Abstract
Salinity is a type of abiotic stress that negatively affects plant growth and development. Textile hemp (Cannabis sativa L.) is an important multi-purpose crop that shows sensitivity to salt stress in a genotype- and developmental stage-dependent manner. The root and shoot biomasses decrease in the presence of NaCl during vegetative growth and several stress-responsive genes are activated. Finding environmentally friendly ways to increase plant health and resilience to exogenous stresses is important for a sustainable agriculture. In this context, the use of beneficial bacteria, collectively referred to as plant growth-promoting bacteria (PGPB), is becoming an attractive and emergent agricultural strategy. In this study, data are provided on the effects of a Pseudomonas isolate (Pseudomonas sp. SVB-B33) phylogenetically closely related to P. psychrotolerans applied via roots to salt-stressed hemp. The application of both living and dead bacteria impacts the fresh weight of the root biomass, as well as the expression of several stress-related genes in roots and leaves. These results pave the way to future investigations on the use of Pseudomonas sp. SVB-B33 in combination with silica to mitigate stress symptoms and increase the resilience to other forms of exogenous stresses in textile hemp.
1. Introduction
The productivity of crops depends on several parameters, both endogenous, such as genotype [1,2], photosynthetic efficiency [3,4], levels of phytohormones [5] and exogenous, namely environmental cues (salinity, heavy metal pollution, temperature, drought, etc.) [6]. Salinity is an abiotic stress that is particularly alarming in coastal areas where the sea level rise caused by global warming leads to seawater intrusion with consequent salinization of the groundwater used for irrigation [7].
The multi-purpose fibre crop hemp (Cannabis sativa L.) is sensitive to salt stress, especially during germination [8]. In the presence of NaCl 200 mM, its leaves show growth arrest due to the increase in lignification and its vascular system is characterized by smaller xylem vessels to limit the incidence of cavitation [9]. The negative effects of salinity in hemp were shown to be ameliorated by the addition of protein hydrolysate [10] and silicon [11]: the former improves seed yield, the average weight and height of stems, as well as photosynthetic activity thanks to its auxin-like effect, while the metalloid, which precipitates in the cell wall and encrusts structures such as trichomes [12], induces the occurrence of xylem vessels with wider lumens in older fan leaves. More recently, the protective effect of a nano-biostimulant composed of hybrid silicon nanoparticles functionalized with quercetin was validated at the phenotypic and gene level in textile hemp leaves exposed to salinity [13].
The use of beneficial bacteria, also known as plant growth-promoting bacteria (PGPB) as a strategy to improve growth and protect against exogenous stresses is gaining increasing attention in sustainable agriculture [14,15]. Their application as biostimulants was indeed shown to improve the growth of different plant species and to protect against the damages caused by (a)biotic stresses [16,17,18,19].
PGPB activate induced systemic resistance, ISR, in plants by favouring a physiological mechanism referred to as “priming” that allows them to respond faster and more effectively to an (a)biotic stressor [20]. ISR takes place via the recognition of microbial compounds (microbial-associated molecular patterns-MAMPs), namely lipopolysaccharides, flagellin, exopolysaccharides and chitin that bind to receptors. These will in turn phosphorylate downstream targets and activate a signalling cascade involving calcium, mitogen-activated protein kinases and oxidative burst [20].
In light of the sustainable nature of PGPB and the beneficial effects they have on the growth of crops, the present study was conceived with the aim of providing additional data on their impact on industrial hemp subjected to salt stress. The beneficial impact of a Pseudomonas strain (SVB-B33) isolated as airborne contamination of agar plates and phylogenetically closely related to the salt-tolerant, silica- and phosphate-solubilizing bacterium Pseudomonas psychrotolerans [21,22] was here assessed on a fibre variety of hemp (Santhica 27). The application of both dead (heat-inactivated) and living bacteria, together with the impact on root/leaf biomass and the expression of a set of target genes were investigated. Heat-inactivated bacteria were investigated in the light of published literature data showing priming effects of defence responses in plants treated with dead bacteria. Indeed, in the 1970s it had already been demonstrated that tobacco leaves infiltrated with heat-inactivated Ralstonia solanacearum showed hypersensitive response inhibition when inoculated with living cells [23]. More recently, heat-killed Agrobacterium tumefaciens cells were shown to suppress to some extent Pseudomonas syringae-triggered symptoms, as well as salicylic acid production [24]. The genes were chosen among cell wall- and stress-related ones, given the implication of the cell wall in the perception and response to exogenous abiotic stimuli [25] and their previously reported transcriptional changes in salt-stressed hemp tissues [9]. More specifically, genes involved in cell wall biosynthesis and lignification (cellulose synthases, fasciclin-like arabinogalactan proteins, laccases) were selected because salinity is an abiotic factor inducing tissue rigidification and growth arrest [9] and since the cell wall is remodelled upon exogenous stresses [26]. Known stress-responsive genes were chosen to provide an indication of stress mitigation by the application of PGPB at the molecular level.
2. Materials and Methods
2.1. Bacterial Isolation, Phylogenetic Analysis and Growth
Pseudomonas sp. (UoA: SVB-B33) was isolated as an airborne contamination of agar plates at the School of Biological Sciences, University of Auckland. It was identified based on its 16S rDNA sequence [27]. The bacterial strain was reactivated from glycerol stocks kept at −80 °C and maintained on nutrient agar (NA) medium (peptone, 5 g·L−1; yeast extract, 3 g·L−1; NaCl, 5 g·L−1; agar, 15 g·L−1; pH 6.8) at 25 °C.
PCRs to amplify the 16S sequence were performed using 2 µL of supernatant obtained after having resuspended a single colony of Pseudomonas sp. SVB-B33 in 25 µL sterile distilled H2O, having heated the suspension to 95 °C for 5 min and having spun it down with a microfuge. For the amplification step, the Q5 Hot Start High-Fidelity 2X Master Mix (New England Biolabs, Leiden, The Netherlands) was used, according to the manufacturer’s instructions. The following primer pairs were used for the 16S amplification: uni-Fwd TGCCAGCAGCCGCGGTA/uni-Rev GACGGGCGGTGTGTACAA [28]. The amplifications were carried out as follows: initial denaturation at 98 °C for 1 min, followed by 35 cycles of denaturation at 98 °C for 10 s, annealing at 58 °C for 1 min, extension at 72 °C for 1 min and a half and a final extension at 72 °C for 5 min with a hold step at 4 °C. The specificity of the PCR products was verified with gel electrophoresis. PCR products were purified using the PCR purification kit from Qiagen (Qiagen, Leusden, The Netherlands) and sent out for sequencing. The 16S sequence was deposited in NCBI with the accession number GenBank: ON148530.
The phylogenetic analysis was constructed after having performed a pair-wise multiple alignment CLUSTAL-Ω (http://www.ebi.ac.uk/Tools/msa/clustalo/, accessed on 15 February 2022) [29]; the alignment was used to build a maximum likelihood phylogenetic tree using the online program W-IQ-TREE [30] (bootstraps in ultrafast mode: 1000; in auto mode, without selecting Free Rate Heterogeneity), available at http://iqtree.cibiv.univie.ac.at (accessed on 15 February 2022). The tree was subsequently visualized using iTOL (available at https://itol.embl.de/, accessed on 15 February 2022).
For the plant growth treatment, Pseudomonas sp. SVB-B33 was cultured in soil extract broth (DSMZ Medium 12 [31]) supplemented with dextrose (1% w/v) at 25 °C for 72 h under continuous agitation at 200 rpm. The resulting bacterial culture was then diluted with sterile soil extract without dextrose to reach approximately 106 colony forming units per mL. In order to determine if any observed effect of bacterial treatment was due to the metabolic activity of bacterial cells rather than their cell wall- or membrane-derived products, products derived from (macro)molecules’ degradation, autoclaved Pseudomonas sp. SVB-B33 culture in soil extract dextrose broth at the same dilution of living cultures was used.
2.2. Silicate Solubilizing Assay
The ability of Pseudomonas sp. SVB-B33 to solubilize silicate minerals was tested in NA plates supplemented with talc (hydrate magnesium silicate) at 0.25% (w/v). The bacterial culture was streaked on the agar media and incubated at 25 °C for 1 week. The clearing zone was then measured [32].
2.3. Plant Growth and Treatments
Plants of C. sativa Santhica 27 were grown as previously detailed [33]. Briefly, seeds were sown in pots containing a mixture of compost/sand (1:1 w/w) and grown under controlled conditions in chambers (Fitotron, Weiss Technik, Reiskirchen, Germany) with 60% humidity and a 16 h light 25 °C/8 h dark 20 °C cycle. When the plantlets were 2 weeks old, the application of 100 mL of freshly diluted dead/living bacteria was started by root amendment (watering in proximity of the soil close to the roots). After 1 week, control plants were watered with 50 mL of tap water, while the others were watered with 50 mL of NaCl 250 mM, as previously described [9,11]. NaCl 250 mM was applied twice a week with a bacterial application in between. The treatment lasted 4 weeks. Three days after the last application of salt, plants were sampled. Three biological replicates (n = 3) composed of a pool of 2 plants each were sampled per treatment, with the exception of non-inoculated plants subjected to stress, for which 1 of the 3 biological replicates consisted only of 1 plant (1 plant died before the day of sampling). The older leaves were harvested (2 leaves per plant) and weighed to determine the fresh weight (FW); they were then put in an oven at 37 °C for 72 h until constant weight to measure the dry weight (DW). Younger leaves (2 fully expanded leaves below the apex) were instead harvested and plunged in liquid nitrogen for gene expression analysis. The roots were sampled, weighed and a portion was snap frozen in liquid nitrogen for subsequent qPCR analysis. The rest of the roots was dried to determine the DW. A schematic representation of the experimental set-up and sampling strategy is shown in Figure 1. The soil was collected from the pots, homogenized and dried at 105 °C for 3 days. Five grams were put in a tube and 25 mL of Milli-Q water (Merck, Kenilworth, NJ, USA) was added. The samples were shaken on a rolling table for one hour at room temperature. The soil electrical conductivity (ECe) and pH were measured with an inoLab Multi 9430 IDS (WTW, Weilheim, Germany), as previously described [9].
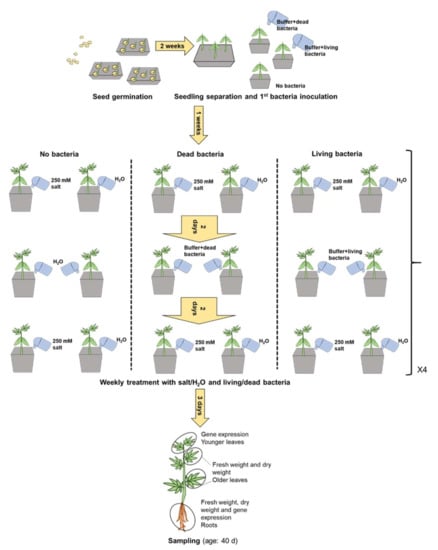
Figure 1.
Schematic representation of the experimental set-up and sampling strategy with details of the timing of the different steps.
2.4. RNA Extraction, cDNA Synthesis, qPCR and Statistical Analyses
Leaf and root samples were ground to a fine powder with a mortar and pestle using liquid nitrogen and they were subsequently processed for RNA extraction using the RNeasy Mini Kit (Qiagen, Leusden, The Netherlands). An on-column DNase I treatment (Qiagen) was carried out to avoid DNA carry-over. RNA was eluted using RNase-free water and the concentrations were measured with a NanoPhotometer NP80 (Implen, Munich, Germany). The RNA Integrity Number (RIN) was evaluated by capillary gel electrophoresis with a 2100 Bioanalyzer (Agilent, Santa Clara, CA, USA) according to the manufacturer’s instructions, using the RNA 6000 nano chip (Agilent). RNA (1 µg) was retro-transcribed and the cDNA was brought to a concentration of 2 ng/µL. Targeted gene expression analyses were performed with the Takyon Low ROX SYBR Green (Eurogentec, Liege, Belgium) in 384 wells reaction plates, as previously reported [11]. Plates were prepared with the aid of an automated dispensing device (epMotion 5073x, Eppendorf, Hambourg, Germany), with 3 technical replicates for each sample. A melt–curve analysis was performed at the end of the amplification to check for a single peak indicating specific amplification. The primers were chosen from published studies and their characteristics were previously reported [9,33,34]. Gene expression was calculated in qBasePLUS (Biogazelle, Ghent, Belgium) [35] with the geNORM tool. Two reference genes were used for data normalization (CDPK and eTIF4E) in roots and three (CDPK, F-box and eTIF4E) in leaves among the four tested (CDPK, eTIF4E, F-box and TIP41 [36]). The log2-transformed normalized relative quantities (NRQs) were used to perform the statistical analyses with IBM SPSS Statistics v20 (IBM SPSS, Chicago, IL, USA). Normality and homogeneity of all the data were checked using a Shapiro–Wilk and Levene’s test, respectively. A one-way ANOVA with a Tukey’s post hoc test was used when parametric tests could be applied, while a Kruskal–Wallis with Dunn’s post hoc test was used for non-normal and/or non-homogeneous data. The principal component analysis (PCA) of the gene expression values was determined with ClustVis [37] (available at https://biit.cs.ut.ee/clustvis/, accessed on 15 March 2022).
3. Results
3.1. Phylogenetic Analysis and Silicate Solubilizing Activity
The isolate Pseudomonas sp. SVB-B33 was analysed using the 16S sequence (100% sequence identity with P. psychrotolerans was obtained based on BLASTn) and the phylogenetic analysis confirmed this result (Figure 2a). P. psychrotolerans has been reported previously as able to solubilize silicate-containing minerals [21] and this ability was checked in the isolate SVB-B33 using nutrient medium supplemented with talc (Figure 2b). Pseudomonas sp. SVB-B33 grew well in the presence of talc and was able to solubilize this silicate effectively, presenting a solubilization clearing zone of 16.2 ± 1.8 mm (n = 3). The halo indicates the ability of the isolate to solubilize crystalline silica and is a proof of its silica bioweathering activity. The Pseudomonas strain here studied is thus a bona fide P. psychrotolerans strain but will be hereafter referred to as Pseudomonas sp. SVB-B33.
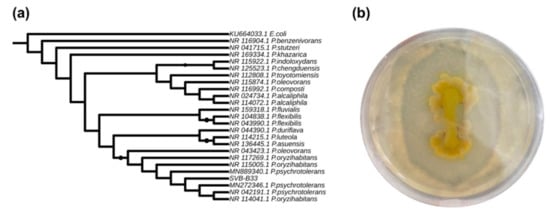
Figure 2.
Phylogenetic analysis of the isolate SVB-B33 based on 16S and silicase activity. (a) Maximum likelihood phylogenetic analysis of SVB-B33 based on 16S. The tree was rooted with Escherichia coli 16S. The accession numbers used to construct the tree are indicated. Bootstrap = 1000. The symbols refer to % of branch support value (the bigger the symbol, the higher the value). Only branch support values >80% are here shown. (b) Pseudomonas sp. SVB-B33 culture grown on nutrient agar supplemented with hydrate magnesium silicate (talc).
3.2. The Application of Living Bacteria to Hemp Increases the Root Biomass under Salinity
In this study, both the application of dead (heat-inactivated) and living bacteria was investigated. In the leaves, the differences in FW recorded among control or salt-stressed samples when comparing the addition of living/dead bacteria to no bacteria are not statistically significant (Figure 3a). However, the DW is significantly higher than the condition without bacteria in the presence of stress, when living bacteria are applied (Figure 3c).
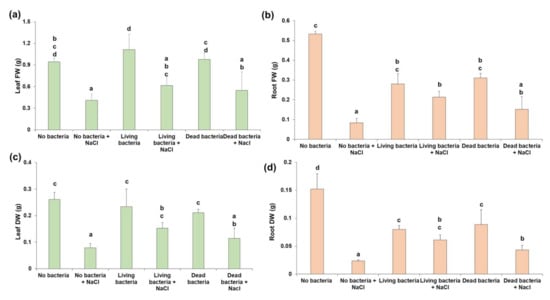
Figure 3.
Fresh and dry weight (FW, DW) of leaves (a,c) and roots (b,d) in the absence and presence of stress in non-inoculated and inoculated roots. The error bars indicate the standard deviation (n = 3). Different letters on the bars indicate statistically significant values (p < 0.05). Statistical parameters: (a) F(5,12) = 9.725, p = 0.001, (b) F(5,12) = 22.036, p = 0.000, (c) F(5,12) = 13.234, p = 0.000, (d) F(5,12) = 33.544, p = 0.000.
In the roots, instead, statistically significant differences are present (Figure 3b,d). While salt provokes a dramatic decrease in the FW and DW under non-inoculated conditions, the application of bacteria mitigates this difference with values that are non-significant in roots treated with living bacteria. When looking at the FW and DW values in roots under salt stress, a statistically significant increase is present in the samples inoculated with living bacteria as compared to non-inoculated ones. It should also be noted that, in the absence of stress, lower FW and DW values are observed in roots inoculated with bacteria, as compared to non-inoculated ones.
The soil pH and electrical conductivity (ECe) were also measured as previously reported [9] (Table 1). No major changes are present in pH when comparing the different treatments under either control or stress conditions. A difference in conductivity is observed, as expected, when comparing non-stressed and stressed conditions, but no statistically significant variations in ECe are present when comparing the soils amended or not with bacteria under a same condition, i.e., control or salt stress.

Table 1.
Electrical conductivity (ECe, measured in mS) and pH of soils in the absence and presence of salt and in non-inoculated and inoculated plants. Different letters indicate statistically significant values (p < 0.05). Statistical parameters: conductivity X2(5) = 14.520, p = 0.013, pH F(5,12) = 1.427, p = 0.283.
Salt stress induces a significant decrease in the length of the roots; it should also be noted that the inoculation with bacteria induces the presence of shorter roots (Supplementary Figures S1 and S2).
The stem length of plants treated with living PGPB was significantly lower under control conditions and higher with dead bacteria; under stress, the height decreased significantly in plants that were not inoculated with PGPB, as well as in those treated with dead bacteria, while the values did not change when living bacteria were supplemented to hemp plants (Supplementary Figure S3).
3.3. The Inoculation with Pseudomonas sp. SVB-B33 Affects Gene Expression in Hemp Roots and Leaves
A targeted gene expression analysis on transcripts involved in stress response and cell wall biosynthesis was performed on roots and leaves treated or not with the bacteria under control and stress conditions. A principal component analysis (PCA) was performed to visualize how well-separated the treatments with/without PGPB were. The PCA of the gene expressions in hemp roots under control conditions shows a clear separation of the replicates into three distinct groups (non-inoculated roots—CNTR, roots inoculated with living bacteria—LIVING and dead bacteria—DEAD) (Figure 4a). Under stress conditions, the three replicates corresponding to the roots inoculated with the living bacteria are more scattered, but the three groups are still well recognizable (Figure 4b). In the leaves under control conditions, the samples inoculated with dead bacteria and non-inoculated ones cluster separately (Figure 4c), while the three replicates corresponding to the leaves treated with living bacteria are scattered. Under stress, a clear separation between the samples inoculated with living bacteria and non-inoculated ones are well separated (Figure 4d).
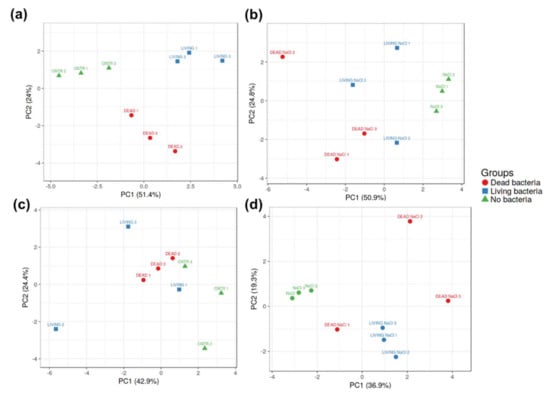
Figure 4.
Principal component analysis (PCA) of the gene expression data in roots (top panels) and leaves (bottom panels) in the absence (a,c) and presence of salt stress (b,d). CNTR: non-stressed/stressed non-inoculated; LIVING: non-stressed/stressed inoculated with living bacteria; DEAD: non-stressed/stressed inoculated with dead bacteria.
Known stress-responsive genes [9] were analysed in this study and, among them, the transcripts coding for the calcium-dependent lipid-binding family proteins CALB4-1, CALB4-2, CALB4-3 and for the heat-shock protein HSP70-2 and HSP81-4 show a statistically significant difference when comparing non-inoculated roots vs. roots treated with living bacteria in the absence of stress (Figure 5a). The expressions are indeed higher in the inoculated roots, with the exception of HSP70-2 for which a higher expression is observed in non-inoculated samples. It should be noted that, for HSP70-2, a statistically significant higher expression is present in roots inoculated with living bacteria than roots treated with dead cells. Under salt stress, no significant differences are observed when comparing non-inoculated roots and roots treated with living bacteria, although a trend denoting lower values can be observed for ERF1, HSP70-2 and HSP81-4, while higher expressions are present for CALB2, CALB4-1 and CALB4-2. Statistically significant differences are instead present for CALB4-3, HSP70-2 and HSP81-4 when comparing roots inoculated with dead bacteria and non-inoculated roots under salt stress.
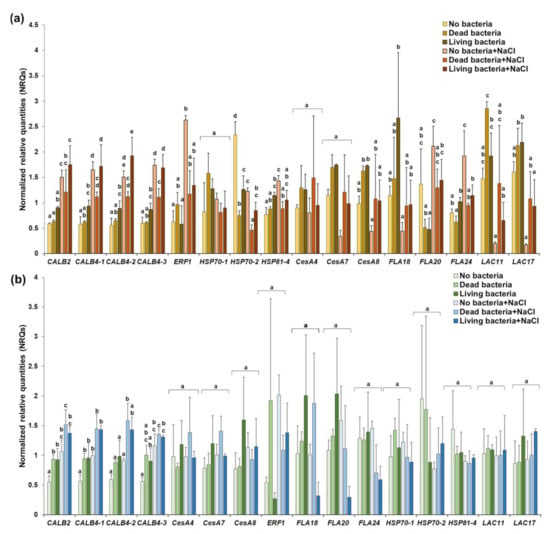
Figure 5.
Bar chart of the gene expression values (Normalized Relative quantities—NRQs) in roots (a) and leaves (b). Different letters indicate statistically significant values (p < 0.05). The statistical parameters can be found in Supplementary Table S1.
In the leaves, higher expression values are observed in the absence of stress for CALB2 and CALB4-1 after inoculation with dead and living cells (Figure 5b). Under stress, CALB4-2 shows a statistically significant higher expression in leaves treated with dead bacteria (Figure 5b).
In addition to stress-related genes, transcripts involved in cell wall biosynthesis were also investigated. The genes chosen code for cellulose synthases (CESAs), fasciclin-like arabinogalactan proteins (FLAs) and laccases (LACs). The reason for focusing on these transcripts is to assess the impact of bacterial inoculation on cell wall-related processes and because these genes are known to respond to salinity in hemp tissues [11].
Although trends in gene expression can be observed for the roots (e.g., lower values for the secondary cell wall CesA7 and CesA8, FLA18 and the laccases), the values are not statistically significant. FLA24 shows significant differences, with a higher expression in the absence of stress in roots treated with living bacteria as compared to dead PGPB and lower expression under salinity in roots treated with dead or living bacteria as compared to samples receiving no PGPB (Figure 5a).
In the leaves, no statistically significant changes are observed in cell wall-related genes (Figure 5b).
4. Discussion
The management of stresses in economically relevant plants by means of eco-friendly approaches is important as a long-term and cost-effective agricultural strategy facing climate change [38]. The use of PGPB has shown beneficial effects in different crops under several (a)biotic stress conditions and has thus emerged as a valuable practice for a sustainable agriculture.
In this study, the impact of the inoculation with both living and dead cells of a Pseudomonas isolate (SVB-B33) was assessed by adopting a gene expression approach focused on key stress- and cell wall-related transcripts known to respond to salinity in textile hemp. The isolate here studied is a bona fide P. psychrotolerans strain based on the phylogenetic analysis (Figure 2a) and on the silica solubilizing activity, as denoted by the agar-based medium assay (Figure 2b). It is known that the production of organic acids is a common method for minerals’ solubilization and it will be relevant to identify the organic acids produced by this isolate during silica solubilization to identify which ones are the most abundant. The protective effects of P. psychrotolerans were previously demonstrated in salt-stressed maize and soybean grown on paddy soils: it reduced the level of the phytohormones abscisic and jasmonic acid in stressed maize plants [22] and it also reduced the endogenous levels of salicylic acid in soybean plants while increasing their shoot height and weight, as well as the chlorophyll content [21]. Hence, the isolate here investigated has potential for use in crop protection.
Textile hemp is a multi-purpose crop providing fibres, as well as added-value phytochemicals [39] and is thus a relevant renewable resource for bioeconomy. It was chosen as model organism to study the effects of the Pseudomonas isolate SVB-B33 inoculation on the response to salinity, a form of abiotic stress previously shown to impact the growth of C. sativa [9,11,13].
Only a handful of reports have studied the use of PGPB in hemp cultivation: a consortium of Gluconacetobacter diazotrophicus, Herbaspirillum seropedicae and Burkholderia ambifaria effectively protected hemp against Fusarium wilt both during pre- and post-emergence [40] and the same consortium, together with Azospirillum brasilense, also contributed to boosting the antioxidant activity and synthesis of secondary metabolites (delta-9-tetrahydrocannabinol, cannabinol and cannabidiol) [41]. Common PGPB associated with both wild and cultivated plants, such as Pseudomonas and Bacillus [42], as well as Cannabis endophytes [43] harbour great potential as beneficial bioinoculants for the protection and improvement of hemp growth.
The present study is novel for three reasons: (1) it provides evidence for an impact at the gene level in hemp of both living and dead Pseudomonas sp. SVB-B33, (2) it opens up future investigations on the molecular impact of the inoculation with dead cells and (3) it provides evidence on the synergistic effects based on the combined application of living bacteria and silica. Concerning future analyses, it will be worthwhile studying the effects over a longer time, as well as of increasing doses (more frequent applications). The experiments here reported were performed on non-sterile soil, which means that a microbiome was already present. Therefore, it will be relevant to also study the effects of the PGPB on sterile soil.
The results obtained show clear and significant effects at the root level under stress (Figure 3b,d): a statistically significant higher biomass is indeed observed in stressed plants treated with living bacteria as compared to non-inoculated ones, a result suggesting that the bacteria positively alter the root traits of hemp in saline soils. Further studies are needed to decipher the pathways affected at the root level; however, from the data collected, it is evident that, under salinity, living bacteria help in the preservation of the root architecture. P. psychrotolerans CS51 induced the production of endogenous indole-3-acetic acid and gibberellins in cucumber under heavy metal stress and the genome sequencing revealed the presence of genes responsible for auxin biosynthesis, nitrogen metabolism, phosphate and sulphate transport system [44]. Hormonomics in hemp tissues will reveal changes in the levels of endogenous phytohormones upon inoculation with the PGPB.
The inoculation with both dead and living bacteria triggers a reduced length of the roots under control conditions (Supplementary Figures S1 and S2) and an increased height under salinity with dead bacteria (Supplementary Figure S3); these results were unexpected for the dead bacteria, but not totally surprising considering that heat-killed Bacillus aryabhattai was shown to exert an action in thale cress and tobacco via the activation of salicylic and jasmonic acid pathways [45]. The inoculation of lettuce plants with cytokinin-producing bacteria resulted in shortened roots [46]; hence, it will be of interest in the future to monitor the levels of cytokinins in hemp plants inoculated with dead Pseudomonas sp. SVB-B33. Heat-inactivation results in the release of components such as lipoteichoic acids, exopolysaccharides, peptidoglycans and the use of the so-called “para-probiotics” or “ghost probiotics” (derived from non-viable cells) in modulating the immune response and positively affecting human health has been evaluated in comparison to viable cells from the point of view of safety risks and shelf-life [47]. Considering these aspects, it will be worthwhile assessing the impact of “ghost microbial biostimulants” in hemp stress protection by characterizing the components present in the heat-inactivated Pseudomonas sp. SVB-B33.
The gene expression analysis confirms an effect of dead cells on hemp roots and leaves: dead bacteria decrease the expression of the stress-responsive gene HSP70-2 under control and salt-stress conditions (Figure 5a), while all the CALB genes show higher expression in the leaves of plants inoculated with heat-inactivated cells (Figure 5b). The lower expression of HSPs in roots inoculated with dead and living PGPB could indicate a protective effect with a resulting lower induction of stress-related genes.
The expression of CALB2, CALB4-1 and CALB4-2 in salt-stressed hemp tends to be higher in roots inoculated with living bacteria (Figure 5a) and this result is in agreement with a previous study in tomato, which reported two CALBs among the up-regulated genes in drought-stressed plants inoculated with Bacillus megaterium strain TG1-E1 [48]. CALBs are involved in Ca2+ signalling and in the transduction pathway controlling stress response and the tendency towards an increased expression under salinity in roots treated with living Pseudomonas sp. SVB-B33 suggests the activation of Ca2+-dependent processes. Living PGPB seem to be necessary to induce the expression of the CALB genes in roots under salinity, since the application of dead bacteria induces lower expression values in stressed roots (Figure 5a).
The cell wall of plants plays an important role in the response to exogenous stresses [25] and to evaluate the impact of the PGPB, genes involved in secondary cell wall biosynthesis (CesA4, CesA7 and CesA8), lignification (LAC11 and LAC17) and cell wall integrity (FLA18, FLA20 and FLA24) were investigated in roots and leaves (Figure 5). Salinity tends to decrease the expression of CesA7 and CesA8, as well as LAC11 and LAC17 in non-inoculated roots; however, trends towards higher expression values are observed in roots inoculated with either dead or living bacteria. These data suggest a preservation of the mechanisms underlying secondary cell wall biosynthesis and lignification under stress in inoculated roots. It should be noted that the adaptation to salinity in thale cress was found to be associated with cell wall reinforcement mediated by lignin deposition [49]. Pseudomonas sp. SVB-B33 may thus contribute to the preservation of the cell wall mechanical features in the roots of hemp subjected to salt stress. It will be of interest to study the effects of biostimulant formulations consisting of living Pseudomonas sp. SVB-B33 and insoluble silica on textile hemp exposed to stresses, as their combination may result in superior protective effects due to the presence of PGPB and silicic acid made available to plants by the silica-solubilizing activity of the microbial cells. A strengthening action of Pseudomonas sp. SVB-B33 on secondary cell wall of hemp roots is to be expected since previous studies reported the ability of C. sativa to take up silicon (Si, as silicic acid), deposit it as silica in the cell wall [12,50] and respond better to abiotic stresses in the presence of the metalloid [51,52].
5. Conclusions
In this study, the beneficial effects of Pseudomonas sp. SVB-B33 are reported, for the first time, in textile hemp exposed to salt stress. The data obtained indicate a protective effect of the living and heat-inactivated bacteria applied to plants via root amendment. Therefore, Pseudomonas sp. SVB-B33 can be used as a PGPB. In the presence of living bacteria and, to a lesser extent, heat-inactivated bacteria, both the fresh and dry root biomasses are higher under salinity. However, the protective effect of living and dead PGPB could be realized in different ways. Indeed, the gene expression analysis shows a statistically significant increase in the expression of some CALB genes in the absence of stress in the roots treated with living PGPB. The application of dead bacteria, instead, decreases the expression of CALB4-3, HSP70-2 and HSP81-4 in the inoculated roots exposed to salinity as compared to non-inoculated ones, while in the leaves it increases the expression of CALB2 and CALB4-1 in the absence of stress and of CALB4-2 under stress condition. These findings warrant further research into the MAMPs mediating these effects in heat-inactivated PGPB, since only few data points are available on the use of inactivated bacteria in crop protection. The use of inactivated PGPB in agriculture would simplify transport and storage and lengthen shelf-life as compared to living bacteria.
From the obtained results, however, differences in the use of living or heat-killed PGPB could be established. The application of living bacteria seems to be necessary to induce changes in the expression of genes involved in the Ca2+-dependent transduction pathway controlling stress response. Indeed, CALB4-1, CALB4-2 and CALB4-3 increase significantly in expression in roots supplemented with living PGPB in the absence of stress, while their abundances tend to decrease under salinity in roots supplemented with heat-inactivated PGPB. The decreased expression of CALB4-3 in roots treated with heat-killed PGPB is significant.
In salt-stressed roots, genes involved in secondary cell wall deposition and lignification tend to be expressed at higher levels after inoculation with living or dead cells. These findings suggest an impact of the PGPB in activating the response to the abiotic stimulus by acting on stress-responsive and cell wall-related genes. It will be interesting, in the future, to study the molecular basis of Pseudomonas sp. SVB-B33 impact on stress tolerance in hemp by adopting a transcriptomics approach coupled to proteomics and metabolomics. These results will help us understand the mechanisms triggering the effects here reported with dead and living cells. Likewise, it will be of relevance to investigate the impact of bacterial inoculation in hemp exposed to other forms of stressors and in the presence of silica, to understand whether the PGPB mediate an improved response by solubilizing silica and making silicic acid available to plants.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/horticulturae8040336/s1, Figure S1: Images of roots from plants grown under control/salt stress conditions in the absence/presence of living or dead bacteria. The scale bar is 5 cm; Figure S2: Length of roots from plants grown under control/salt stress conditions in the absence/presence of living or dead bacteria. Different letters indicate statistically significant values (p < 0.05). Statistical parameters: X2(5) = 73.534, p = 0.000; Figure S3: Length of stems from plants grown under control/salt stress conditions in the absence/presence of living or dead bacteria. Different letters indicate statistically significant values (p < 0.05). Statistical parameters: X2(5) = 43.845, p = 0.000, Table S1: Table showing the statistical parameters of Figure 5. Degree of freedom (df), F or X2 value and significance (p). A parametric test (ANOVA one-way followed by the Tukey’s post hoc test) was applied for normal and homogeneous data, otherwise a non-parametric test (Kruskal–Wallis together with the Dunn’s post hoc test) was performed.
Author Contributions
Conceptualization, R.B., J.-F.H., S.V.-B. and G.G.; methodology, R.B., S.V.-B. and G.G.; validation, R.B., S.V.-B. and G.G.; formal analysis, R.B., S.V.-B. and G.G.; investigation, R.B., S.V.-B. and G.G.; resources, J.-F.H., S.V.-B. and G.G.; data curation, R.B., S.V.-B. and G.G.; writing—original draft preparation, R.B., S.V.-B. and G.G.; writing—review and editing, R.B., J.-F.H., G.G. and S.V.-B. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by the Luxembourg National Research Fund (INTER/FNRS/20/15045745).
Data Availability Statement
All data are made available in the publication.
Acknowledgments
The authors would like to thank Laurent Solinhac for the help with the plant cultures and Margaux Thiry for the assistance in the extraction of RNA and the setup of qPCR reactions.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Beres, B.L.; Hatfield, J.L.; Kirkegaard, J.A.; Eigenbrode, S.D.; Pan, W.L.; Lollato, R.P.; Hunt, J.R.; Strydhorst, S.; Porker, K.; Lyon, D.; et al. Toward a Better Understanding of Genotype × Environment × Management Interactions—A Global Wheat Initiative Agronomic Research Strategy. Front. Plant Sci. 2020, 11, 828. [Google Scholar] [CrossRef] [PubMed]
- Studnicki, M.; Wijata, M.; Sobczyński, G.; Samborski, S.; Gozdowski, D.; Rozbicki, J. Effect of Genotype, Environment and Crop Management on Yield and Quality Traits in Spring Wheat. J. Cereal Sci. 2016, 72, 30–37. [Google Scholar] [CrossRef]
- Long, S.P.; Zhu, X.-G.; Naidu, S.L.; Ort, D.R. Can Improvement in Photosynthesis Increase Crop Yields? Plant Cell Environ. 2006, 29, 315–330. [Google Scholar] [CrossRef] [PubMed]
- Keller, B.; Zimmermann, L.; Rascher, U.; Matsubara, S.; Steier, A.; Muller, O. Toward Predicting Photosynthetic Efficiency and Biomass Gain in Crop Genotypes over a Field Season. Plant Physiol. 2022, 188, 301–317. [Google Scholar] [CrossRef]
- Ciura, J.; Kruk, J. Phytohormones as Targets for Improving Plant Productivity and Stress Tolerance. J. Plant Physiol. 2018, 229, 32–40. [Google Scholar] [CrossRef]
- Fahad, S.; Bajwa, A.A.; Nazir, U.; Anjum, S.A.; Farooq, A.; Zohaib, A.; Sadia, S.; Nasim, W.; Adkins, S.; Saud, S.; et al. Crop Production under Drought and Heat Stress: Plant Responses and Management Options. Front. Plant Sci. 2017, 8, 1147. [Google Scholar] [CrossRef] [Green Version]
- Pereira, C.S.; Lopes, I.; Abrantes, I.; Sousa, J.P.; Chelinho, S. Salinization Effects on Coastal Ecosystems: A Terrestrial Model Ecosystem Approach. Philos. Trans. R. Soc. B Biol. Sci. 2019, 374, 20180251. [Google Scholar] [CrossRef] [Green Version]
- Hu, H.; Liu, H.; Liu, F. Seed Germination of Hemp (Cannabis sativa L.) Cultivars Responds Differently to the Stress of Salt Type and Concentration. Ind. Crops Prod. 2018, 123, 254–261. [Google Scholar] [CrossRef]
- Guerriero, G.; Behr, M.; Hausman, J.-F.; Legay, S. Textile Hemp vs. Salinity: Insights from a Targeted Gene Expression Analysis. Genes 2017, 8, 242. [Google Scholar] [CrossRef] [Green Version]
- Di Mola, I.; Conti, S.; Cozzolino, E.; Melchionna, G.; Ottaiano, L.; Testa, A.; Sabatino, L.; Rouphael, Y.; Mori, M. Plant-Based Protein Hydrolysate Improves Salinity Tolerance in Hemp: Agronomical and Physiological Aspects. Agronomy 2021, 11, 342. [Google Scholar] [CrossRef]
- Berni, R.; Mandlik, R.; Hausman, J.-F.; Guerriero, G. Silicon-Induced Mitigatory Effects in Salt-Stressed Hemp Leaves. Physiol. Plant. 2021, 171, 476–482. [Google Scholar] [CrossRef] [PubMed]
- Guerriero, G.; Deshmukh, R.; Sonah, H.; Sergeant, K.; Hausman, J.-F.; Lentzen, E.; Valle, N.; Siddiqui, K.S.; Exley, C. Identification of the Aquaporin Gene Family in Cannabis sativa and Evidence for the Accumulation of Silicon in Its Tissues. Plant Sci. 2019, 287, 110167. [Google Scholar] [CrossRef] [PubMed]
- Guerriero, G.; Sutera, F.M.; Torabi-Pour, N.; Renaut, J.; Hausman, J.-F.; Berni, R.; Pennington, H.C.; Welsh, M.; Dehsorkhi, A.; Zancan, L.R.; et al. Phyto-Courier, a Silicon Particle-Based Nano-Biostimulant: Evidence from Cannabis sativa Exposed to Salinity. ACS Nano 2021, 15, 3061–3069. [Google Scholar] [CrossRef] [PubMed]
- Orozco-Mosqueda, M.d.C.; Flores, A.; Rojas-Sánchez, B.; Urtis-Flores, C.A.; Morales-Cedeño, L.R.; Valencia-Marin, M.F.; Chávez-Avila, S.; Rojas-Solis, D.; Santoyo, G. Plant Growth-Promoting Bacteria as Bioinoculants: Attributes and Challenges for Sustainable Crop Improvement. Agronomy 2021, 11, 1167. [Google Scholar] [CrossRef]
- Berni, R.; Guerriero, G.; Cai, G. One for All and All for One! Increased Plant Heavy Metal Tolerance by Growth-Promoting Microbes: A Metabolomics Standpoint. In Plant Metallomics and Functional Omics: A System-Wide Perspective; Sablok, G., Ed.; Springer International Publishing: Cham, Switzerland, 2019; pp. 39–54. ISBN 978-3-030-19103-0. [Google Scholar]
- Backes, A.; Charton, S.; Planchon, S.; Esmaeel, Q.; Sergeant, K.; Hausman, J.-F.; Renaut, J.; Barka, E.A.; Jacquard, C.; Guerriero, G. Gene Expression and Metabolite Analysis in Barley Inoculated with Net Blotch Fungus and Plant Growth-Promoting Rhizobacteria. Plant Physiol. Biochem. 2021, 168, 488–500. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, P.; Kanagendran, A.; Samaddar, S.; Pazouki, L.; Sa, T.-M.; Niinemets, Ü. Inoculation of Brevibacterium linens RS16 in Oryza sativa Genotypes Enhanced Salinity Resistance: Impacts on Photosynthetic Traits and Foliar Volatile Emissions. Sci. Total Environ. 2018, 645, 721–732. [Google Scholar] [CrossRef]
- Khan, M.A.; Asaf, S.; Khan, A.L.; Jan, R.; Kang, S.-M.; Kim, K.-M.; Lee, I.-J. Thermotolerance Effect of Plant Growth-Promoting Bacillus cereus SA1 on Soybean during Heat Stress. BMC Microbiol. 2020, 20, 175. [Google Scholar] [CrossRef]
- Ren, X.-M.; Guo, S.-J.; Tian, W.; Chen, Y.; Han, H.; Chen, E.; Li, B.-L.; Li, Y.-Y.; Chen, Z.-J. Effects of Plant Growth-Promoting Bacteria (PGPB) Inoculation on the Growth, Antioxidant Activity, Cu Uptake, and Bacterial Community Structure of Rape (Brassica napus L.) Grown in Cu-Contaminated Agricultural Soil. Front. Microbiol. 2019, 10, 1455. [Google Scholar] [CrossRef]
- Yu, Y.; Gui, Y.; Li, Z.; Jiang, C.; Guo, J.; Niu, D. Induced Systemic Resistance for Improving Plant Immunity by Beneficial Microbes. Plants 2022, 11, 386. [Google Scholar] [CrossRef]
- Lee, K.E.; Kang, S.M.; Adhikari, A.; Lee, I.J. Effect of Silicate Solubilizing Bacteria Pseudomonas psychrotolerans CS51 Treatment on Soybean Crops at Paddy Soil. In Proceedings of the Korean Society of Crop Science Conference; The Korean Society of Crop Science: Suwon, Korea, 2019; p. 46. [Google Scholar]
- Kubi, H.A.A.; Khan, M.A.; Adhikari, A.; Imran, M.; Kang, S.-M.; Hamayun, M.; Lee, I.-J. Silicon and Plant Growth-Promoting Rhizobacteria Pseudomonas psychrotolerans CS51 Mitigates Salt Stress in Zea mays L. Agriculture 2021, 11, 272. [Google Scholar] [CrossRef]
- Lozano, J.C.; Sbqueira, L. Prevention of the hypersensitive reaction in Tobacco leaves by heat-killed bacterial cells. Phytopathology 1970, 60, 875–879. [Google Scholar] [CrossRef]
- Rico, A.; Bennett, M.H.; Forcat, S.; Huang, W.E.; Preston, G.M. Agroinfiltration Reduces ABA Levels and Suppresses Pseudomonas Syringae-Elicited Salicylic Acid Production in Nicotiana tabacum. PLoS ONE 2010, 5, e8977. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Le Gall, H.; Philippe, F.; Domon, J.-M.; Gillet, F.; Pelloux, J.; Rayon, C. Cell Wall Metabolism in Response to Abiotic Stress. Plants 2015, 4, 112–166. [Google Scholar] [CrossRef] [PubMed]
- Tenhaken, R. Cell Wall Remodeling under Abiotic Stress. Front. Plant Sci. 2015, 5. [Google Scholar] [CrossRef] [Green Version]
- Mignard, S.; Flandrois, J.P. 16S rRNA Sequencing in Routine Bacterial Identification: A 30-Month Experiment. J. Microbiol. Methods 2006, 67, 574–581. [Google Scholar] [CrossRef]
- Widmer, F.; Seidler, R.J.; Gillevet, P.M.; Watrud, L.S.; Di Giovanni, G.D. A Highly Selective PCR Protocol for Detecting 16S rRNA Genes of the Genus Pseudomonas (Sensu Stricto) in Environmental Samples. Appl. Environ. Microbiol. 1998, 64, 2545–2553. [Google Scholar] [CrossRef] [Green Version]
- McWilliam, H.; Li, W.; Uludag, M.; Squizzato, S.; Park, Y.M.; Buso, N.; Cowley, A.P.; Lopez, R. Analysis Tool Web Services from the EMBL-EBI. Nucleic Acids Res. 2013, 41, W597–W600. [Google Scholar] [CrossRef] [Green Version]
- Trifinopoulos, J.; Nguyen, L.-T.; von Haeseler, A.; Minh, B.Q. W-IQ-TREE: A Fast Online Phylogenetic Tool for Maximum Likelihood Analysis. Nucleic Acids Res. 2016, 44, W232–W235. [Google Scholar] [CrossRef] [Green Version]
- Available online: https://www.dsmz.de/microorganisms/medium/pdf/DSMZ_Medium12.pdf (accessed on 15 March 2022).
- Vasanthi, N.; Saleena, L.M.; Raj, S.A. Silica Solubilization Potential of Certain Bacterial Species in the Presence of Different Silicate Minerals. Silicon 2018, 10, 267–275. [Google Scholar] [CrossRef]
- Guerriero, G.; Mangeot-Peter, L.; Legay, S.; Behr, M.; Lutts, S.; Siddiqui, K.S.; Hausman, J.-F. Identification of Fasciclin-like Arabinogalactan Proteins in Textile Hemp (Cannabis sativa L.): In Silico Analyses and Gene Expression Patterns in Different Tissues. BMC Genom. 2017, 18, 741. [Google Scholar] [CrossRef] [Green Version]
- Behr, M.; Sergeant, K.; Leclercq, C.C.; Planchon, S.; Guignard, C.; Lenouvel, A.; Renaut, J.; Hausman, J.-F.; Lutts, S.; Guerriero, G. Insights into the Molecular Regulation of Monolignol-Derived Product Biosynthesis in the Growing Hemp Hypocotyl. BMC Plant Biol. 2018, 18, 1. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hellemans, J.; Mortier, G.; De Paepe, A.; Speleman, F.; Vandesompele, J. Open Access Method QBase Relative Quantification Framework and Software for Management and Automated Analysis of Real-Time Quantitative PCR Data. Genome Biol. 2007, 81, R19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- González-Aguilera, K.L.; Saad, C.F.; Chávez Montes, R.A.; Alves-Ferreira, M.; de Folter, S. Selection of Reference Genes for Quantitative Real-Time RT-PCR Studies in Tomato Fruit of the Genotype MT-Rg1. Front. Plant Sci. 2016, 7, 1386. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Metsalu, T.; Vilo, J. ClustVis: A Web Tool for Visualizing Clustering of Multivariate Data Using Principal Component Analysis and Heatmap. Nucleic Acids Res. 2015, 43, W566–W570. [Google Scholar] [CrossRef] [PubMed]
- Fadiji, A.E.; Babalola, O.O.; Santoyo, G.; Perazzolli, M. The Potential Role of Microbial Biostimulants in the Amelioration of Climate Change-Associated Abiotic Stresses on Crops. Front. Microbiol. 2022, 12, 829099. [Google Scholar] [CrossRef] [PubMed]
- Andre, C.M.; Hausman, J.-F.; Guerriero, G. Cannabis sativa: The Plant of the Thousand and One Molecules. Front. Plant Sci. 2016, 7, 19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pellegrini, M.; Ercole, C.; Gianchino, C.; Bernardi, M.; Pace, L.; Del Gallo, M. Fusarium oxysporum f. Sp. Cannabis Isolated from Cannabis sativa L.: In Vitro and In Planta Biocontrol by a Plant Growth Promoting-Bacteria Consortium. Plants 2021, 10, 2436. [Google Scholar] [CrossRef]
- Pagnani, G.; Pellegrini, M.; Galieni, A.; D’Egidio, S.; Matteucci, F.; Ricci, A.; Stagnari, F.; Sergi, M.; Lo Sterzo, C.; Pisante, M.; et al. Plant Growth-Promoting Rhizobacteria (PGPR) in Cannabis sativa ‘Finola’ Cultivation: An Alternative Fertilization Strategy to Improve Plant Growth and Quality Characteristics. Ind. Crops Prod. 2018, 123, 75–83. [Google Scholar] [CrossRef]
- Lyu, D.; Backer, R.; Robinson, W.G.; Smith, D.L. Plant Growth-Promoting Rhizobacteria for Cannabis Production: Yield, Cannabinoid Profile and Disease Resistance. Front. Microbiol. 2019, 10, 1761. [Google Scholar] [CrossRef] [Green Version]
- Scott, M.; Rani, M.; Samsatly, J.; Charron, J.-B.; Jabaji, S. Endophytes of Industrial Hemp (Cannabis sativa L.) Cultivars: Identification of Culturable Bacteria and Fungi in Leaves, Petioles, and Seeds. Can. J. Microbiol. 2018, 64, 10. [Google Scholar] [CrossRef] [Green Version]
- Kang, S.-M.; Asaf, S.; Khan, A.L.; Lubna; Khan, A.; Mun, B.-G.; Khan, M.A.; Gul, H.; Lee, I.-J. Complete Genome Sequence of Pseudomonas psychrotolerans CS51, a Plant Growth-Promoting Bacterium, Under Heavy Metal Stress Conditions. Microorganisms 2020, 8, 382. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Portieles, R.; Xu, H.; Yue, Q.; Zhao, L.; Zhang, D.; Du, L.; Gao, X.; Gao, J.; Portal Gonzalez, N.; Santos Bermudez, R.; et al. Heat-Killed Endophytic Bacterium Induces Robust Plant Defense Responses against Important Pathogens. Sci. Rep. 2021, 11, 12182. [Google Scholar] [CrossRef] [PubMed]
- Arkhipova, T.N.; Prinsen, E.; Veselov, S.U.; Martinenko, E.V.; Melentiev, A.I.; Kudoyarova, G.R. Cytokinin Producing Bacteria Enhance Plant Growth in Drying Soil. Plant Soil 2007, 292, 305–315. [Google Scholar] [CrossRef]
- Taverniti, V.; Guglielmetti, S. The Immunomodulatory Properties of Probiotic Microorganisms beyond Their Viability (Ghost Probiotics: Proposal of Paraprobiotic Concept). Genes Nutr. 2011, 6, 261–274. [Google Scholar] [CrossRef] [Green Version]
- Morcillo, R.J.L.; Vílchez, J.I.; Zhang, S.; Kaushal, R.; He, D.; Zi, H.; Liu, R.; Niehaus, K.; Handa, A.K.; Zhang, H. Plant Transcriptome Reprograming and Bacterial Extracellular Metabolites Underlying Tomato Drought Resistance Triggered by a Beneficial Soil Bacteria. Metabolites 2021, 11, 369. [Google Scholar] [CrossRef]
- Chun, H.J.; Baek, D.; Cho, H.M.; Lee, S.H.; Jin, B.J.; Yun, D.-J.; Hong, Y.-S.; Kim, M.C. Lignin Biosynthesis Genes Play Critical Roles in the Adaptation of Arabidopsis Plants to High-Salt Stress. Plant Signal. Behav. 2019, 14, 1625697. [Google Scholar] [CrossRef]
- Guerriero, G.; Stokes, I.; Valle, N.; Hausman, J.-F.; Exley, C. Visualising Silicon in Plants: Histochemistry, Silica Sculptures and Elemental Imaging. Cells 2020, 9, 1066. [Google Scholar] [CrossRef] [Green Version]
- Luyckx, M.; Hausman, J.-F.; Sergeant, K.; Guerriero, G.; Lutts, S. Molecular and Biochemical Insights Into Early Responses of Hemp to Cd and Zn Exposure and the Potential Effect of Si on Stress Response. Front. Plant Sci. 2021, 12, 711853. [Google Scholar] [CrossRef]
- Luyckx, M.; Hausman, J.-F.; Isenborghs, A.; Guerriero, G.; Lutts, S. Impact of Cadmium and Zinc on Proteins and Cell Wall-Related Gene Expression in Young Stems of Hemp (Cannabis sativa L.) and Influence of Exogenous Silicon. Environ. Exp. Bot. 2021, 183, 104363. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).