriptome and Metabolism Reveals an Inhibitory Effect of Low Light on Anthocyanin Biosynthesis in Purple cai-tai (Brassicarapa L. var. purpurea)" Horticulturae 8, no. 7: 566. https://doi.org/10.3390/horticulturae8070566
APA StyleGuo, J., Wu, T., Fu, M., Li, G., Luo, W., Kang, Y., & Wang, T. (2022). An Integrated Analysis of Transcriptome and Metabolism Reveals an Inhibitory Effect of Low Light on Anthocyanin Biosynthesis in Purple cai-tai (Brassicarapa L. var. purpurea). Horticulturae, 8(7), 566. https://doi.org/10.3390/horticulturae8070566




