Abstract
Apricot (Prunus armeniaca L.) is an important fruit crop and member of the Prunus genus of the Rosaceae family that is planted in many temperate regions worldwide. The quality of fruit is assessed by many pomological parameters which can serve as a decisive factor in apricot breeding, because the introduction of new cultivars is required. These parameters can differ with climate conditions, geographical location or geographic ecological origin. Similarly, another biological characteristic can be measured depending on these terms. The present study was conducted with the aim of estimating pomological traits together with the nuclear DNA content of 35 apricot cultivars with different geographical origins. Only CV values lower than 5% were considered in flow cytometry analysis. All analyzed cultivars were diploid and the genome size value ranged from 0.587 to 0.644 pg/2C, where Turkish apricots reached the highest value (on average 0.628 pg/2C) followed by the European group (on average 0.625 pg/2C). A Spearman-rank correlation was used and the different correlation was found for specific geographical groups of apricot cultivars. The genome size values of apricots and related botanical species P. mume, P. sibirica and P. ansu showed to be very similar values.
1. Introduction
The genus Prunus is divided into the five subgenera: Prunus (plums and apricots), Amygdalus (almonds and peaches), Cerasus (sweet and sour cherries), Laurocerasus (bay-cherries) and Padus (bird cherries) [1,2]. The genus has a considerable economic importance mostly for fruit and seed production, but some species are also grown as ornamentals. The apricot is one of the most popular fruit species planted in many temperate regions worldwide [3]. Three different routes of entry into Europe are known from the Central Asian area of origin. The first is the northern route from China to the Balkans, the second is southern route from Armenia through Syria, Arabia, Greece, Italy and northern Africa and the third is the middle route from the Danube Valley to Germany. Also, Roman soldiers played a major role in apricot distribution in Europe [4]. The fruit with characteristic properties is mainly consumed fresh, but it is also processed in food industry or preserved by drying. In addition, the apricot is an interesting supplement to the human diet, due to its high content of healthy substances as fibers, vitamins, minerals and an entire range of bioactive substances with high antioxidant capacities [5,6]. The apricot quality features that satisfy consumers are affected by many pomological traits [7]. The global world yield of apricot in 2020 was 6613.6 kg/ha with a production area of 562 thousand ha [8]. Apricots are divided into six major eco-geographical groups: Central Asian, East Chinese, North Chinese, Irano-Caucasian, European and Dzungar-Zailij [9]. Nevertheless, the classification is becoming complicated due to the production of new cultivars derived from crossing genotypes of different eco-geographical groups [10]. The breeding of the new cultivars requires information about the available germplasm. In addition, morphological and physiological properties improve the strategy of breeding and new variety selection. Nowadays, researchers are focused on the improvement of germplasm of Prunus to enhance their adaptability in production areas together with biotic and abiotic stresses [11].
Genome size is one of the important attributes of the genome. Knowledge of the nuclear DNA content (2C) is crucial for basic and applied studies involving genome organization, species relationship, gene expression analysis or germplasm improvement. The genome size is nowadays determined at different fruit species, often for the purpose of studying the correlations of the genome size and morphological characteristics [12], origin [12,13,14] or as a supplemental data of genetic information [15,16]. Apricots are mostly diploid (2n = 2x = 16), but cultivars with higher ploidy have been observed as well [17]. The nuclear DNA content of a diploid P. armeniaca has been previously estimated by to be 0.6 pg/2C [14,18,19], which corresponds to a genome size of 293.4 Mbp. It was proven that genome size differs with the climatic condition, and correlations between genome size and altitude, geographical situation, temperature, etc. were found [20]. However, these trends differ for different plant species [14,21].
Over the years, different methods have been used to estimate genome sizes in plants (e.g., Feulgen densitometry, reassociation kinetics, chemical extraction). However, flow cytometry has become the method of choice because of the ease of sample preparation, low cost, possibility to quickly analyze high numbers of particles and high accuracy and resolution [22,23,24,25,26]. The method enables the analysis of the optical properties of individual particles in suspension while they move in a narrow liquid stream through a beam of light. Emitted and scattered light is then evaluated with an optical system. The size of the genome is usually stated with a C value, which determines the DNA content of the unreplicated haploid set of chromosomes [27].
This research is focused study of the variations in the genome sizes of 35 different apricot cultivars and selected botanical species related to P. armeniaca with the aim to investigate if an association exists between genome sizes and the geographical origin and/or the pomological characteristics of the cultivars.
2. Materials and Methods
2.1. Plant Material
35 apricot cultivars growing in the experimental orchard of the Faculty of Horticulture in Lednice Mendel University in Brno (localization 48.80° N/16.80° E, altitude of 172 m) were used for the study. The selected cultivars were divided into five different groups based on their geographical origin: American, Central Asian, East Asian, European and Turkish. In addition, three botanical species related to Prunus armeniaca, P. mume (the Japanese apricot), P. ansu (the Korean apricot), P. sibirica (the Siberian apricot), were analyzed.
2.2. Pomological Analysis
The pomological traits of the fruits were analyzed for four years (2018–2021) in their optimum maturity, which was determined using a DA Meter®. The harvested fruits were analyzed immediately. Average data were obtained from ten fruits of every cultivar according to the “Klasifikátor–Descriptor list for genus Armeniaca Mill” [28]. The weight was determined using laboratory balances. The soluble solid content (°Rf) was carried out with Abbé’s refractometer.
2.3. Genome Size Estimation
For the genome size evaluation, only intact and fully developed leaves of the selected cultivars were collected and analyzed immediately. The average value was determined from three trees for all cultivars. The kit 05-5022 CyStain PI Absolute P® was used for the genome size estimation. Approximately 0.5 cm2 of the leaf tissue of a sample and approximately 0.5 cm2 of the leaf tissue of Raphanus sativus cv. Saxa (2C = 1.11 pg DNA) [29], which served as the internal reference standard, were homogenized together in 1.5 mL of Nuclei Extraction Buffer (pH = 1.8) using a razor blade. After 60 s, the prepared sample was filtered through a 50 μm membrane (Partec CellTrics®) and supplemented with 2 mL of the solution of Staining Buffer (pH = 7.5), 12 µL of propidium iodide (PI) and 6 µL of RNase. After 30 min of incubation in the dark at room temperature, the samples were analyzed in a flow cytometer CyFlow Space–3 equipped with a 532 nm green laser. The maximum acquisition rate was up to 25,000 signals/s and the scatter particle size was 0.1–100 µm. At least 5000 particles were analyzed for three replications of each sample. The speed of flow was carried out at up to 50 particles per second. Only coefficient of variation (CV) values lower than 5% were considered as reliable data [14,26]. The 2C nuclear DNA content was calculated according to following formula: 2C (pg) = DNA content of standard × (mean fluorescence value of sample/mean fluorescence value of standard). The mean nuclear DNA content was then calculated for each genotype from three replications. The results were also expressed as monoploid genome sizes (1Cx) (i.e., DNA content of the non-replicated base set of chromosomes) according to following formula: 1Cx = 2C genome size/ploidy level 0.978 × 109 bp [30].
2.4. Statistical Analysis
STATISTICA 12 was used for the calculation of the homogeneity of variance for pomological traits and genome size of cultivars using the Pearson’s χ2 test with adjusted degrees of freedom (df) using the Kolmogorov-Smirnov test, the average values, the standard deviation and the Kruskal-Wallis test with a post-hoc test of multiple comparisons of mean ranks for all groups of pomological traits.
The distribution of the data for pomological traits and genome size deviated significantly from the Normal distribution (Figure S1). The parameters of the Pearson χ2 test are displayed in the Supplementary Materials (Tables S1 and S2). Based on the results of the preliminary test, the Spearman-rank correlation (correlation coefficient, rs) method was adopted to analyze the correlation between the genome size of the cultivars and the pomological traits. The same correlation analyses were accomplished for genome size and geographical origin.
3. Results
3.1. Pomological Traits
The pomological data of all analyzed cultivars are summarized in Table 1 and Table 2. The ripening date was analyzed only in 2021. The fruit of an East Asian cultivar ‘Liaoning,’ a Central Asian cultivar ‘Gvardejskij’ and three European cultivars ‘Pastyrikz’ ‘Pozde kvetouci’ and ‘Wondercotz’ reached maturity early, in the third week of June. ‘Kechpsar,’ a cultivar from Central Asia, had the latest ripening date on 24 September. All the other cultivars reached maturity during July. The weight of the fruits (Figure 1a) ranged from 22.6 g in the American cultivar ‘Scout’ to 99.9 g in the European cultivar ‘Pastyrik.’ The highest average fruit weight (62.3 g) was recorded in East Asian cultivars, followed by the European cultivars (56.3 g), while the Central Asian cultivars had the lowest average fruit weight (34.6 g). The color of the fruit varied within geographical groups of cultivars, with the exception of Turkish cultivars, in which the yellow color of the skin prevailed. The shape of the fruit had no specific trend. The size of the flesh ranged from 6.5 mm (Scout) to 16.8 mm (Pastyrik). The average values for geographical groups ranged from 9.3 to 11.6 mm with an increasing size from Turkish, Central Asian, American, East Asian to European cultivars. The color of the flesh was mostly light orange or orange. In case of Turkish cultivars, a lighter color than in other geographical groups was determined. The average weight of the stone among geographical groups corresponded with the average fruit weight, ranging from 2.5 g in Central Asian cultivars to 3.9 g in East Asian cultivars.

Table 1.
The pomological traits of apricot cultivars. Data are displayed as the mean ± standard deviation of ten replications in 2018–2021. In the columns, the mean values followed by different letters are significantly different (p < 0.05) based on post-hoc analysis of the Kruskal-Wallis test by multiple comparison of mean ranks for all groups (a…p). * 1—white, 2—cream beige, 3—yellow, 4—light orange, 5—orange, 7—deep orange; ** 1—deformed, 2—flat, 3—rounded, 4—oblong, 5—elliptical, 6—triangular, 7—cordate, 8—strongly elliptical; *** 1—white, 2—light beige yellow, 3—cream, 4—yellow, 5—light orange, 6—orange, 7—deep orange.

Table 2.
The average pomological traits for the geographical groups of apricots. No significant differences (p < 0.05) were detected within the pomological traits based on the Kruskal-Wallis test. * 1—white, 2—cream beige, 3—yellow, 4—light orange, 5—orange, 7—deep orange; ** 1—deformed, 2—flat, 3—rounded, 4—oblong, 5—elliptical, 6—triangular, 7—cordate, 8—strongly elliptical; *** 1—white, 2—light beige yellow, 3—cream, 4—yellow, 5—light orange, 6—orange, 7—deep orange.
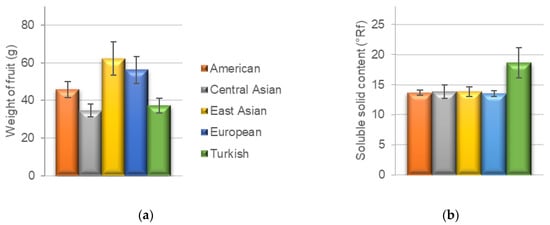
Figure 1.
(a) The average weight of fruit (g); (b) The average soluble solid content of the fruit (°Rf) in the selected geographical groups of apricot cultivars.
From these results, the ratio of the weight of a stone to the whole fruit was calculated and ranged from 6.2 to 7.5%. The values increased in order from the European, East Asian, American and Central Asian to the Turkish geographical groups of cultivars.
The soluble solid content (°Rf) highly affects the taste of the fruit in sense of sweetness; in other words, this parameter is an important qualitative indicator defining the sugar content and affects the selection of the cultivar for the cultivation and use of fruits. Higher values are required if the fruit is processed (e.g., drying or canning). The values of soluble solid content ranged from 11.1 °Rf (Achrori) to 26.1 °Rf (Sekerpare). The average values of the soluble solid content in the different geographical groups (Figure 1b) were very similar, ranging from 13.5 to 13.8 °Rf. The only exception was the group of Turkish cultivars, which highly exceeded the other groups with an average soluble solid content of 18.6 °Rf.
3.2. Genome Size Estimation
The genome sizes of all studied P. armeniaca cultivars and three botanical species related to P. armeniaca were estimated after the flow cytometric analysis of propidium iodide-stained nuclei. All analyses resulted in histograms of relative DNA content with two dominant peaks corresponding to the G1 nuclei of apricots and Raphanus sativus, which served as the internal reference standard (Figure 2), and a smaller peak of the G2 nuclei of apricots. The 2C nuclear DNA content was determined based on the ratio of G1 peaks’ positions and ranged for P. armeniaca from 0.587 pg/2C in ‘Orangered’ to 0.644 pg/2C in ‘Bohuticka.’ The average values of the genome sizes of the analyzed species are shown in Table 3. No significant differences were observed in the geographical groups (Figure 3). The highest average genome size was observed in the Turkish geographical group of cultivars (0.628 pg/2C). In comparison, the botanical species related to P. armeniaca reached values in increasing order from P. mume (0.581 pg/2C) and P. ansu (0.608 pg/2C) to P. sibirica (0.621 pg/2C), and the values were very similar to the nuclear content of P. armeniaca.
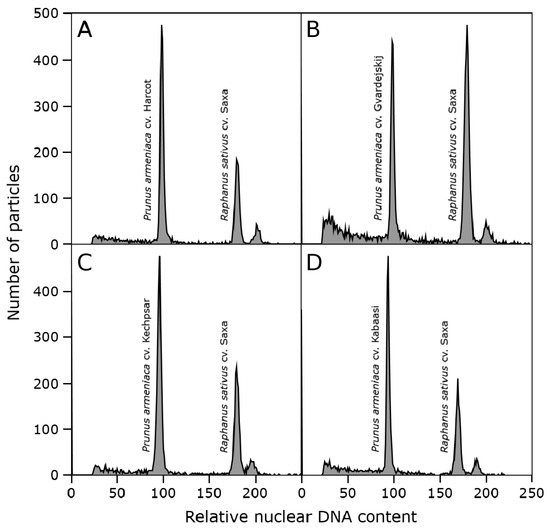
Figure 2.
A histogram of the relative nuclear DNA content obtained after a flow cytometric analysis of propidium iodide-stained nuclei isolated from P. armeniaca ((A)—Harcot, (B)—Gvardejskij, (C)—Kechpsar, (D)—Kabaasi) together with internal standard Raphanus sativus cv. Saxa.

Table 3.
The nuclear DNA content and monoploid genome sizes of the selected cultivars with different geographical origins.
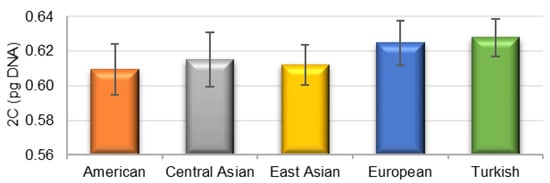
Figure 3.
The average nuclear DNA content (2C) for the geographical groups of apricots. The vertical lines refer to the standard deviation.
3.3. Statistical Analysis
Some studies predicted that genome size variation could be changing together with fruit size, similar to ploidy levels [12,31,32]. The obtained data were evaluated as possible correlations between relative genome size and pomological traits. According to the results of the Pearson χ2 test (Supplementary Material), the non-parametric statistical correlation analysis was used to analyze the correlation between pomological traits, genome size and the geographical origin of apricot cultivars.
The Spearman-rank correlation analysis showed no correlation between relative genome size and weight of fruit (rs = −0.0623), between relative genome size and size of flesh (rs = −0.2141) (Figure 4), between relative genome size and soluble solid content (rs = 0.1750) or between relative genome size and weight of stone (rs = 0.0420).
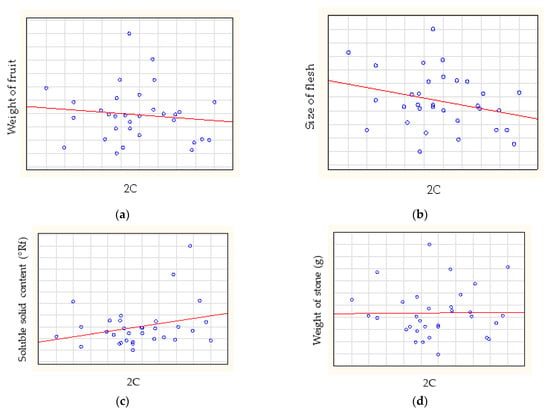
Figure 4.
The correlations between relative genome size and (a) weight of fruit, (b) size of flesh, (c) soluble solid content and (d) weight of stone.
In addition, a Spearman-rank correlation analysis was conducted of the weight of fruit and the relative genome of the analyzed geographical groups (1—American, 2—European, 3—Turkish, 4—Central Asian, 5—East Asian). The normal distribution of the analyzed data showed significant deviation (Table S1), which could be caused by a low number of cultivars in each geographical group.
The Spearman-rank correlation analysis (Figure 4) of weight of fruit and relative genome size showed low negative correlation for American cultivars (rs = −0.2857) and medium correlation for at European cultivars (rs = −0.6786) and Turkish cultivars (rs = −0.7714). A high positive correlation was found for East Asian cultivars (rs = 0.8000) and no correlation was found for Central Asian cultivars (rs = 0.0857). The Spearman-rank correlation analyses showed that the genome size did not correlate with geographical origin (rs = 0.0369) (Figure 5).
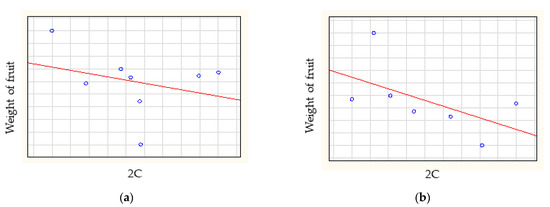
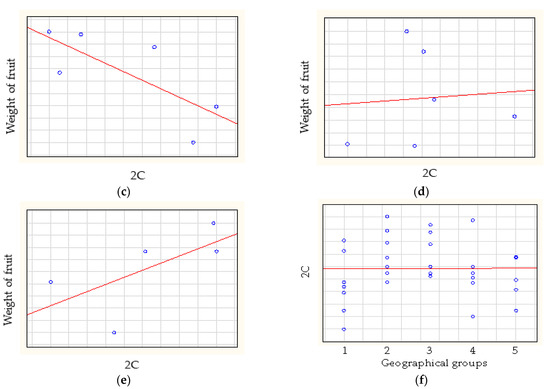
Figure 5.
The correlations between relative genome size and the weight of fruit of (a) American, (b) European, (c) Turkish, (d) Central Asian and (e) East Asian apricot cultivars. (f) The correlation between relative genome size and the geographical origin of apricot cultivars.
4. Discussion
The study of pomological traits is important for the future breeding of fruit species, and similar studies have been introduced worldwide [33,34,35,36,37]. The pomological traits may differ with regard to different geographical conditions. Saridaş et al. (2019) [34] analyzed cultivar ‘Harcot,’ where the weight of the fruit was 69.0 g. Kumar et al. (2015) [35] found that the weight of the fruit of ‘Harcot’ reached 51.73 g. In our study, the value was 47.5 g. Similarly, the soluble solid content may be also affected by climatic conditions. Göttingerová and Nečas (2020) [38] measured the fruit soluble solid content in 16 apricot cultivars planted in the Czech Republic in the range from 10.4 to 19 °Rf, while Gurrieri et al. (2001) [39] determined the soluble solid content in mature apricots from 8.7 to 22.4 °Rf and Saridaş et al. (2019) [34] at Turkish apricots from 5.8 to 18.4 °Rf. In contrast, Gecer et al. (2020) [40] measured Turkish apricots values of soluble solid content from 13.0 to 22.3 °Rf. In our study, the values for soluble solids ranged from 11.1 to 26.1 °Rf and the highest values were measured in the Turkish geographical group of apricots. Thus, our results of soluble solid content were similar to previous studies. The Turkish cultivars exceeded the highest average values with high standard deviations, and thanks to their traits, they are suitable for fruit processing. In previous studies, the pomological traits were supplemented with genome size analysis [41]. This fundamental parameter can be used in other genetic and molecular biological studies [42]. In our study, 35 apricot cultivars were analyzed by flow cytometry. According to Leitch (1998) [43], the relative genome size of P. armeniaca is very small. The alternative method of genome size determination is genome sequencing [44], but by this method, often only a part of genome is analyzed [3,44]. The information about the genomes of specific varieties of apricots are limited so far. In flow cytometry, the measurable value has to be related to a reference with a known genome size [30]. Arumuganathan and Earle (1991) [18] measured by flow cytometry an average value of 0.61 pg/2C of apricot cultivars with chicken red blood cell nuclei as internal standard. Li et al. (2020) [14] focused on 124 apricot varieties from different geographical regions and used Medicago sativa L. cv. Alfalfa queen as the internal standard. In our study, the internal standard Raphanus sativus cv. Saxa was estimated as suitable to use because of the similar value of its genome size. Similar to Li et al. (2020) [14], all analyzed apricot cultivars were diploid in this study. The range of the apricot genome size was 0.597–0.673 pg/2C with an average value of 0.637 pg/2C in Li et al. (2020) [14]. The genome size of apricots in our study ranged from 0.581–0.644 pg/2C with an average value of 0.616 ± 0.008 pg/2C. Results showed that the genome size tended to increase during evolution [45,46]. A great role in the domestication of apricots has the Chinese origins [47]. The apricot cultivars with Asian origin reached the lowest values of relative genome size in this study (0.615 pg/2C). We can compare the value for Central Asian geographical groups, which Li et al. (2020) [14] determined as 0.640 pg/2C. In the opposite, Zhang et al. (2003) indicated the European Ecological Group of apricot cultivars as the youngest ecological group. Li et al. (2020) [14] stated the highest genome size values for the European geographical group of cultivars (0.661 pg/2C). In our study, the European apricot cultivars reached the second highest average value of relative genome size (0.625 pg/2C). The highest values of relative genome size were observed in the Turkish geographic group of cultivars (0.628 pg/2C) which could be partly considered as very related to the European geographical group. In comparison, the old botanical species related to P. armeniaca reached values in increasing order from P. mume (0.581 pg/2C) to P. ansu (0.608 pg/2C) to P. sibirica (0.621 pg/2C). Thus, our results confirmed the tendency of an increasing genome size during evolution. The relative genome size of cultivar ‘Sai mai ti’ was 0.670 pg/2C in the study of Li et al. (2020) [14], while in our study, the genome size reached 0.607 pg/2C. This high discrepancy in genome size of the same cultivar from different studies might be due to the use of different internal standards in each study. Raphanus sativus cv. Saxa was used in our study, while Li et al. (2020) [14] used Medicago sativa L. cv. Alfalfa queen.
Kolano et al. (2012) [20] also mentioned a possible correlation between relative genome size and geographical location (altitude, latitude), temperature and rainfall. Li et al. (2020) [14] found no correlation (very low negative) in wild genotypes of P. armeniaca between genome size and longitude, altitude and latitude. We divided the analyzed apricot cultivars into five groups according to their origin (American, European, Turkish, Central Asian and East Asian), and no correlation between relative genome size and geographical groups were found in this study (rs = 0.0369). The normal distribution of data proved that more cultivars in each geographical group need to be analyzed in a more detailed future study.
In addition, a correlation analysis was used to assess the correlations between the weight of the fruit and genome size (rs = −0.06231) and the size of the flesh and relative genome size (rs = −0.2141). The results indicate no (very low negative) correlation. After dividing the cultivars into geographical groups, the correlation analysis was negative for American, European and Turkish cultivars, and a highly positive correlation was found for East Asian cultivars. The results indicated a difference in relationship between the size of the fruit and the relative genome size with regard to the geographical origin of the apricot cultivars. However, this indication needs to be supported with more detailed analyses.
5. Conclusions
Valuable data and information for a better understanding of pomological traits and genome size variation within Prunus armeniaca cultivars were obtained in this study. The genome size is measurable information used in breeding because of its utilization in studies of the evolutionary relationship between plant species and their varieties. The results of this study supplement the already available information of pomological traits and genome size of known apricot varieties. We observed diversity in pomological traits between and within different geographical groups of apricot cultivars using the Kruskal-Wallis test with a post-hoc multiple comparison of mean ranks for all groups’ analyses. In addition, the correlation between pomological traits and relative genome sizes was analyzed using the Spearman-rank correlation method (correlation coefficient, rs). The obtained values of the relative genome sizes were lower than for cultivars planted in China, but the trend of values was similar. This study can be useful in future research focused on the genetic diversity of apricots.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/horticulturae8030199/s1, Figure S1: Fitting of continuous normal distribution of (a) weight of fruit, (b) size of flesh, (c) weight of stone, (d) soluble solid content, (e) relative genome size of all analysed apricot cultivars, Table S1: Parameters of Pearson χ2 test used for all analysed apricot cultivars, Table S2: Parameters of Pearson χ2 test used for weight of fruit and relative genome size of geographical groups of apricot cultivars (n indicates number of cultivars).
Author Contributions
Conceptualization, E.R., T.N. and J.Č.; methodology, E.R., M.M., T.N. and J.Č.; formal analysis, E.R. and M.M.; investigation, E.R.; resources, T.N. and J.Č.; data curation, E.R., M.M. and J.Č.; writing—original draft preparation, E.R. and J.Č.; writing—review and editing, E.R. and J.Č.; supervision, T.N.; project administration, E.R.; funding acquisition, E.R. and T.N. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by project IGA-ZF/2021-SI1006, study of chromosome ploidy and genome size of selected taxa Prunus armeniaca with different geographical origins by flow cytometry. The results of this research were also obtained with the support of Ministry of Agriculture project NAZV/QK1910137.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
Laboratory equipment was acquired under project CZ.02.1.01/0.0/0.0/16_017/0002334. Research infrastructure for young scientists. Plant materials were provided by activity no. 6.2.10 ref. 51834/2017-MZE-17253, subprogram “National Program of Conservation and Utilization of Plant Genetic Resources and Agrobiodiversity,” which is funded by the Ministry of Agriculture of the Czech Republic.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Rehder, A. Manual of Cultivated Trees and Shrubs, 2nd ed.; MacMillan: New York, NY, USA, 1940; pp. 452–481. [Google Scholar]
- Gharaghani, A.; Solhjoo, S.; Oraguzie, N. A review of genetic resources of almonds and stone fruits (Prunus spp.) in Iran. Genet. Resour. Crop Evol. 2017, 64, 611–640. [Google Scholar] [CrossRef]
- Jiang, F.; Zhang, J.; Wang, S.; Yang, L.; Yinfeng, L.; Gao, S.; Zhang, M.; Wu, S.; Hu, S.; Sun, H.; et al. The apricot (Prunus armeniaca L.) genome elucidates Rosaceae evolution and beta-carotenoid synthesis. Hortic. Res. 2019, 6, 128. [Google Scholar] [CrossRef]
- Löschnig, I.; Passecker, F. Die Marille (Aprikose) und Ihre Kultur; Ost. Agrarverlag: Wienna, Vienna, 1954. [Google Scholar]
- Gatti, E.; Defilippi, G.B.; Predieri, S.; Infante, R. Apricot (Prunus armeniaca L.) quality and breeding perspectives. J. Food. Agric. Environ. 2009, 7, 573–580. [Google Scholar]
- Frantianni, F.; Ombra, M.N.; d’Acierno, A.; Cipriano, L.; Nazzaro, R. Apricots: Biochemistry and functional properties. Curr. Opin. Food Sci. 2018, 19, 23–29. [Google Scholar]
- Milosevic, T.; Milosevic, N.; Glisic, I.; Krska, B. Characteristics of promising apricot (Prunus armeniaca L.) genetic resources in Central Serbia based on blossoming period and fruit quality. Hort. Sci. 2010, 37, 46–55. [Google Scholar]
- Food and Agriculture Organization of the United Nations. Crops and Livestock Products; FAOSTAT. FAO: Rome, Italy, 2022. Available online: https://www.fao.org/faostat/en/#data/QCL (accessed on 5 February 2022).
- Layne, R.E.C.; Bailey, C.H.; Hough, L.F. Apricots. In Fruit Breeding. Vol II: Tree and Tropical Fruits; Janick, J., Moore, J.N., Eds.; John Wiley and Sons: New York, NY, USA, 1996; pp. 79–111. [Google Scholar]
- Faust, M.; Surány, D.; Nyujtó, F. Origin and dissemination of apricot. Hort. Rev. 1998, 22, 225–266. [Google Scholar]
- Childers, N.; Sherman, W. The Peach; Horticultural Publications: Gainesville, FL, USA, 1988. [Google Scholar]
- Kadkhodaei, S.; Arzani, K.; Yadollahi, A.; Karimzadeh, G.; Abdollahi, H. Genetic Diversity and Similarity of Asian and European Pears (Pyrus Spp.) Revealed by Genome Size and Morphological Traits Prediction. Int. J. Fruit Sci. 2021, 21, 619–633. [Google Scholar] [CrossRef]
- Shearer, K.; Ranney, T.G. Ploidy Levels and Relative Genome Sizes of Species, Hybrids, and Cultivars of Dogwood (Cornus spp.). HortScience 2013, 48, 825–830. [Google Scholar] [CrossRef]
- Li, W.; Liu, L.; Wang, Y.; Fan, G.; Zhang, S.; Wang, Y.; Liao, K. Determination of genome size and chromosome ploidy of selected taxa from Prunus armeniaca by flow cytometry. Sci. Hortic. 2020, 261, 417–432. [Google Scholar] [CrossRef]
- Jatt, T.; Lee, M.; Rayburn, A.L.; Jatol, M.A.; Mirani, A.A. Determination of genome size variations among different date palm cultivars (Phoenix dactylifera L.) by flow cytometry. Biotech 2019, 9, 457. [Google Scholar] [CrossRef]
- Tamarzizt, H.B.; Walker, D.; Mustapha, S.B.; Abdallah, D.; Baraket, G.; Hannachi, A.S.; Azzouzi, S.Z. DNA variation and polymorphism in Tunisian plum species (Prunus spp.): Contribution of flow cytometry and molecular markers. Genet. Mol. Res. 2015, 14, 18034–18046. [Google Scholar] [CrossRef] [PubMed]
- Lu, Z.R.; Guo, Z.H.; Xiao, J.Z.; Wang, F.Y. Karyotype analysis of Armeniaca sibirica (L.) Lam. And Armeniaca vulgaris Lam. J. Hebei Agric. Univ. 1986, 6, 14–19. [Google Scholar]
- Arumuganathan, K.; Earle, E.D. Nuclear DNA content of some important plant species. Plant Mol. Biol. Rep. 1991, 9, 208–218. [Google Scholar] [CrossRef]
- Dickson, E.E.; Arumuganathan, K.; Kresovich, S.; Doyle, J.J. Nuclear DNA content variation within the Rosaceae. Am. J. Bot. 1992, 79, 1081–1086. [Google Scholar] [CrossRef]
- Kolano, B.; Siwinska, D.; Gomez Pando, L.; Szymanowska-Pulka, J.; Maluszynska, J. Genome size variation in Chenopodium quinoa (Chenopodiaceae). Plant. Syst. Evol. 2012, 298, 251–255. [Google Scholar] [CrossRef][Green Version]
- Knight, C.A.; Molinary, N.; Petrov, D. The large genome constraint hypothesis:evolution, ecology, and phenotype. Ann. Bot. 2005, 5, 177–190. [Google Scholar] [CrossRef]
- Jadrná, P.; Konza, F.; Plavcová, O. Polyploidization of Pelargonium × hortorum L. H. Bailey in greenhouse conditions. Hort. Sci. 2009, 36, 31–37. [Google Scholar] [CrossRef]
- Dalbato, A.L.; Kobza, F.; Karlsson, L.M. Effect of polyploidy and pollination methods on capsule and seed set of pansies (Viola × wittrockiana Gams). Hort. Sci. 2013, 40, 22–30. [Google Scholar] [CrossRef]
- Müller, L.B.; Zotz, G.; Albach, D.C. Bromeliaceae subfamilies show divergent trends of genome size evolution. Sci. Rep. 2019, 9, 5136. [Google Scholar] [CrossRef]
- Rodrigues, P.S.; Souza, M.M.; Melo, C.A.F.; Pereira, T.N.S.; Corrêa, R.X.C. Karyotype diversity and 2C DNA content in species of the Caesalpinia group. BMC Genet. 2018, 19, 25. [Google Scholar] [CrossRef]
- Sakhanokho, H.F.; Timothy Rinehart, A.; Stringer, S.J.; Islam-Faridi, M.N.; Pounders, C.T. Variation in nuclear DNA content and chromosome numbers in blueberry. Sci. Hortic. 2018, 233, 108–113. [Google Scholar] [CrossRef]
- Swift, H. The constancy of desoxyribose nucleic acid in plant nuclei. Proc. Natl. Acad. Sci. USA 1950, 36, 643–654. [Google Scholar] [CrossRef] [PubMed]
- Nitranský, Š. Klasifikátor—Descriptor List Genus Armeniaca P. Mill. Vyd. Semex ŠP Bratislava; VS Veselé a VÚRV: Praha, Czech Republic, 1992; p. 29. [Google Scholar]
- Doležel, J.; Sgorbatti, S.; Lucretti, S. Comparison of three DNA fluorochromes for flow cytometric estimation of nuclear DNA content in plants. Physiol. Plant. 1992, 85, 625–631. [Google Scholar] [CrossRef]
- Doležel, J.; Liu, L.; Wang, Y.; Fan, G.; Zhang, S.; Wang, Y.; Liao, K. Plant DNA Flow Cytometry and Estimation of Nuclear Genome Size. Ann. Bot. 2005, 95, 99–110. [Google Scholar] [CrossRef]
- Wang, L.; Luo, Z.; Liu, Z.; Zhao, J.; Deng, W.; Wei, H.; Liu, P.; Liu, M. Genome Size Variation within Species of Chinese Jujube (Ziziphus jujube Mill.) and Its Wild Ancestor Sour Jujube (Z. acidojujuba Cheng et Liu). Forests 2019, 10, 460. [Google Scholar] [CrossRef]
- Martínez-Sagarra, G.; Castro, S.; Mota, L.; Loureiro, J.; Devesa, J.A. Genome size, chromosome number and morphological data reveal unexpected infraspecific variability in Festuca (Poaceae). Genes 2021, 12, 906. [Google Scholar] [CrossRef]
- Nečas, T.; Göttingerová, M.; Wolf, J.; Kiss, T.; Rampáčková, E.; Ondrášek, I. New promising apricot hybrids from Faculty of Horticulture in Lednice. Acta Hortic. 2020, 1290, 169–178. [Google Scholar] [CrossRef]
- Saridaş, M.A.; Paydaş Kargı, S.; Kafkas, E. Phenological and pomological characteristics of some important apricot cultivars grown in Turkey. Acta Hortic. 2019, 1242, 553–560. [Google Scholar] [CrossRef]
- Kumar, D.; Lal, S.; Ahmed, N. Morphological and pomological diversity among apricot (Prunus armeniaca) genotypes grown in India. Indian J. Agric. Sci. 2015, 85, 1349–1355. [Google Scholar]
- Ayour, J.; Harrak, H.; Alahyane, A.; Taourirte, M.; Renard, C.M.G.C.; Benichou, M. Pomological and antioxidant properties of apricot (Prunus armeniaca L.): Fruit management and quality analysis. J. Food Microbiol. Saf. Hyg. 2018, 3, 53. [Google Scholar]
- Karatas, N.; Ercisli, S.; Bozhuyuk, M.R. Assessment of Morphological Traits, Nutritional and Nutraceutical Composition in Fruits of 18 Apricot cv. Sekerpare Clones. Sustainability 2021, 13, 11385. [Google Scholar] [CrossRef]
- Göttingerová, M.; Nečas, T. Comparison of selected qualitative characteristic of American, French and Czech apricot cultivars. Acta Hortic. 2020, 1290, 159–168. [Google Scholar]
- Gurrieri, F.; Audergon, J.M.; Albagnac, G.; Reich, M. Soluble sugars and carboxylic acids in ripe apricot fruit as parameters for distinguishing different cultivars. Euphytica 2001, 117, 183–189. [Google Scholar] [CrossRef]
- Gecer, M.K.; Kan, T.; Gundogdu, M.; Ercisli, S.; Ilhan, G.; Sagbas, H.I. Physicochemical characteristics of wild and cultivated apricots (Prunus armeniaca L.) from Aras valley in Turkey. Genet. Resour. Crop Evol. 2020, 67, 935–945. [Google Scholar] [CrossRef]
- Głowacka, A.; Sitarek, M.; Rozpara, E.; Podwyszyńska, M. Pomological Characteristics and Ploidy Levels of Japanese Plum (Prunus salicina Lindl.) Cultivars Preserved in Poland. Plants 2021, 10, 884. [Google Scholar] [CrossRef]
- Baird, W.V.; Estager, A.S.; Wells, J.K. Estimating Nuclear DNA Content in Peach and Related Diploid Species Using Laser Flow Cytometry and DNA Hybridization. J. Am. Soc. Hort. Sci. 1994, 119, 1312–1316. [Google Scholar] [CrossRef]
- Leitch, I.J.; Soltis, D.E.; Soltis, P.S.; Bennet, M.D. Evolution of DNA amounts across land plants (embryophyta). Ann. Bot. 2005, 95, 207–217. [Google Scholar] [CrossRef]
- Zhang, Q.; Chen, W.; Sun, L.; Zhao, F.; Huang, B.; Yang, W.; Tao, Y.; Wang, J.; Yuan, Z.; Fan, G.; et al. The genome of Prunus mume. Nat. Commun. 2012, 3, 1318. [Google Scholar] [CrossRef]
- Vitte, C.; Panaud, O. Formation of solo-LTRs through unequal homologous recombination counterbalances amplifications of LTR retrotransposons in rice Oryza sativa L. Mol. Biol. Evol. 2003, 20, 528–540. [Google Scholar] [CrossRef]
- Maghuly, F.; Fernandez, E.B.; Ruthner, S.; Pedryc, A.; Laimer, M. Microsatellite variability in apricots (Prunus armeniaca L.) reflects their geographic origin and breeding history. Tree Genet. Genomes 2005, 1, 151–165. [Google Scholar] [CrossRef]
- Leitch, I.J.; Chase, M.V.; Bennett, M.D. Phylogenetic analysis of DNA C-values provides evidence for a small ancestral genome size in flowering plants. Ann. Bot. 1998, 82, 85–94. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).