Abstract
Auxin’s role in the post-ripening of peaches is widely recognized as important. However, little is known about the processes by which auxin regulates fruit post-ripening. As one of the early auxin-responsive genes, it is critical to understand the role of small auxin-up RNA (SAUR) genes in fruit post-ripening and softening. Herein, we identified 72 PpSAUR auxin-responsive factors in the peach genome and divided them into eight subfamilies based on phylogenetic analysis. Subsequently, the members related to peach post-ripening in the PpSAUR gene family were screened, and we targeted PpSAUR43. The expression of PpSAUR43 was decreased with fruit post-ripening in melting flesh (MF) fruit and was high in non-melting flesh (NMF) fruit. The overexpression of PpSAUR43 showed a slower rate of firmness decline, reduced ethylene production, and a delayed fruit post-ripening process. The MADS-box gene family plays an important regulatory role in fruit ripening. In this study, we showed with yeast two-hybrid (Y2H) and bimolecular fluorescence complementation (BIFC) experiments that PpSAUR43 can interact with the MADS-box transcription factor PpCMB1(PpMADS2), which indicates that PpSAUR43 may inhibit fruit ripening by suppressing the function of the PpCMB1 protein. Together, these results indicate that PpSAUR43 acts as a negative regulator involved in the peach post-ripening process.
1. Introduction
The fruit is an organism for which metabolic activities are still in order after harvesting [1]. A series of physiological and biochemical processes in the tissues causes the degradation of certain organic macromolecules and changes in cell wall and cell membrane structures. Peaches (Prunus persica (L.) Batsch) are a popular commercial fruit with nutritional and pharmacological properties [2]. According to the Food and Agriculture Organization (FAO), 1,527,052 ha of peaches were cultivated worldwide in 2019, and China is home to the world’s largest peach industry [3]. However, peaches generally soften rapidly, with reduced quality and nutritional value, until they become rotten after harvest. Hence, postharvest softening and the senescence regulatory mechanism have been the focus of much research.
During the period of ripening, peaches undergo textural changes that lead to the loss of tissue firmness [4]. It has already been determined in the existing literature that many different phytohormones play pivotal functions in regulating ripening and softening [5,6,7,8,9]. For example, ethylene has long been regarded as the main regulator of ripening in climacteric fruit [10,11]. For climacteric fruit, ethylene biosynthesis is regulated by two systems, system 1 and system 2. System 1 is only responsible for producing low concentrations of basal ethylene at the pre-climacteric stage, and when the ethylene produced by system 1 reaches a certain level, system 2 begins to produce large levels of ethylene, which is responsible for the ripening process by self-catalysis [12,13,14]. As a climacteric fruit, peach cultivars usually have three types: melting flesh (MF), non-melting flesh (NMF), and stony hard (SH) according to the differences in fruit firmness and texture characteristics [15,16]. Previous studies have found that a high concentration of auxin can stimulate the synthesis of system 2 ethylene through its inductive action on the expression of PpACS1 (1-aminocyclopropane-1-carboxylic acid synthase) in MF peaches during post-ripening. In contrast, the low concentrations of auxin due to the low expression of PpYUC11 suppress PpACS1 expression and ethylene production in SH peaches during post-ripening [17,18,19,20,21]. In fact, the involvement of auxin in peach ripening has long been reported. It has been pointed out earlier that the peak of ethylene during the later stages of peach development coincides with higher levels of auxin, and the application of exogenous auxin could promote ethylene production and peach ripening, suggesting that auxin may be involved in ethylene synthesis [22,23]. Similar findings showing that auxin regulates ripening by modulating ethylene and auxin crosstalk have previously been reported in apples [23], durians [24], papayas [25], and tomatoes [26,27]. Accumulating evidence suggests that both ethylene and auxin interact in the regulation of fruit ripening. In addition to auxin indirectly regulating fruit ripening via modulating ethylene and auxin crosstalk, auxin may also play its own role in the ripening process of peaches. Our predecessors used genomic methods to determine that an increase in auxin has an independent effect on the hormones in the ripening process of climacteric peaches [28]. However, the role of auxin during fruit ripening remains obscure, although auxin accumulation appears to be critical and participates in the regulation of the ripening of fruit.
The fruit ripening regulatory network is not only dependent on the expression of genes directly involved in hormone synthesis and response but also on many transcription factor families (i.e., ERF, NAC, MADS-Box, etc.) that participate in the regulation of fruit ripening by regulating the expression of hormone synthesis and response genes to successfully complete the fruit ripening process [29,30,31,32,33,34]. Among these TFs, MADS-box genes constitute a highly conserved family of TFs and have been shown in numerous studies to be involved in fruit ripening regulation [35]. RIN, which is a member of the SEPALLATA clade (E-class) of MADS-box genes, is essential in regulating tomato fruit ripening and softening because the fruits failed to ripen in the RIN mutants [36]. In apple, a Rin-homologue of MdMADS8/9 was revealed to regulate fruit ripening by directly controlling the auxin levels. MdMADS8/9 suppressed GH3 expression, resulting in increased levels of free-state IAA during apple ripening [37]. In banana, Elitzur et al. determined that the two SEP-like MADS-box genes MaMADS1 and MaMADS2 are essential for fruit ripening because the silencing of either gene resulted in reduced fruit ethylene synthesis and delayed ripening [38]. In peach, Li et al. found that the suppression of PpSEP1 could decrease the transcription of cell wall metabolism genes (i.e., Endo-PG3, and PME1) and ethylene synthesis-related genes (i.e., ACS2 and ACO1) [39]. In other fruits, such as citrus, strawberries, and grapes, MADS-box transcription factors have been demonstrated to be involved in fruit ripening [40,41,42]. PpCMB1(PpMADS2), which belongs to the SEP clade of the MADS-box gene family, is a homolog of SlCMB1 in the tomato MADS-box family [43]. SlCMB1 has been reported to be a potential regulator of ethylene biosynthesis and carotenoid accumulation during maturation [44]. In addition, the homologous genes of PpCMB1 have been reported in apple and cherry as MdMADS6 and PaMADS2, respectively. Both MdMADS6 and PaMADS2 have been shown to play a role in fruit ripening and softening, speculating that the PpCMB1 may have a similar function [45,46]. In peach, PpCMB1 has been found to be highly expressed in fruit and is involved in fleshy fruit development and disease resistance in postharvest fruit [47,48,49].
As one of the three early auxin-responsive gene families, small auxin-up RNA (SAUR) can rapidly respond to auxin applications without protein synthesis and is critically involved in the auxin signaling pathway [50,51]. Since McClure and Guilfoyle [52] identified the first SAUR gene from soybeans, SAUR members have subsequently been identified in several species, including Arabidopsis, rice, tomatoes, and apples [53,54,55,56]. Most SAURs do not contain introns in their coding sequence (CDS), and SAUR proteins are small and contain a conserved SAUR-specific domain (SSD) of approximately 60 residues in the central region [57]. Previous research has shown that SAURs regulate a wide variety of cellular, physiological, and developmental processes that involve the hormonal and environmental control of plant growth and development [58]. For example, a few SAUR proteins have been found to be capable of binding calmodulins (CaM) and CaM-like proteins (CML) [59,60], regulating organ elongation [61] and apical hook development [62], increasing abiotic stress tolerance [63], altering stamen filament elongation [64], and promoting leaf senescence [65,66] and cell expansion [67]. Moreover, SAUR may be related to the polar transport of auxin. In Arabidopsis, both AtSAUR19 overexpression lines and AtSAUR19, 23, and 24 RNA interference lines can change the transport of auxin from the hypocotyl axis to the base [68]. Kant et al. [69] reported that the OsSAUR39 gene negatively regulates auxin synthesis and transport in rice. In addition to auxin, gibberellin [64], abscisic acid [70], brassinosteroid [71], and ethylene [72,73] are all thought to be related to the expression of the SAUR gene, indicating that SAUR may be related to other hormones that regulate plant growth and development. Studies on SAURs involved in ripening have seldom been reported. However, it has been shown that SISAUR69 is an RIN-regulated gene involved in the regulation of ripening in tomatoes. The overexpression of SlSAUR69 represses auxin transport and enhances ethylene sensitivity, thereby resulting in the premature initiation of ripening. Conversely, the downregulation of SlSAUR69 delays the initiation of fruit ripening [74]. However, the precise biological roles of SAUR in the context of peach ripening still need to be clarified.
At present, reducing the fruit softening rate has become an important breeding objective. A previous study developed by Carrasco-Valenzuela et al. [75] identified PpSAUR43 (Prupe.8G079500) as a key candidate gene for the rate of peach softening by integrating quantitative trait loci (QTL) analyses. Additionally, the expression profile of the differentially expressed gene coding for PpSAUR43 has been experimentally confirmed by RT-qPCR analyses and has been shown to be upregulated in low-softening-rate progenies. However, there are no direct reports on the regulation of the fruit post-ripening process by PpSAUR in peaches. Therefore, this study aimed to identify the regulatory pathway of PpSAUR43 during the post-ripening of peaches, which will help to further understand the molecular mechanism of peach ripening and softening.
2. Materials and Methods
2.1. Plant Materials
Fruits of the melting flesh (MF) peach ‘Yu Hua Lu’ (‘YHL’) and the non-melting flesh (NMF) peach ‘Babygold 5’ (‘B5’) were collected as the experimental materials. The ‘YHL’ (27 June 2021) and ‘B5’ (24 July 2021) fruits were harvested in the commodity maturation period and stored at 25.0 ± 1 °C in the laboratory, from which samples were taken at two-day intervals for ‘YHL’ and three-day intervals for ‘B5’, which were then stored at −80 °C. In this study, the fruits were harvested from the orchard of the Experimental Station of the College of Horticulture of Northwest A&F University, Yangling, Shaanxi, China (34°20′ N, 108°24′ E), which contains five-year-old trees.
Transgenic tobacco (Nicotiana benthamiana), containing a nuclear localization signal (NLS-mCherry), used for subcellular localization analysis, was grown in a phytotron (25.0 °C; photoperiod: 16 h/8 h light/dark). Arabidopsis, used for a bimolecular fluorescence complementation (BiFC) assay, was grown in a growth chamber at 22.0 °C under a 16 h/8 h light/dark photoperiod.
2.2. Measurement of Fruit Firmness
Fruit firmness was measured by a GY-4 penetrometer (Top Instrument Co., Ltd., Hangzhou, China) equipped with a 7.9 mm probe. From each group, 10 fruits were randomly selected, and fruit firmness was measured around the injection site after peeling.
2.3. Determination of Ethylene Production
For ethylene production, nine fruit samples were weighed and sealed in a jar for 1 h (temperature 25.0 ± 1 °C and relative humidity of 75–85%; 0.03% of CO2 and 21% of O2.). Then, the sample gases (1 mL) were withdrawn from the jar and measured using a gas chromatograph (Trace GC Ultra; Thermo Fisher Scientific, Milan, Italy). The working conditions of gas chromatography were: chromatographic column (Rtx-1701) of 30 m × 0.32 mm, column temperature of 80 °C, carrier gas of nitrogen (N2), detector (FID) temperature of 130 °C, inlet temperature of 120 °C, hydrogen flow rate of 40 mL·min–1, air flow rate of 400 mL·min–1, pressure of 100 kpa, tail gas flow of 30 mL·min–1, column flow rate of 1.19 mL·min–1, injection volume of 1 mL, and splitting ratio of 5.0. There were three replicates per treatment, and three values per replicate.
where c is the converted ethylene content after the gas chromatographic determination in the sample gas (μL·L−1), V is the volume of the closed space of the desiccator (mL), t is the measurement time (h), and m is the fruit quality (g).
2.4. Determination of IAA Concentration
The extraction and purification of IAA in the injection site after the peeling of the fruits were carried out according to the method of Tatsuki et al. [17]. The content of IAA in each sample was determined using the LC-MS/MS analysis method as described by Müller et al. [76].
2.5. Identification of the PpSAUR Genes in Peaches
To identify the SAUR family genes in the peach species, we first acquired the SAUR family member CDS sequences and protein sequences of Arabidopsis thaliana [53] and Solanum lycopersicum [55]. Then, the protein sequences of Arabidopsis thaliana and Solanum lycopersicum were used as queries in the Genome Database for Rosaceae (Prunus persica v2.0.a1 genome) to identify the potential SAUR protein sequences in peaches (GDR: https://www.rosaceae.org/species/prunus/all (accessed on 18 August 2020)). The protein candidate sequences were inspected using SMART (http://smart.embl-heidelberg.de/ (accessed on 18 August 2020)) and Pfam (the protein families database: http://pfam.xfam.org/ (accessed on 18 August 2020)) to confirm that they conserved the PF02519 domain. Subsequently, we used Expasy (https://www.expasy.org/ (accessed on 4 September 2020)) to estimate the physicochemical parameters (i.e., length, molecular weight (MW), and isoelectric point (pI)) of the PpSAURs.
2.6. Phylogenetic Analyses of the SAUR Genes in Peaches and Arabidopsis
In order to analyze the phylogenetic organization of the SAUR family, we performed multiple sequence alignments for all available SAUR full-length protein sequences of Arabidopsis and peaches using DNAMAN 9.0 (Lynnon Biosoft, San Ramon, CA, USA). A phylogenetic tree based on the neighbor-joining (NJ) method was constructed for SAUR proteins by using the software MEGA 7.0 (with a Jones–Taylor–Thornton (JTT) model and complete deletion). A bootstrap test was carried out using 1000 replicates to construct the reliability of the tree.
2.7. RNA Isolation and Quantitative Real-Time Polymerase Chain Reaction (RT-qPCR)
‘YHL’ fruits were taken after 0, 2, 4, 6, and 8 days of storage (25.0 ± 1 °C) for the RT-qPCR analysis. ‘B5’ fruits were taken after 0, 3, 6, 9, and 12 days of storage (25.0 ± 1 °C) for RT-qPCR analysis. Three biological replicates were designed, and each repetition was carried out by mixing flesh samples from three fruits.
The RNA extraction of the peach samples (3.5 g) was carried out using cetyltrimethylammonium bromide according to the method of Xing et al. [77]. Then, 1 μg of RNA was used to synthesize the cDNA with a PrimeScriptTM RT reagent Kit with a gDNA Eraser (Perfect Real Time) (TaKaRa, Beijing, China). Then, quantitative reverse transcription RT-qPCR was performed in volumes of 10 μL of reaction buffer containing 5 μL of SYBR® Premix Ex TaqTM II (Tli RNaseH Plus) (2×) (TaKaRa), 1 μL of cDNA, 0.2 μL of each primer, and 3.4 μL of deionized water. The PCR program was as follows: 95 °C for 1 min, then 40 cycles of 95 °C for 15 s, 60 °C for 20 s, and 72 °C for 20 s. After this, the mixed sample was heated to 95 °C for 10 s and cooled to 65 °C for 15 s. The sample was then heated up to 95 °C at a rate of 0.1 °C·s–1 for melting curve analyses. RT-qPCR was run on the CFX Connect Real-Time System (Bio-Rad Laboratories, Inc., Hercules, CA, USA). The peach PpTUA5 (Prupe.6G004100) was used as an internal control and the 2−∆∆CT method was used to calculate the transcript accumulation [78]. Each sample was analyzed in triplicate. The primers are listed in Table S1.
2.8. Subcellular Localization Analysis
The coding regions of PpSAUR43 and PpCMB1 without the stop codon were amplified by PCR using Takara LA Taq high-fidelity DNA polymerase (TaKaRa). PCR primers (Table S1) were designed using Primer 6.0 software (PREMIER Biosoft International, CA, USA). Then, we inserted them into the pCAMBIA2300-green fluorescent protein (GFP) vector to produce the respective fusion constructs of the pCAMBIA2300-PpSAUR43-green fluorescent protein (GFP) and pCAMBIA2300-PpCMB1-green fluorescent protein (GFP) using a Seamless Cloning Kit (Sangon Biotech, Shanghai, China). The fusion plasmids and the control vector (pCAMBIA2300-GFP) were separately transformed into the Agrobacterium tumefaciens GV3101 chemically competent cell. Transgenic tobacco plant material containing a nuclear localization signal (NLS-mCherry) was used in this experiment [79]. The Agrobacterium tumefaciens GV3101 strain was agroinfiltrated into five-week-old leaves of Nicotiana tabacum with different vectors. Transformed leaves were analyzed at 48 h after the infection of the lower epidermal cells. Confocal imaging was performed using an inverted Leica TCS-SP8 SR laser scanning microscope. For the imaging expression of GFP constructs, excitation lines of a solid-state laser of 488 nm were used with a 498/510-nm bandpass filter in the single-track facility of the microscope. For the imaging co-expression of the GFP and mCherry constructs, excitation lines of a solid-state laser of 488 nm for GFP and 552 nm for mCherry were used alternately with line switching using the sequential scanning mode of the microscope.
2.9. Peach Injection Assays
The overexpressed PpSAUR43 construct was generated by cloning the CDS of PpSAUR43 into the pCAMBIA2300 vector. The empty pCAMBIA2300 vector was used as a control. The recombinant plasmids and empty vectors were respectively introduced into the Agrobacterium tumefaciens GV3101 chemically competent cell. The agroinfiltration of ‘YHL‘ fruits was performed according to Jia et al. [80]. The fruits were harvested when they were commercially mature (i.e., 110 days after full bloom). At least 200 fruits per treatment were randomly collected with three biological replicates for an injection test of Agrobacterium tumefaciens in peaches. The infiltrated peach fruits were stored at 25.0 ± 1 °C. The ethylene production was determined with whole fruits, while the positions on fruits for the firmness measurement and for sampling with gene expression analysis were the injection site after the peeling of the fruits. The injection sites after the peeling of the fruits were then snap-frozen in liquid nitrogen and stored at −80 °C.
2.10. Yeast Two-Hybrid (Y2H) Assay
PpSAUR43 was amplified using LA Taq high-fidelity DNA polymerase (TaKaRa) and inserted into the pGBKT7 vector (Clontech, Mountain View, CA, USA) to generate pGBKT7-PpSAUR43. The CDS of PpCMB1 was inserted into the pGADT7 vector (Clontech) to generate the pGADT7-PpCMB1 recombinant plasmid using a Seamless Cloning Kit (Sangon Biotech). The recombinant plasmid pGBKT7-PpSAUR43 was separately co-transformed with pGADT7-PpCMB1 into the Y2H-Gold yeast (Clontech) chemically competent cell. The pGADT7 empty vector was used as a negative control. A cDNA library was constructed from the RNA extracted from ‘Qianjianbai’ fruit during post-ripening (constructed by Takara Bio Company). The interaction assay consisted of three biological replicates. The yeast transformation experiment was conducted using the Yeastmaker Yeast Transformation System 2 User Manual (Clontech). The transformed Y2H-Gold yeast strain was cultured on SD/-Leu/-Trp and SD/-Trp/-Leu/-Ade/-His media with the indicator x-α-gal (5-Bromo-4-chloro-3-indolyl-α-D-galactoside) and cultured at 28.0 °C for 3 days.
2.11. Bimolecular Fluorescence Complementation (BiFC) Assay
The CDSs of PpSAUR43 and PpCMB1, excluding the stop codons, were inserted into both pSPYCE and pSPYNE using a Seamless Cloning Kit (Sangon Biotech), respectively. Different combinations of plasmids (pSPYCE-PpSAUR43 + pSPYNE-PpCMB1 and pSPYCE-PpSAUR43 + pSPYNE) were transformed into Arabidopsis protoplasts according to the polyethylene glycol (PEG) mediation method, as reported by Yoo et al. [81]. The protoplasts were then cultured at 22.0 °C in an incubator. After 18–22 h, the yellow fluorescent protein (YFP) signal was detected by confocal microscopy (Leica TCS-SP8 SR) at a wavelength of 514 nm.
2.12. Statistical Analysis
All data are presented as the mean and standard deviation (SD) and were derived from at least three biological replicates. An ANOVA with Duncan’s test was conducted using IBM SPSS Statistics 23.0 (SPSS Inc., Chicago, IL, USA). A pairwise comparison was computed using the Duncan test. A single asterisk (*) and double asterisks (**) in the figures indicate significant differences of p < 0.05 and p < 0.01, respectively. Duncan’s test was used, and different letters indicate significant differences (p < 0.05). Origin 7.0 (OriginLab, Northampton, MA, USA) was used to prepare the figures.
3. Results
3.1. Screening and Identification of the PpSAUR Family Genes
We identified 72 SAUR genes from the peach genome, and all of them contained the domains (PF02519) of the SAUR gene family (Table 1). According to the sequence of their chromosomes or scaffold positions, they were numbered from top to bottom and named PpSAUR1–PpSAUR72 (Figure S1). These members are distributed among eight chromosomes, but the number of members on different chromosomes varies widely. For example, chromosome 8 contains 40 members, while chromosome 4 contains only one member (Figure S1). These SAUR gene family members encode 73–206 amino acids (aa), with an isoelectric point (Theoretical Isoelectric Point, pI) of 4.53–11.12, and a molecular weight (MW) of 8.77–23.31 KDa (Table 1). The calculated MW and pI were nearly identical to those determined previously in other plant species [53,54,55,56].

Table 1.
Small auxin-up RNA (SAUR) gene family in peach.
3.2. Sequence and Phylogenetic Analysis
Using the protein sequences of 72 peach SAUR family members and 72 Arabidopsis SAUR family members, we constructed a phylogenetic tree to better understand the evolutionary relationship between the PpSAUR and AtSAUR proteins (Figure 1). The peach SAUR family was divided into groups (I–VIII) according to the 72 SAUR members of Arabidopsis (Figure 1). The results show that the number of family members in different groups varied greatly. For example, the largest group I contained 29 members, while the smallest group VI contained only 8 members. In group I, only one member was from Arabidopsis and the rest were from peach. PpSAUR43 and PpSAUR47 belong to paralogous genes and showed a closer relationship with AtSAUR1.
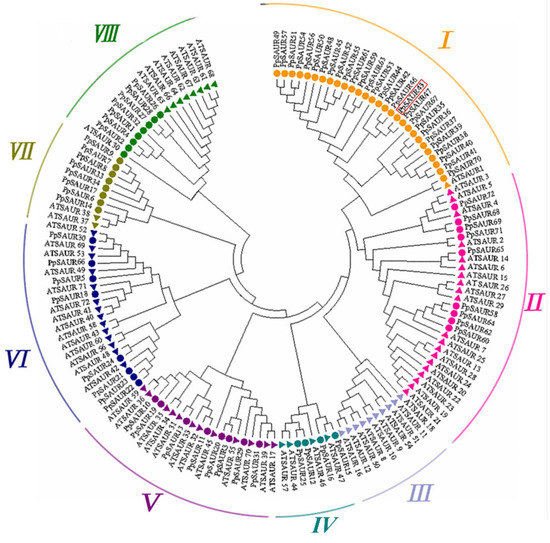
Figure 1.
Phylogenetic analysis of SAUR from peaches (PpSAUR) and Arabidopsis (AtSAUR). The unrooted tree was generated using the MEGA7 program by the neighbor-joining method with the following parameters: A bootstrap analysis with 1000 replicates was carried out. Gene IDs corresponding to the peach SAUR proteins listed in the tree are presented in Table 1.
3.3. Expression Patterns of PpSAUR43 in the MF and NMF Cultivars during Peach Post-Ripening
Previous studies have shown that the expression profile of the differentially expressed gene PpSAUR43, as a candidate for the fruit softening rate gene, experimentally confirmed by RT-qPCR, is upregulated in low-softening-rate siblings [75]. We were interested in PpSAUR43, which has been predicted by previous studies during peach post-ripening. Therefore, we selected the fruit of the MF peach ‘YHL’ and the NMF peach ‘B5’ for study at the post-ripening stage. Previous studies have shown that the firmness of the NMF fruit ‘B5’ has a declining tendency in the post-ripening stage compared to the MF fruit ‘YHL’ [82]. Meanwhile, the ethylene peak for the ‘YHL’ fruit was higher than that found for the ‘B5’ fruit [82]. We detected the expression patterns of PpSAUR43 in the fruit of the MF peach ‘YHL’ and the NMF peach ‘B5’ (Figure 2). In total, PpSAUR43 expression was decreased during post-ripening. In addition, we observed a high expression level of PpSAUR43 in ‘B5’ and a low expression level in ‘YHL’. These results suggest that PpSAUR43 may be involved in peach post-ripening.
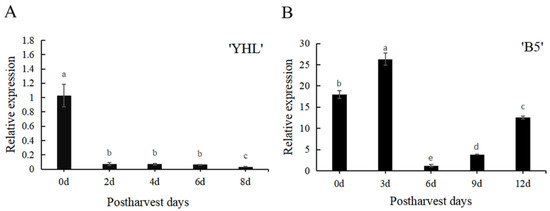
Figure 2.
Expression level of PpSAUR43 of ‘YHL’ and ‘B5’ in the post-ripening stage. (A) Expression level of PpSAUR43 of ‘YHL’. (B) Expression level of PpSAUR43 of ‘B5’. The fruits were stored at 25.0 ± 1 °C after harvesting. Three biological replicates were used and each repetition was carried out by mixing flesh samples from three fruits. The expression of PpSAUR43 in ‘YHL’ at day 0 was used as the standard for normalization. Duncan’s test was used with the expression of PpSAUR43 at day 0 as a control, with different letters indicating significant differences (p < 0.05).
3.4. PpSAUR43 Proteins Are Localized in the Cell Membrane and Nucleus
To determine the subcellular localization of PpSAUR43, a 35S-PpSAUR43-GFP recombinant plasmid was generated (Figure 3A). Here, the 35S-PpSAUR43-GFP recombinant plasmid was transformed into transgenic tobacco containing a nuclear localization signal (NLS-mCherry). At the same time, empty pCAMBIA2300 was used as a control. The results show that the PpSAUR43 signal overlapped the nuclear localization signal, suggesting that PpSAUR43 was located in the nucleus (Figure 3B). At the same time, we also detected a GFP signal on the cell membrane. Therefore, PpSAUR43 may be localized in the membrane and nucleus.
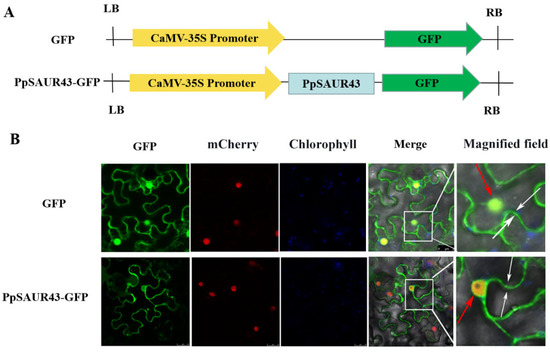
Figure 3.
Subcellular localization of PpSAUR43 proteins. (A) Schematic diagram of GFP constructs. (B) Subcellular localization of proteins in Nicotiana tabacum leaf mesophyll cells. The nucleus was indicated by mCherry carrying a nuclear localization signal. The GFP fluorescence, chlorophyll autofluorescence, and mCherry were merged. The magnified field showed GFP signals in different parts of cells. White and red arrows indicate the presence of GFP signals in the cell membrane and nucleus, respectively.
3.5. PpSAUR43 Impacts Peach Fruit Post-Ripening and Softening as an Inhibitor
To gain insight into the role of PpSAUR43 in peach fruit post-ripening, we investigated the effects of the transient overexpression of PpSAUR43 on peach fruit post-ripening by performing fruit peel injection assays. As shown in Figure 4, the infested portion of the pCAMBIA2300–PpSAUR43 infiltrated fruit surface was dark green, while the surface of the empty pCAMBIA2300 vector-infiltrated control fruit was light green or normal red (Figure 4A). The firmness of the peach tended to decrease during storage at 25.0 ± 1 °C, and the transient overexpression of PpSAUR43 significantly reduced the rate of decline in fruit firmness after the fourth day of storage (Figure 4B). In addition, we also measured the ethylene release rate of overexpressed fruits. During the storage period, the ethylene release rate in PpSAUR43-overexpressing fruits and the corresponding control fruits had the same trend, both of which increased first and then decreased, reaching the peak ethylene release at four days. The ethylene release rate for the PC2300-PpSAUR43 fruit showed no significant difference when compared with the control fruit (Figure 4B). Subsequently, we selected the agroinfiltrated parts of the fruit to analyze the changes in the free-state IAA content (Figure 4B). The results show that the free-state IAA content in the PpSAUR43 overexpressing fruits was significantly lower than that of the control on days 0, 2, 4, and 6 of storage. We confirmed that PpSAUR43 expression is effectively overexpressed at the molecular level using RT-qPCR analyses at seven days post-inoculation. The levels of PpSAUR43 expression at the injection site of the overexpressed fruit were significantly higher than the control by approximately 10-, 14-, 2-, and 45-fold on days 0, 2, 4, and 6 of storage. At day 8 of storage, the transient overexpression effect decayed, and the expression of PpSAUR43 at the injection site of the overexpressed and control fruits was not significantly different. This indicates that the transient overexpression of PpSAUR43 in peaches is significant (Figure 4C). To further understand the role of PpSAUR43 in the regulation of peach fruit post-ripening, we analyzed several ripening-related genes in the PpSAUR43 overexpressing fruits and control fruits by RT-qPCR analyses: cell wall metabolism-related gene (polygalacturonase, PpPG), 1-amino cyclopropane-1-carboxylic acid synthase gene (PpACS1), and 1-amino cyclopropane-1-carboxylic acid oxidase gene (PpACO1). The transient overexpression of PpSAUR43 led to the downregulation of the ethylene synthesis gene PpASC1 and the cell wall degradation gene PpPG, and no significant difference in PpACO1, except that it was significantly lower than the control at four days. We additionally measured the expression levels of PpCMB1, a member of the MADS-box family that is closely related to fruit ripening. The transcript abundance of PpCMB1 in the PpSAUR43 overexpressing fruits was significantly lower than that of the control fruit on days 0, 2, and 4. These results indicate that PpSAUR43 is essential for peach fruit post-ripening by acting as an inhibitor. To elucidate the mechanism of PpSAUR43 overexpression on the delay of peach post-ripening and softening, we also estimated the transcript levels of a set of genes related to auxin transport and homeostasis, including the polar auxin transport gene (PpPIN1), the IAA-amido synthetase gene (Gretchen-Hagen 3 (PpGH3.1)), and the indole-3-acetic acid (IAA)-amino hydrolase gene (PpILR1). The expressions of PpPIN1 and PpGH3.1 were upregulated more in the transient overexpression of the PpSAUR43 overexpressing fruit compared to those of the control fruit, while the expression of PpILR1 was markedly more downregulated than in the control fruit (Figure 4C), indicating that PpSAUR43 might promote the transport of auxin and the conversion of the free state to the conjugate state.
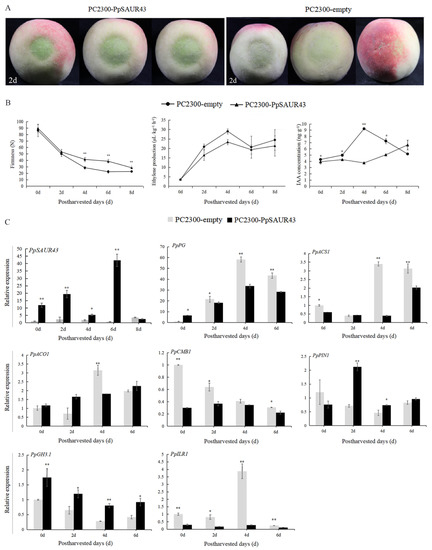
Figure 4.
Transient overexpression of PpSAUR43 in ‘YHL’ fruit. (A) Phenotypes of infiltrated fruits. Empty pCAMBIA2300 vectors (PC2300-empty) were used as a control. (B) Changes in fruit firmness, ethylene production, and IAA concentration during fruit storage. (C) Gene expression of PpSAUR43, PpPG, PpACS1, PpACO1, PpCMB1, PpPIN1, PpGH3.1, and PpILR1. Gene expression was measured relative to that of PpTUA5. Fruit was stored at 25.0 ± 1 °C after harvest. At the same storage days, PC2300-PpSAUR43 was compared with controls. Error bars represent the SD of the measurements from three independent repeat experiments. Duncan’s test was used to compare the differences. * p < 0.05; ** p < 0.01.
3.6. PpSAUR43 Interacts with PpCMB1
To identify the proteins that interact with PpSAUR43, we performed a yeast two-hybrid (Y2H) library assay using PpSAUR43 as bait. We first validated the self-activation of PpSAUR43. It turned out that neither the control nor the experimental groups showed a blue color on the SD/-Trp/-Leu/-His/x-α-gal plates, indicating that PpSAUR43 does not have transcriptional activation activity (Figure S3). Subsequently, we co-transformed the yeast strain with BD-PpSAUR43 and the library plasmid, coated the co-transformed bacteria on SD/-Ade/-His/-Leu/-Trp/x-α-gal plates, and after three days, the blue spots were subjected to PCR. The products with bands greater than 500 bp were selected for sequencing and for blast screening of the possible interacting proteins with the NCBI website (Table S2 and Figure S4). Notably, we identified PpCMB1, which has been shown to regulate fruit ripening and softening in tomatoes, as a potential PpSAUR43-interacting protein [44]. To further validate the interaction between PpSAUR43 and PpCMB1, we inserted the PpCMB1 full-length CDS into the pGADT7 vector to obtain AD-PpCMB1 and co-transformed AD-PpCMB1 and BD-PpSAUR43 into the Y2H-Gold strain (Figure 5A). As expected, the Y2H-Gold strain harboring BD-PpSAUR43/AD-PpCMB1 grew and turned blue on SD/-Leu/-Trp/-His/-Ade medium supplemented with X-α-gal (Figure 5A). The results confirm the interaction between PpSAUR43 and PpCMB1 in yeast. We further examined the subcellular localization of PpCMB1 by transforming Nicotiana tabacum leaves with Agrobacterium tumefaciens-containing 35S-PpCMB1-GFP vectors. The results show that PpCMB1 was localized in the cell membrane and nucleus. (Figure S5), which was the same for PpSAUR43. Furthermore, a BIFC assay confirmed this interaction in Arabidopsis protoplasts. Different combinations, including pSPYCE-PpSAUR43/pSPYNE-PpCMB1 and pSPYCE-PpSAUR43/pSPYNE, were co-transformed into Arabidopsis protoplasts (Figure 5B). The YFP signal that was detected in the nucleus could be detected only when pSPYCE-PpSAUR43 was co-transformed with pSPYNE-PpCMB1. These results demonstrate that PpSAUR43 interacts physically with PpCMB1 in the nucleus (Figure 5C).
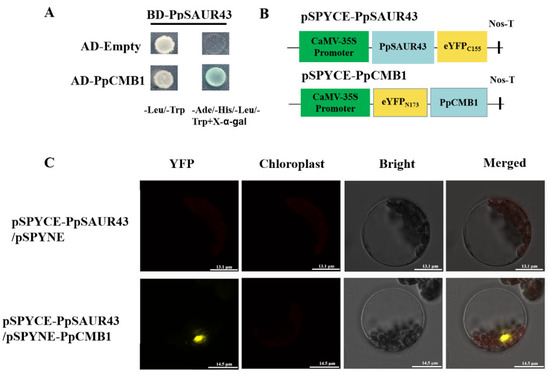
Figure 5.
Analysis of the interaction between PpSAUR43 and PpCMB1. (A) The Y2H assay confirmed the interaction between PpSAUR43 and PpCMB1 in yeast. (B) Schematic diagrams of the vectors used in the BIFC assay. (C) The BIFC assay confirmed the interaction between PpSAUR43 and PpCMB1 Arabidopsis protoplasts.
4. Discussion
Fruit ripening is a complex process involving metabolic and physiological changes that depend on the coordinated role of multiple hormone signals, microRNAs, epigenetic maintenance, epigenetic modifying genes, and other major regulatory factors [83,84,85]. As is already known, ethylene is regarded as the major regulator of climacteric fruit ripening [84,86]. However, an increasing body of evidence has pointed toward a more complex role of auxin in climacteric fruit ripening [28]. Primary auxin-responsive genes, the SAUR gene family, are present as large gene families in diverse plant species and have been well established in plant growth and development, such as cell elongation [87] and leaf senescence [65,66].
In this study, we identified 72 SAUR genes in peaches and named them according to their positions on the chromosome site. All of the members were encoded with the SAUR domains. The number of SAUR family members in peaches is almost the same as that of Arabidopsis (81) [53], rice (58) [54], and tomatoes (99) [55]. Based on their chromosomal distribution, it was found that the distribution of SAUR family members is often concentrated in tandem clusters on parts of the chromosome (Figure S1). SAUR gene clusters are also commonly found in other species. For example, four SAUR gene clusters have been identified in Arabidopsis, located on chromosomes 1, 3, 4, and 5, while five SAUR gene clusters have been identified in rice, located on chromosomes 1, 2, 3, 9, and 12 [53,54]. There were two SAUR genes clusters on chromosome 8 in the peach genome. We deduced that this may be due to gene duplication in the family. Gene duplication is a major way of increasing the number of gene families in which genes are usually similar, such as unequal exchange, retro-transposition, or whole-gene duplication, to produce genes or base sequences similar to the original gene [88]. It was reported that rapid gene amplification is prevalent in gene families associated with morphogenesis and stress responses, and gene duplication plays an important role in plant evolution [89]. To further investigate the evolutionary relationships of SAUR family genes, we constructed a phylogenetic tree of PpSAUR and divided it into eight subfamilies. We found a high degree of sequence similarity among the gene cluster members. The gene cluster members located on chromosome 8 are part of the first subgroup and are genetically close to AtSAUR1 (AT4G34770) and are presumed to have similar functions and to be important in hormone crosstalk [90]. Furthermore, we analyzed the gene structure of the SAUR family members (Figure S2). We found that 67 of the 72 SAUR families lacked intron structures, a feature also found in SAUR gene families of other species, such as rice and tomatoes [90,91]. This may be due to the fact that the genes that respond quickly to auxin tend to be more likely to have genes without introns [92].
Previous studies developed by Carrasco-Valenzuela et al. [75] used quantitative trait locus (QTL) and expression QTL (eQTL) analysis to screen the candidate gene, indole-3-acetic acid-induced protein (PpSAUR43), which is related to the fruit softening rate. Moreover, PpSAUR43 was identified to be differentially expressed in peaches with different softening rates by RNA-seq and RT-qPCR analysis, so we put the spotlight on PpSAUR43 (the RNA-seq data are available at the NCBI’s Sequence Read Achieve (available at: http://www.ncbi.nlm.nih.gov/sra (accessed on 22 June 2020)) with SRA accession number SRP186384). First, we analyzed the expression pattern of PpSAUR43 in different textured peach cultivars during post-ripening. Interestingly, the expression of PpSAUR43 was negatively correlated with the post-ripening process in NMF and MF fruits, and the expression of PpSAUR43 was high in the NMF varieties and low in the MF varieties. These results are concordant with the previous findings that the expression of PpSAUR43 is higher in the fruits of low-softening-rate cultivars than in high-softening-rate cultivars [75]. It is speculated that PpSAUR43 plays a negative role in peach fruit post-ripening and softening.
Some studies have shown that SAUR proteins may be localized in subcellular structures such as the nucleus, cytoplasm, and plasma membrane (PM), which may imply that these proteins have multiple functions in plants [50,93]. Our data show that PpSAUR43 is located in the nucleus and cell membrane. This dual localization hints at its function in transmitting auxin signals between the cell membrane and the nucleus. To further validate the role of SAUR in peach fruit post-ripening and softening, we characterized the biological function of PpSAUR43 in peaches by transient gene overexpression. The overexpression of PpSAUR43 significantly inhibits peach post-ripening, as evidenced by noticeable increases in the fruit firmness and delays in the fruit softening. A more noticeable metabolic change in fruit ripening is the softening caused by changes in the structure of the cell wall [94]. The main component of the gel layer in the cell wall is pectin, which plays an essential role in intercellular adhesion [95,96]. The degradation of pectin causes a rapid reduction in cell viscosity and leads to a reduction in fruit hardness. As polygalacturonase is the predominant enzyme that degrades pectin in the cell wall (PG), we measured the expression of the PpPG gene, a key gene that regulates fruit softening, to assess the degree of cell wall degradation between the overexpression of PpSAUR43 and the control fruit [97]. We found that although there was no significant difference between the experimental and the control groups on day 0 of storage, the overexpression of PpSAUR43 led to a decrease in the transcript level of PpPG in the fruit as the storage days increased. Ethylene is known to play a key role in fruit ripening. Hence, we also measured the amount of ethylene produced. Unfortunately, no significant differences were found between the overexpressed and control fruit. We speculate that the reason for this may be that we used whole peaches to measure the ethylene content and the small area of the injection site did not result in a significant difference. Subsequently, we further determined the expression of PpACS1, which is related to ethylene synthesis in the fruit, and found that the overexpression of PpSAUR43 resulted in a significantly lower expression of PpACS1 than the control. Recent studies have shown that a low level of auxin in peaches inhibits the expression of the PpACS1 gene [17,18,19]. Furthermore, we measured the transcript levels of PpACO1, a key ACC oxidase gene involved in peach ethylene synthesis, and found no significant change in PpACO1 expression after the overexpression of PpSAUR43, except for days. Based on the above data, we speculate that PpSAUR43 affects the transcription of PpACS1, probably due to reduced auxin levels in the fruit. The ability of SAUR proteins to alter auxin levels in vivo has been reported in other species for a long time. In Arabidopsis, AtSAUR19 overexpression in seedling hypocotyls increases translocation to auxin [68]. Similar findings have been obtained with OsSAUR39 in rice [98]. We measured the content of free-state IAA in the PpSAUR43 overexpressing fruit and control fruits and found that the overexpression of PpSAUR43 significantly reduced the level of IAA in vivo. Reducing auxin levels can be achieved through auxin transport and auxin conjugation or degradation mechanisms. The PIN-FORMED (PIN) gene family of transmembrane proteins as auxin efflux carriers plays a crucial role in polar auxin transport [99]. It has been suggested that PpPIN1 may regulate the distribution of auxin involved in peach ripening [22]. The expression of PpPIN1 was significantly upregulated in response to the overexpression of PpSAUR43, suggesting that PpSAUR43 facilitates the transport of auxin. GH3 genes, which catalyze the conversion of free-form auxin to the conjugated state, play a role in the maintenance of cellular auxin levels in a range of plant species [100]. Our results show that the expression of PpGH3.1 was elevated in comparison to the control. Interestingly, we then analyzed the transcript levels of the indole-3-acetic acid (IAA)-amino hydrolase PpILR1 and showed that the expression of PpILR1 was significantly lower when overexpressed by PpSAUR43. As previously reported, PpILR1 acts as a key gene for IAA-amino hydrolase and is positively involved in the regulation of peach fruit ripening [101]. This supports our speculation that PpSAUR43 is associated with auxin homeostasis and reduces the level of free-state auxin in vivo by promoting the formation of the auxin-conjugated state.
To further investigate the regulatory mechanism of PpSAUR43 on fruit softening, we demonstrated the specific interaction of PpSAUR43 with PpCMB1, a MADS-box protein of the SEP class, by Y2H and BIFC analysis. In our study, PpSAUR43 and PpCMB1 were both localized and interacted in the cell nucleus and membrane, which provided the possibility for their physical interaction. Some research indicates that SlCMB1, which shares high homology with RIN, SlMADS1, and SlMBP21, is a new component of the current model of the regulatory network that regulates ethylene biosynthesis during tomato fruit ripening [44]. Then, RT-qPCR analysis showed that PpCMB1 was downregulated during post-ripening in PpSAUR43 overexpressed fruits. These results imply that there is an antagonistic relationship between PpSAUR43 and PpCMB1 in the regulation of peach post-ripening and that PpSAUR43 may act as a negative regulator, while PpCMB1 is a positive regulator. This coincides with previous studies on tomatoes that have shown that the suppression of SlCMB1 results in delayed fruit ripening by affecting ethylene biosynthesis and signal transduction [44]. Notably, promoter analysis by website prediction showed that the PpCMB1 and PpSAUR43 gene promoter region contained the auxin-responsive element AuxRR-Core (GGTCCAT) and the ethylene-responsive element ERE motif (ATTTTAAA/ATTTCATA) (Table S3) [102,103]. These results suggest that PpSAUR43 and PpCMB1 act as modulators of the optimal balance between auxin and ethylene actions. Hence, on the one hand, overexpressing PpSAUR43 inhibits the transcriptional activation of PpCMB1, thereby suppressing the expression of downstream ethylene synthesis and signal transduction genes. On the other hand, PpSAUR43 could work in a regulation loop to decrease the free auxin level, which, in turn, would decrease the expression of PpACS1 via the auxin signal transduction pathway.
5. Conclusions
In conclusion, our study reports 72 SAUR family members in peaches after we performed bioinformatics analysis. The functions of peach PpSAUR43 were then identified by transient overexpression in peaches, and a new mechanism was found for the interaction between PpSAUR43 and PpCMB1 regulating peach post-ripening and softening. The findings of this study will help to develop methods to precisely control peach post-ripening and extend shelf life.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/horticulturae8050379/s1, Figure S1: Chromosomal localization of SAUR family members; Figure S2: Gene structure of SAUR family members; Figure S3: Transcriptional activation identification of the full length of PpSAUR43; Figure S4: The interacting confirmation between PpSAUR43 and PpCMB1, PpMY66, PpWHY1, and PpPP2C5 in yeast two-hybrid system; Figure S5: Subcellular localization of GFP constructs of PpCMB1 proteins; Table S1: Primers for cloning and RT-qPCR in this study; Table S2: Result of partial candidate positive clones by yeast two-hybrid (Y2H) library assay in peach; Table S3: Cis-acting regulatory elements were predicted in the promoter regions of PpSAUR43, PpCAMB1 related to fruit development and ripening in peach.
Author Contributions
C.Z. conceived and designed the research. J.W., W.S., K.L. and Z.X. conducted the experiments. J.M., D.Z. and C.Z. contributed reagents and gave advice. J.W. and Y.H. wrote the manuscript. Y.H. and K.S. edited the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research and the APC were funded by the National Key R&D Program of China (Grant No. 2019YFD1000203).
Institutional Review Board Statement
This research content of the manuscript does not involve ethical issues.
Informed Consent Statement
This research content of the manuscript does not involve humans.
Data Availability Statement
All results are included with in the article.
Acknowledgments
We thank Jing Zhang (Horticulture Science Research Center, Northwest A&F University, Xianyang, China) for providing professional technical assistance with LC-MS/MS analysis.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Grierson, D.; Kader, A.A. Fruit ripening and quality. In The Tomato Crop; Rudich, J., Ed.; Springer: London, UK, 1986; pp. 241–280. [Google Scholar]
- Li, Y.; Wang, L. Genetic Resources, Breeding Programs in China, and Gene Mining of Peach: A Review. Hortic. Plant J. 2020, 6, 205–215. [Google Scholar] [CrossRef]
- Food and Agricultural Organization. Available online: www.fao.org (accessed on 18 December 2021).
- Yoshida, M. Genetical studies on the fruit quality of peach varieties. III. Texture and keeping quality. Bull. Fruit Tree Res. Stn. 1976, 3, 1–16. [Google Scholar]
- Given, N.K.; Venis, M.A.; Gierson, D. Hormonal regulation of ripening in the strawberry, a non-climacteric fruit. Planta 1988, 174, 402–406. [Google Scholar] [CrossRef]
- Fuentes, L.; Figueroa, C.R.; Valdenegro, M. Recent Advances in Hormonal Regulation and Cross-Talk during Non-Climacteric Fruit Development and Ripening. Horticulturae 2019, 5, 45. [Google Scholar] [CrossRef] [Green Version]
- Obroucheva, N.V. Hormonal regulation during plant fruit development. Russ. J. Dev. Biol. 2014, 45, 14. [Google Scholar] [CrossRef]
- Wang, P.; Lu, S.; Zhang, X.; Hyden, B.; Qin, L.; Liu, L.; Bai, Y.; Han, Y.; Wen, Z.; Xu, J.; et al. Double NCED isozymes control ABA biosynthesis for ripening and senescent regulation in peach fruits. Plant Sci. 2021, 304, 110739. [Google Scholar] [CrossRef]
- Tan, B.; Lian, X.; Cheng, J.; Zeng, W.; Zheng, X.; Wang, W.; Ye, X.; Li, J.; Li, Z.; Zhang, L.; et al. Genome-wide identification and transcriptome profiling reveal that E3 ubiquitin ligase genes relevant to ethylene, auxin and abscisic acid are differentially expressed in the fruits of melting flesh and stony hard peach varieties. BMC Genom. 2019, 20, 892. [Google Scholar] [CrossRef] [Green Version]
- Burg, S.P. Ethylene action and the ripening of fruits. Science 1965, 148, 1190–1196. [Google Scholar] [CrossRef]
- Alexander, L.; Grierson, D. Ethylene biosynthesis and action in tomato: A model for climacteric fruit ripening. J. Exp. Bot. 2002, 377, 2039–2055. [Google Scholar] [CrossRef]
- Lelièvre, J.M.; Latchè, A.; Jones, B.; Hall, M.A. Ethylene and fruit ripening. Physiol. Plant. 1997, 101, 727–739. [Google Scholar] [CrossRef]
- Zarembinski, T.I.; Theologis, A. Ethylene biosynthesis and action: A case of conservation. Plant Mol. Biol. 1994, 26, 1579–1597. [Google Scholar] [CrossRef] [PubMed]
- Jerie, P.H.; Hall, M.A.; Jones, B. Aspects of the role of ethylene in fruit ripening. Acta Hortic. 1978, 80, 325–332. [Google Scholar] [CrossRef]
- Haji, T.; Yaegaki, H.; Yamaguchi, M. Inheritance and expression of fruit texture melting, non-melting and stony hard in peach. Sci. Hortic. 2005, 105, 241–248. [Google Scholar] [CrossRef]
- Bailey, J.S.; French, A.P. The inheritance of certain characters in the peach. Proc. Am. Soc. Hortic. Sci. 1932, 29, 127–130. [Google Scholar]
- Tatsuki, M.; Yamaguchi, H.M. The involvement of 1-aminocyclopropane-1-carboxylic acid synthase isogene, PpACS1, in peach fruit softening. J. Exp. Bot. 2006, 57, 1281–1289. [Google Scholar] [CrossRef]
- Tatsuki, M.; Haji, T.; Yamaguchi, M. The peach 1-aminocyclopropane-1-carboxylic acid synthase isogene, Pp-ACS1, is required for fruit softening. In Advances in Plant Ethylene Research; Springer: Dordrecht, Germany, 2007; pp. 227–228. [Google Scholar] [CrossRef]
- Tatsuki, M.; Nakajima, N.; Fujii, H.; Shimada, T.; Nakano, M.; Hayashi, K.-I.; Hayama, H.; Yoshioka, H.; Nakamura, Y. Increased levels of IAA are required for system 2 ethylene synthesis causing fruit softening in peach (Prunus persica L. Batsch). J. Exp. Bot. 2013, 64, 1049–1059. [Google Scholar] [CrossRef]
- Pan, L.; Zeng, W.; Liang, N. PpYUC11, a strong candidate gene for the stony hard phenotype in peach (Prunus persica L. Batsch), participates in IAA biosynthesis during fruit ripening. J. Exp. Bot. 2015, 22, 7031–7044. [Google Scholar] [CrossRef] [Green Version]
- Zeng, W.; Ding, Y.; Pan, L.; Wang, X.; Niu, L.; Lu, Z.; Cui, G.; Wang, Z. A CACTA transposable element in a PpYUC11 gene promoter is associated with the stony hard phenotype in peach. J. Fruit Sci. 2017, 4, 1239–1248. [Google Scholar] [CrossRef]
- Tadiello, A.; Ziosi, V.; Negri, A.S.; Noferini, M.; Fiori, G.; Busatto, N.; Espen, L.; Costa, G.; Trainotti, L. On the role of ethylene, auxin and a GOLVEN-like peptide hormone in the regulation of peach ripening. BMC Plant Biol. 2016, 16, 44. [Google Scholar] [CrossRef] [Green Version]
- Busatto, N.; Tadiello, A.; Trainotti, L.; Costa, F. Climacteric ripening of apple fruit is regulated by transcriptional circuits stimulated by cross-talks between ethylene and auxin. Plant Signal. Behav. 2016, 12, e1268312. [Google Scholar] [CrossRef] [Green Version]
- Khaksar, G.; Sirikantaramas, S. Auxin Response Factor 2A Is Part of the Regulatory Network Mediating Fruit Ripening Through Auxin-Ethylene Crosstalk in Durian. Front. Plant Sci. 2020, 11, 543747. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Li, W.; Xie, R.; Xu, L.; Zhou, Y.; Li, H.; Yuan, C.; Zheng, X.; Xiao, L.; Liu, K. CpARF2 and CpEIL1 interact to mediate auxin–ethylene interaction and regulate fruit ripening in papaya. Plant J. 2020, 103, 1318–1337. [Google Scholar] [CrossRef]
- Hao, Y.; Hu, G.; Breitel, D.; Liu, M.; Mila, I.; Frasse, P.; Fu, Y.; Aharoni, A.; Bouzayen, M.; Zouine, M. Auxin Response Factor SlARF2 Is an Essential Component of the Regulatory Mechanism Controlling Fruit Ripening in Tomato. PLoS Genet. 2015, 11, e1005649. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Chen, L.; Pang, S.; Zheng, Q.; Quan, S.; Liu, Y.; Xu, T.; Liu, Y.; Qi, M. Function Analysis of the ERF and DREB Subfamilies in Tomato Fruit Development and Ripening. Front. Plant Sci. 2022, 13, 849048. [Google Scholar] [CrossRef] [PubMed]
- Trainotti, L.; Tadiello, A.; Casadoro, G. The involvement of auxin in the ripening of climacteric fruits comes of age: The hormone plays a role of its own and has an intense interplay with ethylene in ripening peaches. J. Exp. Bot. 2007, 58, 3299–3308. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gu, T.; Jia, S.; Huang, X.; Wang, L.; Fu, W.; Huo, G.; Gan, L.; Ding, J.; Li, Y. Transcriptome and hormone analyses provide insights into hormonal regulation in strawberry ripening. Planta 2019, 250, 145–162. [Google Scholar] [CrossRef]
- Liu, G.S.; Li, H.L.; Grierson, D.; Fu, D.Q. NAC transcription factor family regulation of fruit ripening and quality: A Review. Cells 2022, 11, 525. [Google Scholar] [CrossRef]
- Xie, F.; Hua, Q.; Chen, C.; Zhang, Z.; Zhang, R.; Zhao, J.; Hu, G.; Chen, J.; Qin, Y. Genome-Wide Characterization of R2R3-MYB Transcription Factors in Pitaya Reveals a R2R3-MYB Repressor HuMYB1 Involved in Fruit Ripening through Regulation of Betalain Biosynthesis by Repressing Betalain Biosynthesis-Related Genes. Cells 2021, 10, 1949. [Google Scholar] [CrossRef]
- Jiang, G.; Zeng, J.; Li, Z.; Song, Y.; Yan, H.; He, J.; Jiang, Y.; Duan, X. Redox Regulation of the NOR Transcription Factor Is Involved in the Regulation of Fruit Ripening in Tomato. Plant Physiol. 2020, 183, 671–685. [Google Scholar] [CrossRef]
- Deng, H.; Chen, Y.; Liu, Z.; Liu, Z.Q.; Shu, P.; Wang, R.; Hao, Y.; Su, D.; Pirrello, J.; Liu, Y.; et al. SlERF.F12 modulates the transition to ripening in tomato fruit by recruiting the co-repressor TOPLESS and histone deacetylases to repress key ripening genes. Plant Cell. 2022, 4, 1250–1272. [Google Scholar] [CrossRef]
- Wang, X.; Pan, L.; Wang, Y.; Meng, J.; Deng, L.; Niu, L.; Liu, H.; Ding, Y.; Yao, J.-L.; Nieuwenhuizen, N.J.; et al. PpIAA1 and PpERF4 form a positive feedback loop to regulate peach fruit ripening by integrating auxin and ethylene signals. Plant Sci. 2021, 313, 111084. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Chen, K.; Grierson, D. A critical evaluation of the role of ethylene and MADS transcription factors in the network controlling fleshy fruit ripening. New Phytol. 2019, 221, 1724–1741. [Google Scholar] [CrossRef] [Green Version]
- Vrebalov, J.; Ruezinsky, D.; Padmanabhan, V.; White, R.; Medrano, D.; Drake, R.; Schuch, W.; Giovannoni, J. A MADS-Box Gene Necessary for Fruit Ripening at the Tomato Ripening-Inhibitor (Rin) Locus. Science 2002, 296, 343–346. [Google Scholar] [CrossRef] [PubMed]
- Schaffer, R.J.; Ireland, H.S.; Ross, J.J.; Ling, T.J. SEPALLATA1/2-suppressed mature apples have low ethylene, high auxin and reduced transcription of ripening-related genes. AOB Plants 2013, 5, 047. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Elitzur, T.; Yakir, E.; Quansah, L.; Zhangjun, F.; Vrebalov, J.; Khayat, E.; Giovannoni, J.J.; Friedman, H. Banana MaMADS Transcription Factors Are Necessary for Fruit Ripening and Molecular Tools to Promote Shelf-Life and Food Security. Plant Physiol. 2016, 171, 380–391. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Li, F.; Qian, M.; Han, M.; Liu, H.; Zhang, D.; Ma, J.; Zhao, C. Characteristics and regulatory pathway of the PrupeSEP1 SEPALLATA gene during ripening and softening in peach fruits. Plant Sci. 2017, 257, 63–73. [Google Scholar] [CrossRef] [Green Version]
- Pi, M.; Hu, S.; Cheng, L.; Zhong, R.; Cai, Z.; Liu, Z.; Yao, J.-L.; Kang, C. The MADS-box gene FveSEP3 plays essential roles in flower organogenesis and fruit development in woodland strawberry. Hortic. Res. 2021, 8, 247. [Google Scholar] [CrossRef]
- Lu, S.; Ye, J.; Zhu, K.; Zhang, Y.; Zhang, M.; Xu, Q.; Deng, X. A fruit ripening-associated transcription factor CsMADS5 positively regulates carotenoid biosynthesis in citrus. J. Exp. Bot. 2021, 72, 3028–3043. [Google Scholar] [CrossRef]
- Slugina, M.A. Transcription Factor Ripening Inhibitor and Its Homologs in Regulation of Fleshy Fruit Ripening of Various Plant Species. Russ. J. Plant Physiol. 2021, 68, 783–799. [Google Scholar] [CrossRef]
- Xu, Y.; Lin, Z.; Hua, X.; Zhang, Y.Q.; Oliveira, M.M. Expression analysis and genetic mapping of three SEPALLATA-like genes from peach (Prunus persica (L.) Batsch). Tree Genet. Genomes 2008, 4, 693–703. [Google Scholar] [CrossRef]
- Zhang, J.; Hu, Z.; Yao, Q.; Guo, X.; Nguyen, V.; Li, F.; Chen, G. A tomato MADS-box protein, SlCMB1, regulates ethylene biosynthesis and carotenoid accumulation during fruit ripening. Sci. Rep. 2018, 8, 3413. [Google Scholar] [CrossRef]
- Li, Q.; Wang, T.; Xu, C.; Li, M.; Tian, J.; Wang, Y.; Zhang, X.; Xu, X.; Han, Z.; Wu, T. MdMADS6 Recruits Histone Deacetylase MdHDA19 to Repress the Expression of the Carotenoid Synthesis-Related Gene MdCCD1 during Fruit Ripening. Plants 2022, 11, 668. [Google Scholar] [CrossRef]
- Qi, X.; Liu, C.; Song, L.; Li, M. PaMADS7, a MADS-box transcription factor, regulates sweet cherry fruit ripening and softening. Plant Sci. 2020, 301, 110634. [Google Scholar] [CrossRef] [PubMed]
- Tani, E.; Polidoros, A.; Flemetakis, E.; Stedel, C.; Kalloniati, C.; Demetriou, K.; Katinakis, P.; Tsaftaris, A.S. Characterization and expression analysis of AGAMOUS-like, SEEDSTICK-like, and SEPALLATA-like MADS-box genes in peach (Prunus persica) fruit. Plant Physiol. Biochem. 2009, 47, 690–700. [Google Scholar] [CrossRef]
- Li, M.; Galimba, K.; Xiao, Y.; Dardick, C.; Mount, S.M.; Callahan, A.; Liu, Z. Comparative transcriptomic analysis of apple and peach fruits: Insights into fruit type specification. Plant J. 2022, 109, 1614–1629. [Google Scholar] [CrossRef]
- Li, C.; Lei, C.; Wang, K.; Tan, M.; Xu, F.; Wang, J.; Zheng, Y. A novel MADS-box gene regulated a priming defence in postharvest peach through SA- and ABA-signaling collaboration. J. Exp. Bot. 2022, erac099, accepted manuscript. [Google Scholar] [CrossRef] [PubMed]
- Abel, S.; Oeller, P.W.; Theologis, A. Early auxin-induced genes encode short-lived nuclear proteins. Proc. Natl. Acad. Sci. USA 1994, 91, 326–330. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stortenbeker, N.; Bemer, M. The SAUR gene family: The plant’s toolbox for adaptation of growth and development. J. Exp. Bot. 2018, 70, 17–27. [Google Scholar] [CrossRef] [Green Version]
- Mcclure, B.A.; Guilfoyle, T. Characterization of a class of small auxin-inducible soybean polyadenylated RNAs. Plant Mol. Biol. 1987, 9, 611–623. [Google Scholar] [CrossRef] [PubMed]
- Hagen, G.; Guilfoyle, T. Auxin-responsive gene expression: Genes, promoters and regulatory factors. Plant Mol. Biol. 2002, 49, 373–385. [Google Scholar] [CrossRef]
- Jain, M.; Tyagi, A.K.; Khurana, J.P. Genome-wide analysis, evolutionary expansion, and expression of early auxin-responsive SAUR gene family in rice (Oryza sativa). Genomics 2006, 88, 360–371. [Google Scholar] [CrossRef] [Green Version]
- Wu, J.; Liu, S.; He, Y.; Guan, X.; Zhu, X.; Cheng, L.; Wang, J.; Lu, G. Genome-wide analysis of SAUR gene family in Solanaceae species. Gene 2012, 509, 38–50. [Google Scholar] [CrossRef]
- Wang, P.; Lu, S.; Xie, M.; Wu, M.; Ding, S.; Khaliq, A.; Ma, Z.; Mao, J.; Chen, B. Identification and expression analysis of the small auxin-up RNA (SAUR) gene family in apple by inducing of auxin. Gene 2020, 750, 144725. [Google Scholar] [CrossRef]
- Park, J.E.; Kim, Y.S.; Yoon, H.K.; Park, C.M. Functional characterization of a small auxin-up RNA gene in apical hook development in Arabidopsis. Plant Sci. 2007, 172, 150–157. [Google Scholar] [CrossRef]
- Hong, R.; Gray, W.M. SAUR proteins as effectors of hormonal and environmental signals in plant growth. Mol. Plant 2015, 8, 1153–1164. [Google Scholar] [CrossRef] [Green Version]
- Kathare, P.K.; Dharmasiri, S.; Arellano, I.; Dharmasiri, N. Interaction of SAUR53 and its close homologs with calmodulin may play a role in early development in Arabidopsis. Plant Mol. Biol. Rep. 2020, 38, 343–351. [Google Scholar] [CrossRef]
- Yang, T.; Poovaiah, B.W. Molecular and biochemical evidence for the involvement of calcium/calmodulin in auxin action. J. Biol. Chem. 2000, 275, 3137. [Google Scholar] [CrossRef] [Green Version]
- Kathare, P.K.; Dharmasiri, S.; Dharmasiri, N. SAUR53 regulates organ elongation and apical hook development in Arabidopsis. Plant Signal. Behav. 2018, 13, 1–7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Sun, N.; Zhang, F.; Yu, R. Differential regulation of Arabidopsis PP2C-D1 by SAUR17 and SAUR50 in apical hook development and cotyledon opening. Plant Cell 2020, 32, 3792–3811. [Google Scholar] [CrossRef]
- Guo, Y.; Jiang, Q.; Hu, Z.; Sun, X.; Fan, S.; Zhang, H. Function of the auxin-responsive gene TaSAUR75 under salt and drought stress. Crop. J. 2018, 6, 181–190. [Google Scholar] [CrossRef]
- Gastaldi, V.; Lucero, L.E.; Ariel, F.D.; Gonzalez, D.H. Class-I TCP transcription factors activate the SAUR63 gene subfamily in gibberellin-dependent stamen filament elongation. Plant Physiol. 2020, 182, 01501. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, J.; Wen, Z.W.; Mei, Y.Y.; Gonzalez, D.H. The mechanism underlying the role of SAUR72 in Arabidopsis leaf senescence regulation. Plant Physiol. J. 2018, 54, 379–385. [Google Scholar] [CrossRef]
- Wen, Z.; Mei, Y.; Zhou, J.; Cui, Y.; Wang, D.; Wang, N.N. SAUR49 Can Positively Regulate Leaf Senescence by Suppressing SSPP in Arabidopsis. Plant Cell Physiol. 2019, 61, 644–658. [Google Scholar] [CrossRef]
- Wong, J.H.; Spartz, A.K.; Park, M.Y.; Gonzalez, D.H. Mutation of a conserved motif of PP2C.D phosphatases confers SAUR immunity and constitutive activity. Plant Physiol. 2019, 181, 353–366. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Strader, L.; Ak, S.; Sang, H.L.; Wenger, J.P. Faculty Opinions recommendation of The SAUR19 subfamily of SMALL AUXIN UP RNA genes promote cell expansion. Plant J. 2012, 70, 978–990. [Google Scholar] [CrossRef]
- Kant, S.; Rothstein, S. Auxin-responsiveSAUR39gene modulates auxin level in rice. Plant Signal. Behav. 2009, 4, 1174–1175. [Google Scholar] [CrossRef] [Green Version]
- He, Y.; Liu, Y.; Li, M.; Lamin-Samu, A.T.; Yang, D.; Yu, X.; Izhar, M.; Jan, I.; Ali, M.; Lu, G. The Arabidopsis SMALL AUXIN UP RNA32 Protein Regulates ABA-Mediated Responses to Drought Stress. Front. Plant Sci. 2021, 12, 625493. [Google Scholar] [CrossRef]
- Goda, H.; Sawa, S.; Asami, T.; Fujioka, S.; Shimada, Y.; Yoshida, S. Comprehensive Comparison of Auxin-Regulated and Brassinosteroid-Regulated Genes in Arabidopsis. Plant Physiol. 2004, 134, 1555–1573. [Google Scholar] [CrossRef] [Green Version]
- Li, Z.G.; Chen, H.W.; Li, Q.T.; Shimada, Y. Three SAUR proteins SAUR76, SAUR77 and SAUR78 promote plant growth in Arabidopsis. Sci. Rep. 2015, 5, 12477. [Google Scholar] [CrossRef] [Green Version]
- Zhai, Y. Functional analysis of ERF19 and the ERF19 downstream gene SAUR32 in Arabidopsis. Ph.D. Thesis, Institute of Biotechnology, National Chung Hsing University, Taiwan, China, 2016. [Google Scholar]
- Shin, J.Y.; Mila, I.; Liu, M.; Rodrigues, M.A. The RIN-regulated small auxin-up RNA SAUR69 is involved in the unripe-to-ripe phase transition of tomato fruit via enhancement of the sensitivity to ethylene. New Phytol. 2019, 222, 820–836. [Google Scholar] [CrossRef]
- Carrasco-Valenzuela, T.; Muñoz-Espinoza, C.; Riveros, A.; Pedreschi, R.; Arús, P.; Campos-Vargas, R.; Meneses, C. Expression QTL (eQTLs) Analyses Reveal Candidate Genes Associated with Fruit Flesh Softening Rate in Peach [Prunus persica (L.) Batsch]. Front. Plant Sci. 2019, 10, 1581. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Müller, M.; Munné-Bosch, S. Rapid and sensitive hormonal profiling of complex plant samples by liquid chromatography coupled to electrospray ionization tandem mass spectrometry. Plant Methods 2011, 7, 37. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xing, L.; Zhang, D.; Li, Y.; Zhao, C.; Zhang, S.; Shen, Y.; An, N.; Han, M. Genome-wide identification of vegetative phase transition-associated microRNAs and target predictions using degradome sequencing in Malus hupehensis. BMC Genom. 2014, 15, 1125. [Google Scholar] [CrossRef] [Green Version]
- Livak, K.J.; Schmittgen, T. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Yang, Q.; Niu, Q.; Li, J.; Zheng, X. PpHB22, a member of HD-Zip proteins, activates PpDAM1 to regulate bud dormancy transition in ‘suli’ pear (Pyrus pyrifolia White Pear Group). Plant Physiol. Biochem. 2018, 127, 355–365. [Google Scholar] [CrossRef]
- Jia, H.-F.; Chai, Y.-M.; Li, C.-L.; Qin, L.; Shen, Y.-Y. Cloning and Characterization of the H Subunit of a Magnesium Chelatase Gene (PpCHLH) in Peach. J. Plant Growth Regul. 2011, 30, 445–455. [Google Scholar] [CrossRef] [Green Version]
- Yoo, S.D.; Cho, Y.H.; Sheen, J. Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat. Protoc. 2007, 2, 1565–1572. [Google Scholar] [CrossRef] [Green Version]
- Qian, M.; Xu, Z.; Zhang, Z.; Li, Q.; Yan, X.; Liu, H.; Han, M.; Li, F.; Zheng, J.; Zhang, D.; et al. The downregulation of PpPG21 and PpPG22 influences peach fruit texture and softening. Planta 2021, 254, 22. [Google Scholar] [CrossRef]
- Gray, J.; Picton, S.; Shabbeer, J.; Schuch, W.; Grierson, D. Molecular biology of fruit ripening and its manipulation with antisense genes. Plant Mol. Biol. 1992, 19, 69–87. [Google Scholar] [CrossRef]
- Liu, M.; Pirrello, J.; Chervin, C.; Roustan, J.-P.; Bouzayen, M. Ethylene control of fruit ripening: Revisiting the complex network of transcriptional regulation. Plant Physiol. 2015, 169, 2380–2390. [Google Scholar] [CrossRef] [Green Version]
- Adams-Phillips, L.; Barry, C.; Giovannoni, J. Signal transduction systems regulating fruit ripening. Trends Plant Sci. 2004, 9, 331–338. [Google Scholar] [CrossRef] [PubMed]
- Iqbal, N.; Khan, N.A.; Ferrante, A.; Trivellini, A.; Francini, A.; Khan, M.I.R. Ethylene Role in Plant Growth, Development and Senescence: Interaction with Other Phytohormones. Front. Plant Sci. 2017, 8, 475. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Du, M.; Spalding, E.P.; Gray, W.M. Rapid auxin-mediated cell expansion. Annu. Rev. Plant Biol. 2020, 71, 1–24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, J. Evolution by gene duplication: An update. Trends Ecol. Evol. 2003, 18, 292–298. [Google Scholar] [CrossRef] [Green Version]
- Hanada, K.; Zou, C.; Lehti-Shiu, M.D.; Shinozaki, K.; Shiu, S.-H. Importance of Lineage-Specific Expansion of Plant Tandem Duplicates in the Adaptive Response to Environmental Stimuli. Plant Physiol. 2008, 148, 993–1003. [Google Scholar] [CrossRef] [Green Version]
- Nemhauser, J.L.; Hong, F.; Chory, J. Different plant hormones regulate similar processes through largely nonoverlapping transcriptional responses. Cell 2006, 126, 467–475. [Google Scholar] [CrossRef] [Green Version]
- Boivin, S.; Fonouni-Farde, C.; Frugier, F. How Auxin and Cytokinin Phytohormones Modulate Root Microbe Interactions. Front. Plant Sci. 2016, 7, 1240. [Google Scholar] [CrossRef] [Green Version]
- Jeffares, D.C.; Penkett, C.J.; Jürg, B. Rapidly regulated genes are intron poor. Trends Genet. 2008, 24, 375–378. [Google Scholar] [CrossRef]
- Guo, Y.; Xu, C.-B.; Sun, X.-J.; Hu, Z.; Fan, S.-J.; Jiang, Q.-Y.; Zhang, H. TaSAUR78 enhances multiple abiotic stress tolerance by regulating the interacting gene TaVDAC1. J. Integr. Agric. 2019, 18, 2682–2690. [Google Scholar] [CrossRef]
- Bank, A.D. Cell wall disassembly in ripening fruit. Funct. Plant Biol. 2006, 33, 103–119. [Google Scholar] [CrossRef]
- Orfila, C.; Huisman, M.M.; Willats, W.G.; van Alebeek, G.-J.W.; Schols, H.A.; Seymour, G.B.; Knox, P.J. Altered cell wall disassembly during ripening of Cnr tomato fruit: Implications for cell adhesion and fruit softening. Planta 2002, 215, 440–447. [Google Scholar] [CrossRef] [PubMed]
- Payasi, A.; Mishra, N.N.; Chaves, A.L.S.; Singh, R. Biochemistry of fruit softening: An overview. Physiol. Mol. Biol. Plants 2009, 15, 103–113. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qian, M.; Zhang, Y.; Yan, X.; Han, M.; Li, J.; Li, F.; Li, F.; Zhang, D.; Zhao, C. Identification and Expression Analysis of Polygalacturonase Family Members during Peach Fruit Softening. Int. J. Mol. Sci. 2016, 17, 1933. [Google Scholar] [CrossRef] [Green Version]
- Kant, S.; Bi, Y.M.; Tong, Z. SAUR39, a small auxin-up RNA gene, acts as a negative regulator of auxin synthesis and transport in rice. Plant Signal. Behav. 2009, 151, 691–701. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Palme, K.; Gälweiler, L. PIN-pointing the molecular basis of auxin transport. Curr. Opin. Plant Biol. 1999, 2, 375–381. [Google Scholar] [CrossRef]
- Zeng, W.F.; Pan, L.; Niu, L.; Lu, Z.H. Bioinformatics analysis and expression of the nectarine indole-3-aceticacid-amido synthase (GH3) gene family during fruit development. Acta Hortic. Sin. 2015, 42, 833–842. [Google Scholar] [CrossRef]
- Wang, X.; Meng, J.; Deng, L.; Wang, Y.; Liu, H.; Yao, J.-L.; Nieuwenhuizen, N.J.; Wang, Z.; Zeng, W. Diverse Functions of IAA-Leucine Resistant PpILR1 Provide a Genic Basis for Auxin-Ethylene Crosstalk During Peach Fruit Ripening. Front. Plant Sci. 2021, 12, 655758. [Google Scholar] [CrossRef]
- Guilfoyle, T.J.; Hagen, G. Auxin response factors. Curr. Opin. Plant Biol. 2007, 10, 453–460. [Google Scholar] [CrossRef]
- Shinshi, O.T. Ethylene-inducible DNA binding proteins that interact with an ethylene-responsive element. Plant Cell 1995, 7, 173–182. [Google Scholar] [CrossRef] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).