Abstract
CCT genes play vital roles in flowering, plant growth, development, and response to abiotic stresses. Although they have been reported in many plants, the characterization and expression pattern of CCT genes is still limited in R. sativus. In this study, a total of 58 CCT genes were identified in R. sativus. Phylogenetic tree, gene structure, and conserved domains revealed that all CCT genes were classified into three groups: COL, CMF, and PRR. Genome-wide identification and evolutionary analysis showed that segmental duplication expanded the CCT gene families considerably, with the LF subgenome retaining more CCT genes. We observed strong purifying selection pressure for CCT genes. RsCCT genes showed tissue specificity, and some genes (such as RsCCT22, RsCCT36, RsCCT42 and RsCCT51) were highly expressed in flowers. Promoter cis-elements and RNA-seq data analysis showed that RsCCT genes could play roles in controlling flowering through the photoperiodic pathway and vernalization pathway. The expression profiles of RsCCT genes under Cd, Cr, Pb, and heat and salt stresses revealed that many RsCCT genes could respond to one or more abiotic stresses. Our findings could provide essential information for further studies on the function of RsCCT genes.
1. Introduction
The CCT genes play essential roles in plant growth, development and morphogenesis various processes, including flowering time regulation, circadian rhythm response, light signal induction and abiotic stress response [1,2]. The proteins encoded by CCT genes contain a conserved domain composed of 43–45 amino acids, which has a nuclear localization function [3,4,5]. It was first proved in the C-terminal of the AtCO (CONSTANS) protein, subsequently, in CO-LIKE proteins and TOC1 (TIMING OF CAB1) proteins. Since it is conserved, it is, therefore, called the CCT domain [6,7,8].
According to the differences in the motif composition of CCT genes, the CCT gene family can be divided into three subfamilies: COL subfamily (CONSTANS-LIKE, containing one CCT and B-box domain), PRR subfamily (Pseudo Response Regulator, containing one CCT and receiver-like domain), and CMF subfamily (CCT Motif Family, containing only one CCT domain) [5,6]. Some CCT genes also contain a B-box domain or receiver-like domain (RLD). The B-box is related to light signal transduction, and the RLD is related to circadian rhythm [9,10].
The genes that control flowering are relatively conserved in plants, but each plant has evolved a unique mechanism to induce flowering under optimal conditions [11]. In Arabidopsis thaliana, AtCO mutants delayed flowering under long-day conditions, while overexpression of AtCO led to flowering early under long- and short-day conditions [3,12]. COL3 was a positive regulatory factor in the light signal transduction pathway. COL3-deficient mutants showed early flowering, longer hypocotyls, and fewer lateral branches under short daylight [13]. TOC1 functioned in the circadian clock central oscillator, while the TOC1 itself was regulated by circadian rhythm and participated in feedback loops to control its expression [9,14]. Hd1, a direct homolog of AtCO, was involved in heading regulation interacting with Ghd7 [15]. Furthermore, CCT genes may also be involved in the regulation of abiotic stress response. Hd2, another homolog of AtCO in rice, regulated leaf senescence and drought resistance [16]. Although CCT genes regulate flowering time in most species, whether these genes have evolved new functions and the evolutionary relationship between CCT genes, needs to be studied further.
Radish (Raphanus sativus L., 2n = 2x = 18) is an important Brassicaceae vegetable crop widely cultivated and consumed globally. Approximately 6.5 MYA (million years ago), R. sativus and B. nigra simultaneously emerged and differentiated. Subsequently, the divergence of B. rapa and B. oleracea occurred approximately 4.6 MYA [17,18]. A quadrilateral model with four vertices, Brassica rapa (A genome), B. nigra (B genome), B. oleracea (C genome), and R. sativus (R genome), can better reveal the evolutionary relationship of the Brassicaceae (18). The identification and analysis of CCT genes have been reported in detail in some species, such as A. thaliana, Oryza sativa, Medicago truncatula, and Aegilops tauschii [19,20,21]. However, CCT genes in Brassicaceae species have still not been studied.
In this study, we aim to uncover the evolution and expression patterns of RsCCT genes. We first identified CCT genes in B. rapa, B. nigra, B. oleracea, and R. sativus, respectively. Then, the gene structure, chromosomal location, and evolutionary relationships of CCT genes were analyzed. The spatiotemporal expression patterns and the expression profile of flowering regulation along with the abiotic stress response of the RsCCT genes were further analyzed and verified by RT-qPCR. Our results provide new insights into the evolution of CCT genes and pave the way for further functional studies of RsCCT genes.
2. Materials and Methods
2.1. Plant Materials, Growth Conditions, and Stress Treatments
We selected radish inbred lines ‘19CJ012′, ‘19CJ025′, and ‘19CJ093′ to analyze the expression of RsCCT genes under different treatments. They were grown in a greenhouse under 16 h-light/8 h-dark conditions at 24 °C. The radish ‘19CJ093′ (long cylindrical with white skin and white flesh) was used to analyze the expression of different tissues. Artificial pollination was carried out after flowering, then the leaf, stem, flower, bud, and siliques were sampled 10 days after flowering. The radish ‘19CJ025′ (white skin, white flesh, middle column) was used for vernalization and salt stress treatment. For vernalization treatments, the 21-day-old seedlings were vernalized under 3–10 °C for 30 days and with the ones at a room temperature of 24 °C as the control, the leaves were then sampled. For salt stress treatment, the germinated seeds were grown in plastic containers with nutrient solution. One week later, seedlings were treated with 100 mM and 200 mM sodium chloride (NaCl), respectively. The NaCl-free solution was used as the control. Radish roots were sampled after 24 h of NaCl treatments. The radish ‘19CJ012′ (red skin, white flesh, round) was used for heat stress treatment. The 30-day-old seedlings were incubated in a growth chamber at 40 °C light/35 °C dark for 9 days, leaf samples were then collected, and 24 °C light/20 °C dark was used as the control. Three biological replicates were performed for all samples which were immediately frozen with liquid nitrogen and ground to extract total RNA.
2.2. Identification and Phylogenetic Analysis of CCT Genes
From the BRAD V3.0 database (http://brassicadb.org/brad/, accessed on 2 October 2021) and TAIR10 website (http://www.arabidopsis.org, accessed on 2 October 2021), the whole genome and protein sequence of B. rapa, B. oleracea, B. nigra, and A. thaliana were downloaded, respectively [22,23]. The whole genome and protein sequence of R. sativus were obtained from the GenomeWarehouse (GWH) database of the National Genomics Data Center (BIG Data Center) (https://bigd.big.ac.cn, accessed on 2 October 2021). The CCT proteins were isolated using the HMMER3.0 software (http://hmmer.org/, accessed on 2 October 2021) based on the Hidden Markov Model (HMM) file of the CCT motif (PF06203). Each predicted CCT protein was subsequently compared and verified, with an E-value < 1×10−5 and the removal of redundant sequences through the public databases, including NCBI CDD (https://www.ncbi.nlm.nih.gov/cdd/, accessed on 2 October 2021), Pfam (http://pfam.xfam.org/search#tabview=tab1, accessed on 2 October 2021), and SMART (http://smart.embl.de/, accessed on 2 October 2021), to confirm its membership in the CCT family [24].
A phylogenetic tree was constructed using Molecular Evolutionary Genetics Analysis MEGA 7.0 software (https://mega.software.informer.com/7.2/, accessed on 3 October 2021), with the Maximum Likelihood method and 1000 bootstrap replicates method to investigate and analyze the phylogenetic relationship of CCT proteins. Phylogenetic tree visualization was performed in the online tool-Interactive Tree Of Life (iTOL), version 5.6.1 (https://www.itol.embl.de/, accessed on 5 October 2021).
2.3. Collinearity and Selection Pressure Analysis of CCT Genes
The collinearity blocks were analyzed using the Multiple Collinearity Scan tool kit (MCscanx) across the whole genome. Duplicate segment pairs with CCT genes were extracted to draw a synteny map using the Dual Systeny Plotter software (https://github.com/CJ-Chen/TBtools, accessed on 5 October 2021). The rates of non-synonymous (Ka) and synonymous (Ks) substitutions, along with their ratios, were calculated by a Ka/Ks calculator via TBtools v1.0971. This analysis was performed to estimate the selection pressure of duplicated CCT gene pairs [25]. Multiple CCT protein sequences were aligned using ClustalW2.0, and sequence logos were created using the Web logo platform (https://weblogo.berkeley.edu/logo.cgi, accessed on 5 October 2021).
2.4. Characteristic and Chromosomal Localization of RsCCT Genes
The physical and chemical properties of RsCCT proteins among the species, such as sequence length, molecular weight (M.W.), and theoretical isoelectric points (pI), were predicted by online tools from the ExPASy website (https://web.expasy.org/protparam/) [26]. The subcellular localization of RsCCT proteins was analyzed by pSORT (https://wolfpsort.hgc.jp/, accessed on 6 October 2021) and CELLO v.2.5 (http://cello.life.nctu.edu.tw/, accessed on 6 October 2021). The chromosome distribution and physical location of CCT genes were drafted with TBtools according to the gene position in the genome.
2.5. Conserved Motifs, Gene Structure, and Cis-Elements Analysis of RsCCT Genes
The online gene structure display server (GSDs) (http://gsds.cbi.pku.edu.cn/, accessed on 6 October 2021) was used to obtain the exon–intron structures of RsCCT genes, based on CD and genome files. The conserved motifs of RsCCT proteins were predicted using the online MEME software (http://meme-suite.org/tools/meme, accessed on 6 October 2021). The exon–intron structures and conserved motifs of RsCCT genes were visualized using the TBtools. The upstream 2000 bp sequences of each RsCCT gene promoter were extracted from the genome sequence of radish. The cis-regulatory elements in the promoter regions of RsCCT genes were predicted and analyzed by the PlantCARE Database (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/, accessed on 6 October 2021) [27].
2.6. Expression Analysis of RsCCT Genes
The RNA sequencing data were obtained from the NCBI Sequence Read Archive to analyze the expression of RsCCT genes (SRA, http://www.ncbi.nlm.nih.gov/Traces/sra/, accessed on 16 October 2021) and then visualized using TBtools software (https://github.com/CJ-Chen/TBtools, accessed on 16 October 2021). Additionally, the transcription data for five R. sativus tissues (flower, siliques, leaf, stem, and callus) were selected to analyze the tissue-specific expression. The transcription data under Cd, Cr, Pb, and heat and salt stresses were used to analyze the responses to abiotic stresses. The differentially expressed genes associated with different developmental stages and responded to vernalization were used to analyze the role of RsCCT genes in the regulation of bolting and flowering.
2.7. RNA Isolation and Real-Time Quantitative PCR (RT-qPCR) Analysis
The total RNA of each sample was isolated with AG RNAex Pro Reagent (Accurate Biotechnology, AG21102, Changsha, China) and treated with RNase-free DNase I. Then, the first-strand cDNA was synthesized using the RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific, K1622, MA, USA). RT-qPCR was conducted according to the instructions of TB Green Premix Ex Taq II (Takara, RR820Q, Dalian, China). The gene-specific primers were designed in NCBI Primer-BLAST and the RsActin gene was selected as the reference gene (Supplementary Table S1).
3. Results
3.1. Identification and Distribution of the CCT Genes
A set of 58 putative CCT proteins were identified in R. sativus. The RsCCT genes were named RsCCT1 to RsCCT58 based on their positions on the chromosome (Supplementary Table S2). RsCCT genes had a wide range of gene lengths from 582 (RsCCT48) to 5442 (RsCCT32) bps and encoding protein lengths from 193 (RsCCT48) to 1813 (RsCCT32) aa. The molecular weights of the RsCCT proteins ranged from 21,941.88 (RsCCT45) to 202,008.43 (RsCCT32) Da. The isoelectric point values of RsCCT proteins were between 4.42 (RsCCT26) and 9.76 (RsCCT41), and most were less than 7, indicating that RsCCT proteins were rich in acidic amino acids (Supplementary Table S1). Subcellular localization prediction showed that 49 RsCCT proteins were localized in the nucleus and 3 RsCCT proteins were localized in chloroplasts.
In R. sativus, chromosomes R04 and R09 harbored the most RsCCT genes (9 genes), followed by chromosome R01, R02, and R05 (8 genes). Chromosome R03 harbored the least RsCCT genes (2 genes) (Figure 1a). Similar to that in RsCCT genes, a total of 196 CCT genes (64, 66, and 66 from B. rapa, B. nigra, and B. oleracea, respectively) were unevenly distributed on 27 chromosomes (10, 8, and 9 from B. rapa, B. nigra, and B. oleracea, respectively) (Supplementary Figure S1). It indicates that some CCT genes have likely been duplicated or lost due to genome recombination.
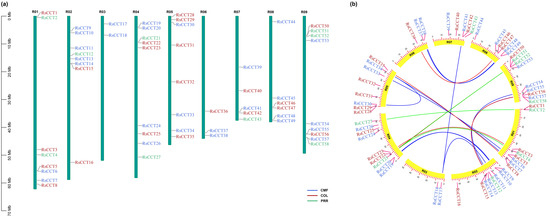
Figure 1.
Chromosome localization (a) and duplication analyses (b) of RsCCT genes. Gene codes represented different groups with different colors in (a). Different colored lines in (b) were used to connect synteny genes of different groups.
Brassicaceae species have undergone whole-genome duplication events in the evolutionary history, especially the Brassica genus-specific whole-genome triplication (WGT) [28]. The genomes of B. rapa, B. nigra, B. oleracea, and R. sativus were divided into triplicated genome blocks (LF, MF1, MF2). There were 26 BrCCTs, 25 BoCCTs, 20 RsCCTs, and 25 BnCCTs in the LF subgenome; 19 BrCCTs, 22 BoCCTs, 21 RsCCTs, and 22 BnCCTs in the MF1 subgenome; 19 BrCCTs, 19 BoCCTs, 15 RsCCTs, and 19 BnCCTs in the MF2 subgenome (Supplementary Figure S2; Supplementary Table S3). The difference of CCT gene loss among subgenomic blocks was evident. Compared with the MF1 and MF2 subgenomes, the LF subgenome retained more CCT genes.
3.2. Phylogenetic Analysis of CCT Genes
To better understand the phylogenetic relationship of CCT genes among Brassicaceae species, we constructed an unrooted phylogenetic tree (Figure 2) of full-length CCT protein sequences among A. thaliana, B. rapa, B. oleracea, B. nigra, and R. sativus.
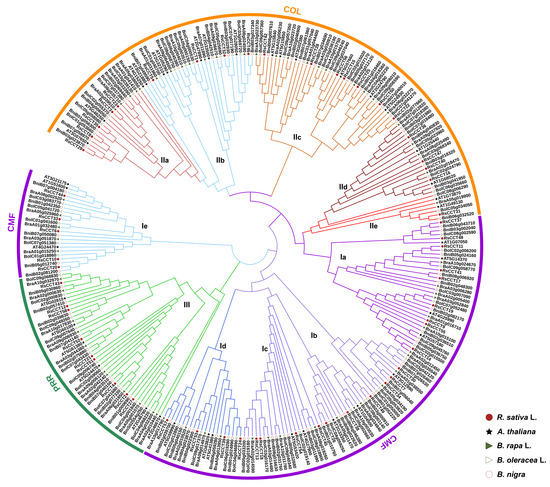
Figure 2.
Phylogenetic relationships among CCT motif family genes in A. thaliana, B. rapa, B. nigra, B. oleracea, and R. sativus. The phylogenetic tree was constructed using MEGA7.0, and the bootstrap was 1000 replicates. The gene names were shown in Supplementary Table S4. The different colors of the circle represented proteins of different groups.
According to the classification criteria and predicted function of AtCCT genes [29,30], three major groups named CMF, COL, and PRR were classified in the phylogenetic tree (Figure 2). A total of 128 CCT genes belong to the CMF group, which was divided into five subgroups, named Ia, Ib, Ic, Id, and Ie. A total of 120 CCT genes belong to the COL group, which was divided into five subgroups, named IIa, IIb, IIc, IId, and IIe. The PRR group showed the smallest number of CCT genes (46 CCT genes), and was classified into one subgroup, named III. The number of CCT genes from Brassica species and R. sativus in each subgroup was balanced, indicating that CCT genes had similarly expanded in these genomes. Notably, we found that all clades contained CCT genes from R. sativus and A. thaliana, indicating that these original CCT genes existed before the differentiation of R. sativus and A. thaliana.
The distinct motif and exon–intron structure of the RsCCT genes further corroborated the reliability of the phylogenetic tree. Twenty conserved motifs were appraised in RsCCT genes and named motif 1–20 (Figure 3, Supplementary Table S5). All RsCCT genes contained motif 1 and motif 3, which may constitute the CCT conserved domain. Motif 2 and motif 5 only appeared in COL genes. Furthermore, motif 4 formed the RLD domain and only appeared in PRR genes (Figure 3). These results suggested that these motifs could be used as markers to identify different CCT genes. CCT genes with some specific motifs might have multiple functions.
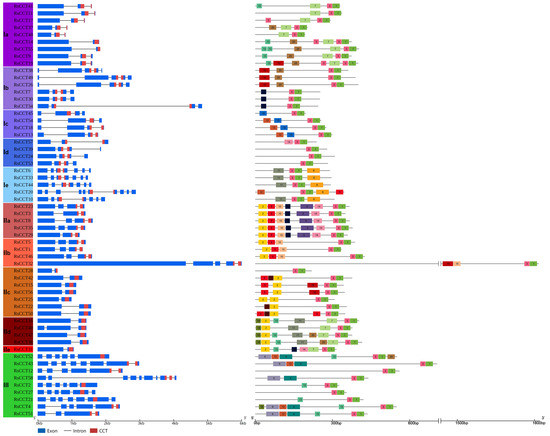
Figure 3.
Gene structure and conserved motifs of RsCCT. Different subgroups of RsCCT genes are highlighted in different colors. The gene structure of RsCCT genes was generated from the GSDS website. The blue block represented the coding sequence (CDS) while the black line indicated the introns. The red square represented the location of the CCT conserved domain. The conserved motifs in RsCCT proteins were identified through the MEME software. Different colored blocks represented different motifs.
In COL genes of R. sativus, IIa, IIb, and IIc had from 2–4 exons, and had both motif 2 and motif 5 (Figure 3). Ia (part of CMF), IId, and IIe were grouped on one evolutionary branch (Figure 2). They contained motif 7 and had from 2–3 exons (Figure 3). Interestingly, Ie and III had from 7–10 exons. Ie (part of CMF), away from the rest of the CMF, had a closer relationship with III in PRR (Figure 2). Family members in the same subgroups shared similar motifs and gene structure, indicating that they may share the same evolution and function.
Multiple alignments were carried out to investigate sequence conservation of the CCT domain. Residues at positions 1, 11, 15, 25, 28, 32, 37, 40, and 42 of the CCT domain were highly conserved (Supplementary Figure S3a–d). The conservation of the CCT domain among different species was similar. Therefore, we suggested that the CCT domains were highly conserved within species.
3.3. Comparative Synteny Analyses of CCT Genes
The duplication events of CCT genes were also studied. The syntenic relationship mainly comes from WGT, tandem duplication (TD), and segmental duplication (SD) events [31,32]. Tandem duplication events were detected in A. thaliana (chromosome Chr5, AT5G15840, and AT5G15850) and their homologous genes in B. rapa (chromosome A10, BraA10g023720, and BraA10g023730), and in B. oleracea (chromosome C09, BolC09g057350, and BolC09g057360). However, the same tandem replication event did not occur in B. nigra and R. sativus.
The synteny map showed that 55, 40, 46, and 34 CCT gene pairs (Supplementary Table S6) were formed and mapped to corresponding genomic chromosomes in B. rapa, B. nigra, B. oleracea, and R. sativus, respectively (Figure 1b; Supplementary Figure S1a–c). This indicated that the amplification of most CCT genes was mainly caused by segmental duplication. Both R. sativus and Brassica have undergone similar ancestral WGT processes, and recent WGT events occurred approximately 16 million years ago (Ks~0.3) [21]. Four duplicated gene pairs of RsCCTs could be derived from the latest WGT event with Ks values less than 0.3 (Supplementary Table S7).
To further investigate the phylogenetic relationships between the CCT genes, we compared syntenic relationships of a total of 294 CCT genes among the selected five species. RsCCT44, RsCCT45, and RsCCT52 had a one-to-one relationship between R. sativus and A. thaliana, B. rapa, B. oleracea, B. nigra (Supplementary Table S8), indicating that these genes might be of great significance in CCT gene evolution. Interestingly, there were five copies of the AT3G07650 ortholog in B. rapa, B. nigra, B. oleracea, and R. sativus, respectively. There were 17 collinear blocks (Figure 4, Supplementary Table S9), involving 24 AtCCTs, 39 RsCCTs, 41 BrCCTs, 38 BoCCTs, and 37 BnCCTs, forming a huge interspecific syntenic network. AT5G24470 and AT5G53420 had three copies of syntenic genes in the B. rapa, B. nigra, B. oleracea, and R. sativus genomes, indicating that these genes might have duplicated and then retained completely in CCT gene evolution. The extensive synteny among B. rapa, B. nigra, B. oleracea, and R. sativus supported their close evolutionary relationship among Brassicaceae species. A series of synteny events indicated that many CCT genes shared an ancestor before lineage differentiation of the A. thaliana, R. sativus, and Brassica species.
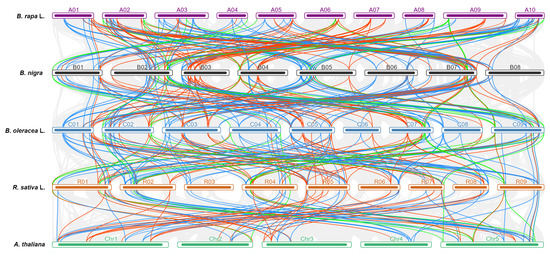
Figure 4.
Syntenic relationship among A. thaliana, B. rapa, B. nigra, B. oleracea, and R. sativus constructed according to orthologous gene pair positions. Different color curves linked AtCCT genes with their orthologous genes in B. rapa, B. oleracea, and R. sativus.
3.4. Cis-Elements in RsCCT Promoter
The cis-elements of the 2000 bp upstream sequence of 58 RsCCT gene coding regions were identified and analyzed by PlantCARE. The identified cis-elements were classified into four categories: (i) light-responsive, (ii) phytohormone-responsive, (iii) stress-responsive, and (iv) plant growth and development-responsive. There were 28 cis-elements in the light-responsive category, 10 cis-elements in the phytohormone-responsive category, and 6 cis-elements in the stress-responsive category (Supplementary Table S10). In the light-responsive category, the major cis-elements corresponded to G-box and Box-4 cis-elements. There were multiple G-box repeats in RsCCT2, RsCCT5, RsCCT11, RsCCT41, and RsCCT49 (Figure 5, Supplementary Table S11). Other light-responsive elements, such as the GTAT-motif, GT1-motif, and TCT-motif, were enriched in the Ib, IIc, and IId subgroups. In the phytohormone-responsive category, the top three cis-elements in RsCCT genes were the abscisic acid (ABA) response element (ABRE), methyl jasmonate (MeJA) response element (CGTCA-motif and TGACG-motif) (Figure 5, Supplementary Table S11). Interestingly, some ABRE also coexisted with the G-box cis-element. In the stress-responsive category, some elements were found to be related to different stress responses, such as drought, wounding, oxidation, and low temperature. ARE and the GC-motif were responsive to anaerobic conditions, TC-rich repeats were related to defense and stress responsiveness, LTR was responsive to low-temperature, and MBS to drought tolerance (Supplementary Table S11). In the plant growth and development category, the O2-site involved in zein metabolism regulation was found in many genes. Additional elements were also found, such as CAT-box related to meristem expression, MSA-like cis-elements involved in cell cycle regulation, and the circadian cis-element involved in circadian control (Supplementary Table S11).
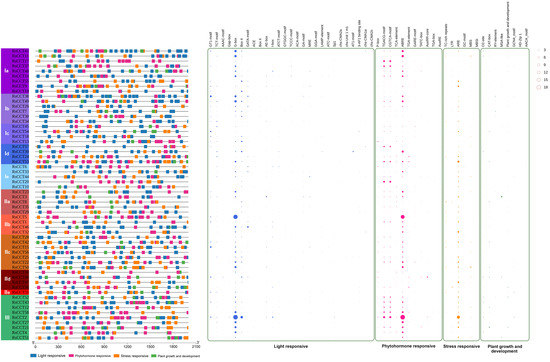
Figure 5.
Cis-element analysis in RsCCT genes. Different color squares represented the distribution of different cis-elements in the RsCCT gene promoter. Circles of different sizes represent the number of different promoters of cis-elements in RsCCT genes.
3.5. Expression Profiles of RsCCT Genes in Different Tissues
A heat map (Figure 6) was drawn based on the RNA-Seq data from five radish tissues (leaf, flower, silique, callus, and stem) to investigate the expression patterns of RsCCT genes [33]. Gene expression patterns were closely related to gene function. The RsCCT genes exhibited different expression patterns in different tissues and showed tissue specificity. Generally, the COL genes were expressed in various tissues, mainly in the leaf and flower. For example, RsCCT15 and RsCCT25 were abundantly expressed in all tissues, RsCCT22, RsCCT36, and RsCCT50 were expressed at high levels in the leaf, flower, silique, and stem, while RsCCT5 was only expressed in the flower (Figure 6). RT-qPCR analyzed the expression levels of eight RsCCT genes in five tissues (stem, leaf, flower, bud, and silique) (Figure 7a). Compared with other tissues, RsCCT14, RsCCT22, RsCCT36, RsCCT42, and RsCCT51 showed higher expression levels in the leaf, while RsCCT15, RsCCT21, and RsCCT56 showed higher expression levels in the flower.
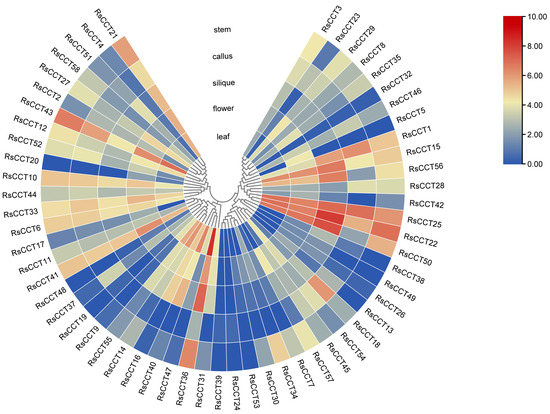
Figure 6.
Tissue-specific expression pattern of RsCCT genes. The heatmap was constructed based on the log2 (FPKM + 1) values of RsCCT genes. Red and blue, respectively, indicate higher and lower transcript abundance.
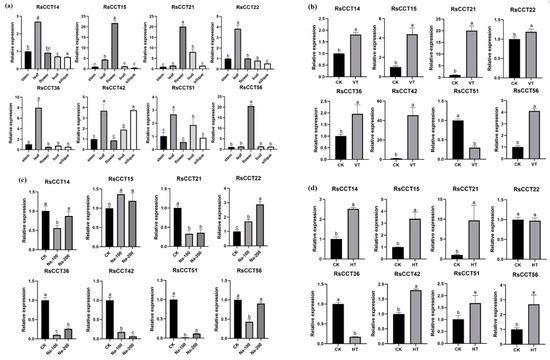
Figure 7.
The expression levels of representative RsCCT genes (a) of five radish tissues (stem, leaf, flower, bud, and siliques) under (b) vernalization treatment, (c) salt stress, (d) heat stress. Each error bar indicates standard deviation among three biological replicates. The value with different letter indicates significant difference at p < 0.05.
3.6. Expression Profiles of RsCCT Genes in Response to Vernalization and Its Potential Role in Flowering
The expression of RsCCT genes from the vegetative stage to the reproductive stage (Figure 8) was analyzed according to available RNA-seq data [34]. Most RsCCT genes showed differential expression patterns at the two stages. The expression level of IIc subgroup genes, except RsCCT42, were upregulated at the reproductive stage, suggesting that these genes may be closely related to the bolting and flowering of radish. While RsCCT37 and RsCCT48 were highly expressed only at the vegetative stage (Figure 8).
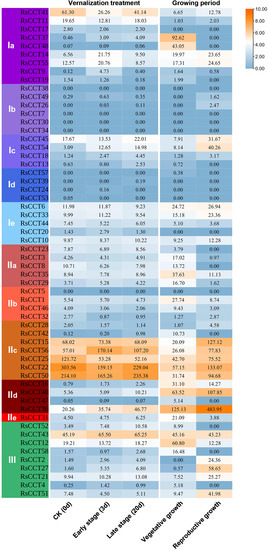
Figure 8.
Expression profiles of RsCCT genes at different stages of vernalization and growing period. The heatmap was constructed based on the log2 (FPKM + 1) values of RsCCT genes. The value in the rectangle was FPKM of each RsCCT gene.
The expression patterns of RsCCT genes (Figure 8) under different stages of vernalization were investigated based on RNA-seq data [35]. The expression patterns of some COL genes, such as RsCCT22 and RsCCT50, were downregulated in the early stage of vernalization and upregulated in the late stage of vernalization. Furthermore, RsCCT36 and RsCCT54 had continuously upregulated expression during the vernalization process (Figure 8), indicating their essential roles during flowering. RT-qPCR (Figure 7b) showed that the expression trend of eight RsCCT genes under vernalization treatment was similar to that of RNA-Seq data. Some members showed strong sensitivity to vernalization treatment. For example, the expression of RsCCT15, RsCCT21, RsCCT42, and RsCCT56 were significantly upregulated under vernalization treatment. By contrast, the expression of RsCCT51 was significantly downregulated.
3.7. Expression Profiles of RsCCT Genes in Response to Various Abiotic Stress
To investigate the expression patterns of RsCCT under various abiotic stress treatments, we compared the differences in the expression changes of RsCCT genes under Cd, Cr, Pb, and salt stresses, using published RNA-seq data [36,37,38,39], as well as under heat stress, using measured RNA-seq data (Figure 9). The expression patterns of most RsCCT genes were different under the various abiotic stresses. Specifically, RsCCT1 exhibited upregulated expression under three heavy metal (Cd, Cr, and Pb) stresses. The expression pattern of RsCCT15 was upregulated, and the expression pattern of RsCCT28 was downregulated under Pb stress. The expression pattern of RsCCT21 was upregulated under Cd stress, while the expression pattern of RsCCT22 was downregulated under Cr stress. Under salt stresses, the expression patterns of RsCCT16 and RsCCT40 were upregulated and the expression patterns of RsCCT50 and RsCCT51 were downregulated. Under heat stress, the expression patterns of RsCCT22, RsCCT36, RsCCT40, and RsCCT50 were downregulated (Figure 9). The changes in RsCCT expression profiles indicate that they might play important roles during abiotic stress response.
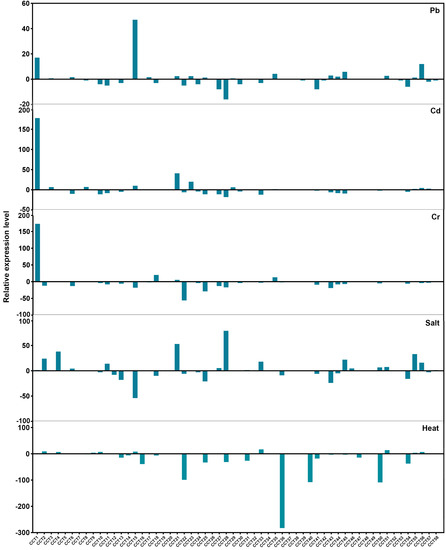
Figure 9.
Expression profiles of RsCCT genes under Pb, Cd, Cr, and salt and heat stresses, obtained from RNA-seq data. The expression level of the RsCCT gene was standardized with log2 (FPKM + 1).
We analyzed the expression profiles of eight RsCCT genes under heat stress and salt stress conditions by RT-qPCR (Figure 7c–d). Under heat stress, the expression levels of RsCCT14, RsCCT15, RsCCT21, and RsCCT56 increased exponentially, and the expression levels of RsCCT42 and RsCCT51 increased moderately (Figure 7d). The expression levels of RsCCT36 were significantly downregulated. In addition, we analyzed the expression level of RsCCT genes after 24 h of 100 mM and 200 mM salt stress (Figure 7c). The expression level of RsCCT21, RsCCT36, RsCCT42, and RsCCT51 were significantly downregulated under 100 mM and 200 mM salt stress treatment. The expression level of RsCCT22 was significantly upregulated under the same stress treatment. Moreover, the expression level of RsCCT36 and RsCCT51 were significantly downregulated under 100 mM salt stress treatment and there was no significant change under 200 mM salt stress treatment.
4. Discussion
CCT proteins have drawn more attention in recent years because of their multiple functions in plant growth, development, morphogenesis, and abiotic stress response [29,30]. To date, CCT genes have been extensively analyzed in different plant species [5,18,30,40,41]. However, there are no related reports on R. sativus. Brassicaceae species had similar flowering habits with various flowering times and responded differently to the day’s temperature and length [28]. Here, we systematically analyzed the CCT genes of B. rapa, B. nigra, B. oleracea, and R. sativus to provide more information about the evolution of CCT genes.
4.1. Characterization and Evolutionary Relationship of CCT Genes
Sequence alignment, phylogenetic analysis, and the syntenic relationship of all CCT proteins from these five species were clustered into three groups and divided into eleven subgroups (Figure 2 and Figure 3). The group classifications were the same as that in previous reports [8], indicating that our results were reliable. The phylogenetic tree showed that CCT genes in R. sativus have similar evolutionary features as Brassica species. It demonstrated that the Brassicaceae CCT genes may have been rapidly and independently amplified by a common ancestor in the process of evolution. Levels of predicted tandem and segmental duplications in gene families are negatively correlated, and segmental duplication events usually cause scattered family genes [42]. We detected 55, 40, 46, and 34 pairs of CCT genes in B. rapa, B. nigra, B. oleracea, and R. sativus, respectively (Figure 1b; Supplementary Figure S2), suggesting that segmental duplication events were the major force of the expansion of CCT genes. Ka/Ks values of all CCT gene pairs were less than 1 (Supplementary Table S8), indicating that these genes mainly underwent purifying selection and tended to have a conserved original function [43].
During evolution, members of the polygenic family often undergo functional differentiation, acquiring or losing specific conserved domains. In this study, the phylogenetic tree showed that Ia of CFM genes were closer to COL genes than the other CFM genes (Figure 2). In CFM genes, the gene structure of Ia genes differed from that of Ib, Ic, and Id, while Ia had similar gene structures with that of IId and IIe (COL) (Figure 3). These results supported that COL and CMF genes had an evolutionary correlation, and CMF genes may have evolved from the COL genes [5]. It was also consistent with previous report that the CCT domain was more conserved than the B-box domain [44].
Differential retention, deletion, and mutation of gene structure are critical for species domestication and adaptability enhancement [5]. Plant evolution was related to intron number and distribution. According to the gene structure of RsCCT genes, COL and PRR genes contain more introns than CMF genes, indicating that the COL and PRR genes might have complex differentiation and numerous functional discrepancies.
The CCT genes in R. sativus showed similar evolutionary characteristics as that in three Brassica [21], with the LF subgenome retaining more genes than the MF1 and MF2 subgenomes (Supplementary Figure S2). Subgenome dominance was a common phenomenon in Brassica, and also occurred in R. sativus [45]. The functions of RsCCT genes in the LF subgenome may be relatively conservative, while their homologous genes in MF1 and MF2 subgenomes may accumulate more functional mutations.
4.2. Potential Roles of RsCCT Genes in Bolting and Flowering
The long-term evolution and selection have resulted in the substantial divergence of functional and regulation methods of the CCT genes. There is considerable evidence that the CCT genes widely participate in the regulation of flowering time. However, the regulatory mechanism of RsCCT genes and their specific relationship with flowering time has not been extensively explored.
Recent research has demonstrated that COL genes regulate plant flowering by mediating light signals of plant development. PRR genes regulated flowering time by responding to the circadian rhythm and participating in the photoperiodic pathway [46,47]. In the present study, 27 light-responsive elements (such as G-box and Box 4) were identified in the promoter of RsCCT genes (Figure 6). For example, some CMF genes such as RsCCT11, RsCCT41, RsCCT38 and RsCCT49, COL genes such as RsCCT5, and PRR genes RsCCT2, RsCCT21, RsCCT27, and RsCCT58 contained more than eight copies of G-box (Supplementary Table S11). We proposed that these RsCCT genes may play crucial roles in controlling flowering through the photoperiodic pathway. Homology analysis effectively finds candidate flowering genes and constructs flowering time regulatory networks. Based on the functional conservation of homologous genes in A. thaliana, RsCCT genes in the Ie and III subgroups would play similar roles in regulating the circadian rhythm (Figure 2). In A. thaliana, ZIM, ZML1, and ZML2 were expressed in flowers and flower buds at the reproductive stage [48]. In our study, the expression pattern of RsCCT33, the homolog of ZML1 and ZML2, was similar to ZIM, indicating the potential regulation related to flower development.
Vernalization is the process of perceiving prolonged exposure to winter cold leading to spring flowering [49]. In A. thaliana, a VRN gene-mediated vernalization pathway promoted flowering upon exposure to long-term cold temperatures [50,51]. In this study, it was found that the expression of some RsCCT genes (Figure 8), such as RsCCT10, RsCCT11, and RsCCT50, changed in the early stage of vernalization and returned to the original expression in the late stage of vernalization. This indicated that these genes may play important roles during the vernalization pathway.
4.3. Potential Role of RsCCT Genes in Abiotic Stress Responses
Increasing studies have revealed that CCT genes participate in the photoperiod pathways and the circadian clock and are involved in stress response and many other related traits [29,30].
Some RsCCT genes have been found to be involved in a series of abiotic stress responses, and their transcriptional expressions were significantly altered by Cd, Cr, Pb, and heat and salt stresses, suggesting their contribution to multiple stresses in radish. Under Cd, Cr, and Pb stress treatments (Figure 9), the expression of RsCCT1, RsCCT15, RsCCT22, RsCCT27, and other RsCCT genes were either induced or inhibited. These results were consistent with various cis-regulatory elements (Figure 5), such as TGACG-motif, CGTCA-motif, ABRE, and ARE responsive to phytohormones and stress responses presented in their gene promoters. The expression of IIa COL genes such as RsCCT16 and RsCCT40 were upregulated, while IIe COL genes such as RsCCT15 and RsCCT56 were downregulated. These results supposed the co-existence of CCT, and the B-box domain might synergize abiotic stress [52].
Most genes related to abiotic stress response were controlled by the biological clock. In A. thaliana, PRR5 was directly involved in the regulation of flowering time and cold stress response by regulating transcription factors [53,54]. RsPRR5 (RsCCT51) was upregulated under Cd, Cr, Pb, and heat and salt stresses (Figure 9). Phytohormones mediate abiotic stress processes in many plants. For example, abscisic acid (ABA) accumulates rapidly under salt stresses [55]. In this study, the ABRE cis-element involved in the ABA responsiveness was repeated multiple times in RsCCT14, RsCCT21, and RsCCT51 (Figure 5); these genes were significantly downregulated under salt stress and were significantly upregulated under heat stress. Overall, RsCCT genes have evolved their own way to respond to abiotic stress, which could be valuable for improving the adaptability to adverse environments.
5. Conclusions
In summary, we first performed a whole-genome identification and analyzed the evolutionary relationship of CCT genes among B. rapa, B. nigra, B. oleracea, and R. sativus. A total of 254 CCT genes were identified and clustered into three groups. Then, the gene structures, the promoter cis-elements, and the expression patterns were analyzed, which provided useful information on the function of RsCCT genes in bolting, flowering, and abiotic stress tolerance. Our findings laid a foundation for further studies on the function of RsCCT genes.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/horticulturae8050381/s1, Figure S1: Chromosome localization and duplication analyses of CCT genes Chromosome localization and duplication analyses of CCT genes in B. rapa (a), B. nigra (b), and B. oleracea (c). Gene codes represented different groups in different colors. Different colored lines connecting two chromosomal regions indicate synteny blocks in the genome; Figure S2: The number of CCT genes in the three subgenomes (LF, MF1, MF2) of B. rapa, B. nigra, B. oleracea and R. sativus; Figure S3: (a–d) Web logos of CCT domain. Comparative analysis of CCT domains among B. rapa, B. oleracea, R. sativus, and B. nigra, respectively. The x-axis indicates the conserved domain sequences. The y-axis presents the relative entropy, which reflects the conservation rate of each amino acid; Table S1: Primers used for qRT-PCR verification of differentially expressed genes; Table S2: Properties and locations of the predicted RsCCT proteins; Table S3: All species CCT genes name control list; Table S4: Subgenome distribution of CCT genes; Table S5: Motif sequence comparison table; Table S6: Segmental duplication CCT genes; Table S7: Ka/Ks values of duplicated RsCCT gene pairs; Table S8: Ka/Ks values of duplicated CCT gene pairs; Table S9: List of orthologous gene groups among five Brassicaceae species; Table S10: cis-element function of RsCCT genes; Table S11: cis-element statistics of RsCCT genes.
Author Contributions
Q.T.: Data curation, Formal analysis, Writing—original draft, Writing—review and editing. Z.H.: Writing—review and editing. S.M.: Conceptualization, Validation, Funding acquisition, Writing—review and editing. J.Z.: Conceptualization, Validation, Funding acquisition, Writing—review and editing. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the National Key Research and Development Program of China (2017YFD0101806), the Agricultural Talents Program of the Chinese Academy of Agricultural Sciences (CAASQNYC-KYYJ-38), the Natural Science Foundation of Hunan Province (2020JJ5642), and the Central Public-interest Scientific Institution Basal Research Fund (No. 1610242021008).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare that there are no conflict of interest.
References
- Song, Y.H.; Shim, J.S.; Kinmonth-Schultz, H.A.; Imaizumi, T. Photoperiodic flowering: Time measurement mechanisms in leaves. Annu. Rev. Plant Biol. 2015, 66, 441–464. [Google Scholar]
- Liu, H.; Zhou, X.; Li, Q.; Wang, L. CCT domain-containing genes in cereal crops: Flowering time and beyond. Theor. Appl. Genet. 2020, 133, 1385–1396. [Google Scholar] [CrossRef] [PubMed]
- Putterill, J.; Robson, F.; Lee, K.; Simon, R.; Coupland, G. The CONSTANS gene of Arabidopsis promotes flowering and encodes a protein showing similarities to zinc finger transcription factors. Cell 1995, 80, 847–857. [Google Scholar] [CrossRef] [Green Version]
- Wenkel, S.; Turck, F.; Singer, K.; Gissot, L.; Gourrierec, J.L.; Samach, A.; Coupland, G. CONSTANS and the CCAAT box binding complex share a functionally important domain and interact to regulate flowering of Arabidopsis. Plant Cell 2006, 18, 2971–2984. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- James, C.; Thomas, T.; Burkhard, S.; Nils, S.; Stefan, T.; Bailey, P.C.; O’Sullivan, D.M.; Blazquez, M.A. Genome dynamics explain the evolution of flowering time CCT domain gene families in the Poaceae. PLoS ONE 2012, 7, e45307. [Google Scholar]
- Robson, F.; Costa, M.M.; Hepworth, S.R.; Vizir, I.; Pineiro, M.; Reeves, P.H.; Putterill, J.; Coupland, G. Functional importance of conserved domains in the flowering-time gene CONSTANS demonstrated by analysis of mutant alleles and transgenic plants. Plant J. 2001, 28, 619–631. [Google Scholar] [CrossRef] [PubMed]
- Suarez-Lopez, P.; Wheatley, K.; Robson, F.; Onouchi, H.; Valverde, F.; Coupland, G. CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis. Nature 2001, 410, 1116–1120. [Google Scholar] [CrossRef] [PubMed]
- Griffiths, S.; Dunford, R.P.; Coupland, G.; Laurie, D.A. The evolution of CONSTANS-Like gene families in barley, rice, and Arabidopsis. Plant Physiol. 2003, 131, 1855–1867. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murakami, M.; Ashikari, M.; Miura, K.; Yamashino, T.; Mizuno, T. The Evolutionarily conserved OsPRR quintet: Rice pseudo-response regulators implicated in circadian rhythm. Plant Cell Physiol. 2003, 44, 1229–1236. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chia, T.Y.P.; Müller, A.; Jung, C.; Mutasa-Göttgens, E.S. Sugar beet contains a large CONSTANS-LIKE gene family including a CO homologue that is independent of the early-bolting (B) gene locus. J. Exp. Bot. 2008, 59, 2735–2748. [Google Scholar] [PubMed] [Green Version]
- Kikuchi, R.; Kawahigashi, R.; Oshima, M.; Ando, T.; Handa, H. The differential expression of HvCO9, a member of the CONSTANS-like gene family, contributes to the control of flowering under short-day conditions in barley. J. Exp. Bot. 2012, 63, 773–784. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Simon, S.; Rühl, M.; Montaigu, A.; Wötzel, S.; Coupland, G. Evolution of CONSTANS regulation and function after gene duplication produced a photoperiodic flowering switch in the Brassicaceae. Mol. Biol. Evol. 2015, 32, 2284–2301. [Google Scholar] [CrossRef] [Green Version]
- Datta, S.; Hettiarachchi, G.H.; Deng, X.W.; Holm, M. Arabidopsis CONSTANS-LIKE3 is a positive regulator of red light signaling and root growth. Plant Cell 2006, 18, 70–84. [Google Scholar] [CrossRef] [Green Version]
- Takeshi, M.; Norihito, N. Pseudo-Response Regulators (PRRs) or True Oscillator Components (TOCs). Plant Cell Physiol. 2005, 46, 677–685. [Google Scholar]
- Nemoto, Y.; Nonoue, Y.; Yano, M.; Izawa, T. Hd1, a CONSTANS ortholog in rice, functions as an Ehd1 repressor through interaction with monocot-specific CCT-domain protein Ghd7. Plant J. 2016, 86, 221–233. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Shen, J.; Yan, X.; Li, X.; Xiao, J.; Xiong, L. Ghd2, a constans-like gene, confers drought sensitivity through regulation of senescence in rice. J. Exp. Bot. 2016, 67, 5785–5798. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mitsui, Y.; Shimomura, M.; Komatsu, K.; Namiki, N.; Shibata-Hatta, M.; Imai, M.; Katayose, Y.; Mukai, Y.; Kanamori, H.; Kurita, K. The radish genome and comprehensive gene expression profile of tuberous root formation and development. Sci. Rep. 2015, 5, 10835. [Google Scholar] [CrossRef] [Green Version]
- Cheng, F.; Liang, J.; Cai, C.; Cai, X.; Wu, J.; Wang, X. Genome sequencing supports a multi-vertex model for Brassiceae species. Curr. Opin. Plant Biol. 2017, 36, 79–87. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Li, X.; Ge, C.; Chang, J.; Shi, M.; Chen, J.; Qiao, L.; Chang, Z.; Zhang, J.; Zhang, J. Characterization of the CCT family and analysis of gene expression in aegilops tauschii. PLoS ONE 2017, 12, e0189333. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, L.; Yi, D.; Yang, J.; Liu, X.; Pang, Y. Genome-wide identification, expression analysis and functional study of CCT gene family in medicago truncatula. Plants 2020, 9, 513. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Fan, X.; Hu, Y.; Zhou, X.; He, Q.; Liang, L.; Xing, Y. Global analysis of CCT family knockout mutants identifies four genes involved in regulating heading date in rice. J. Integr. Plant Biol. 2021, 63, 913–923. [Google Scholar] [CrossRef]
- Chen, H.; Wang, T.; He, X.; Cai, X.; Lin, R.; Liang, J.; Wu, J.; King, G.; Wang, X. BRAD V3.0: An upgraded Brassicaceae database. Nucleic Acids Res. 2022, 50, D1432–D1441. [Google Scholar] [CrossRef] [PubMed]
- Lamesch, P.; Berardini, T.Z.; Li, D.; Swarbreck, D.; Wilks, C.; Sasidharan, R.; Muller, R.; Dreher, K.; Alexander, D.; Garcia-Hernandez, M.; et al. The Arabidopsis Information Resource (TAIR): Improved gene annotation and new tools. Nucleic Acids Res. 2012, 40, 1202–1210. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Doerks, T.; Bork, P. SMART 7: Recent updates to the protein domain annotation resource. Nucleic Acids Res. 2012, 40, 302–305. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Elisabeth, G.; Alexandre, G.; Christine, H.; Ivan, I.; Appel, R.D.; Amos, B. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003, 31, 3784–3788. [Google Scholar]
- Lescot, M. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Liu, S.; Liu, Y.; Yang, X.; Tong, C.; Edwards, D.; Parkin, I.A.; Zhao, M.; Ma, J.; Yu, J.; Huang, S. The Brassica oleracea genome reveals the asymmetrical evolution of polyploid genomes. Nat. Commun. 2014, 5, 3930. [Google Scholar] [CrossRef]
- Li, Y.P.; Xu, M.L. CCT family genes in cereal crops: A current overview. Crop J. 2017, 5, 449–458. [Google Scholar] [CrossRef]
- Mengarelli, D.A.; Zanor, M.I. Genome-wide characterization and analysis of the CCT motif family genes in soybean (Glycine max). Planta 2020, 253, 15. [Google Scholar] [CrossRef] [PubMed]
- Bowers, J.E.; Chapman, B.A.; Rong, J.; Paterson, A.H. Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature 2003, 422, 433–438. [Google Scholar] [CrossRef] [PubMed]
- Semon, M.; Wolfe, K.H. Consequences of genome duplicate on. Curr. Opin. Genet. Dev. 2007, 17, 505–512. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Yue, Z.; Mei, S.; Qiu, Y.; Yang, X.; Chen, X. A de novo genome of a Chinese radish cultivar. Hortic. Plant J. 2015, 1, 155–164. [Google Scholar]
- Nie, S.; Li, C.; Wang, Y.; Xu, L.; Muleke, E.M.; Tang, M.; Sun, X.; Liu, L. Transcriptomic analysis identifies differentially expressed genes (DEGs) associated with bolting and flowering in radish (Raphanus sativus L.). Front. Plant Sci. 2016, 7, 682. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, C.; Wang, S.; Xu, W.; Liu, X. Genome-wide transcriptome profiling of radish (Raphanus sativus L.) in response to vernalization. PLoS ONE 2017, 12, e0177594. [Google Scholar] [CrossRef]
- Xu, L.; Wang, Y.; Liu, W.; Wang, J.; Zhu, X.; Zhang, K.; Yu, R.; Wang, R.; Xie, Y.; Zhang, W.; et al. De novo sequencing of root transcriptome reveals complex cadmium-responsive regulatory networks in radish (Raphanus sativus L.). Plant Sci. 2015, 236, 313–323. [Google Scholar] [CrossRef]
- Xie, Y.; Ye, S.; Wang, Y.; Xu, L.; Zhu, X.; Yang, J.; Feng, H.; Yu, R.; Karanja, B.; Gong, Y.; et al. Transcriptome-based gene profiling provides novel insights into the characteristics of radish root response to Cr stress with next-generation sequencing. Front. Plant Sci. 2015, 6, 202. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Xu, L.; Chen, Y.; Shen, H.; Gong, Y.; Limera, C.; Liu, L. Transcriptome profiling of radish (Raphanus sativus L.) root and identification of genes involved in response to lead (Pb) stress with next generation sequencing. PLoS ONE 2013, 8, e66539. [Google Scholar] [CrossRef]
- Sun, X.; Xu, L.; Wang, Y.; Luo, X.; Zhu, X.; Karanja, B.; Nie, S.; Feng, H.; Li, C.; Liu, L. Transcriptome-based gene expression profiling identifies differentially expressed genes critical for salt stress response in radish (Raphanus sativus L.). Plant Cell Rep. 2016, 35, 329–346. [Google Scholar] [CrossRef]
- Zhang, L.; Li, Q.; Dong, H.; He, Q.; Liang, L.; Tan, C.; Han, Z.; Yao, W.; Li, G.; Zhao, H.; et al. Three CCT domain-containing genes were identified to regulate heading date by candidate gene-based association mapping and transformation in rice. Sci. Rep. 2015, 5, 7663. [Google Scholar] [CrossRef] [Green Version]
- Jin, M.; Liu, X.; Jia, W.; Liu, H.; Li, W.; Peng, Y.; Du, Y.; Wang, Y.; Yin, Y.; Zhang, X.; et al. ZmCOL3, a CCT gene represses flowering in maize by interfering with the circadian clock and activating expression of ZmCCT. J. Integr. Plant Biol. 2018, 60, 465–480. [Google Scholar] [CrossRef]
- Cannon, S.B.; Mitra, A.; Baumgarten, A.; Young, N.D.; May, G. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 2004, 4, 10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, J.; Zhang, Z.; Vang, S.; Yu, J.; Wong, G.K.; Wang, J. Correlation between Ka/Ks and Ks is related to substitution model and evolutionary lineage. J. Mol. Evol. 2009, 68, 414–423. [Google Scholar] [CrossRef]
- Shalmani, A.; Jing, X.Q.; Shi, Y.; Muhammad, I.; Zhou, M.R.; Wei, X.Y.; Chen, Q.Q.; Li, W.Q.; Liu, W.T.; Chen, K.M. Characterization of B-BOX gene family and their expression profiles under hormonal, abiotic and metal stresses in Poaceae plants. BMC Genom. 2019, 20, 27. [Google Scholar] [CrossRef] [PubMed]
- Jeong, Y.M.; Kim, N.; Ahn, B.O.; Oh, M.; Chung, W.H.; Chung, H.; Jeong, S.; Lim, K.B.; Hwang, Y.J.; Kim, G.B.; et al. Elucidating the triplicated ancestral genome structure of radish based on chromosome-level comparison with the Brassica genomes. Theor. Appl. Genet. 2016, 129, 1357–1372. [Google Scholar] [CrossRef]
- Errum, A.; Rehman, N.; Khan, M.R.; Ali, G.M. Genome-wide characterization and expression analysis of pseudo-response regulator gene family in wheat. Mol. Biol. Rep. 2021, 48, 2411–2427. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.Y.; Chen, X.H.; Yu, H.W.; Tian, Q.L.; Lu, L.M. Identification and characterization of CONSTANS-like genes from Curcuma alismatifolia. Hortic. Environ. Biotechnol. 2021, 62, 279–286. [Google Scholar] [CrossRef]
- Shikata, M.; Matsuda, Y.; Ando, K.; Nishii, A.; Takemura, M.; Yokota, A.; Kohchi, T. Characterization of Arabidopsis ZIM, a member of a novel plant-specific GATA factor gene family. J. Exp. Bot. 2004, 55, 631–639. [Google Scholar] [CrossRef] [PubMed]
- Sung, S.; He, Y.; Eshoo, T.W.; Tamada, Y.; Johnson, L.; Nakahigashi, K.; Goto, K.; Jacobsen, S.E.; Amasino, R.M. Epigenetic maintenance of the vernalized state in Arabidopsis thaliana requires Like Heterochromatin Protein 1. Nat. Genet. 2006, 38, 706–710. [Google Scholar] [CrossRef] [PubMed]
- Gendall, A.R.; Levy, Y.Y.; Wilson, A.; Dean, C. The VERNALIZATION 2 gene mediates the epigenetic regulation of vernalization in Arabidopsis. Cell 2001, 107, 525–535. [Google Scholar] [CrossRef] [Green Version]
- Levy, Y.Y.; Mesnage, S.; Mylne, J.S.; Gendall, A.R.; Dean, C. Multiple roles of Arabidopsis VRN1 in vernalization and flowering time control. Science 2002, 297, 243–246. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Li, R.; Dai, Y.; Chen, X.; Wang, X. Genome-wide identification and expression analysis of the B-box gene family in the Apple (Malus domestica Borkh.) genome. Mol. Genet. Genom. 2018, 293, 303–315. [Google Scholar] [CrossRef] [PubMed]
- Santosh, S.B.; Yamashino, T.; Okada, R.; Nomoto, Y.; Mizuno, T.; Tezuka, Y.; Itoh, T.; Tomita, M.; Otsuki, S.; Aoki, S. Pseudo-Response Regulator (PRR) homologues of the moss physcomitrella patens: Insights into the evolution of the PRR family in land plants. DNA Res. 2011, 18, 39–52. [Google Scholar]
- Nakamichi, N.; Kiba, T.; Kamioka, M.; Suzuki, T.; Yamashino, T.; Higashiyama, T. Transcriptional repressor PRR5 directly regulates clock-output pathways. Proc. Natl. Acad. Sci. USA 2012, 109, 17123–17128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Agarwal, P.K.; Jha, B. Transcription factors in plants and aba dependent and independent abiotic stress signalling. Biol. Plant. 2010, 54, 201–212. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).