Identification of Suitable Reference Genes for qRT-PCR Normalization in Kiwifruit
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials and Treatments
2.2. RNA Isolation and cDNA Synthesis
2.3. qRT-PCR Primer Design of Candidate Reference Genes
2.4. Quantitative Real-Time PCR Analysis
2.5. Standard Curve of Candidate Reference Genes
2.6. Data Analysis
3. Results
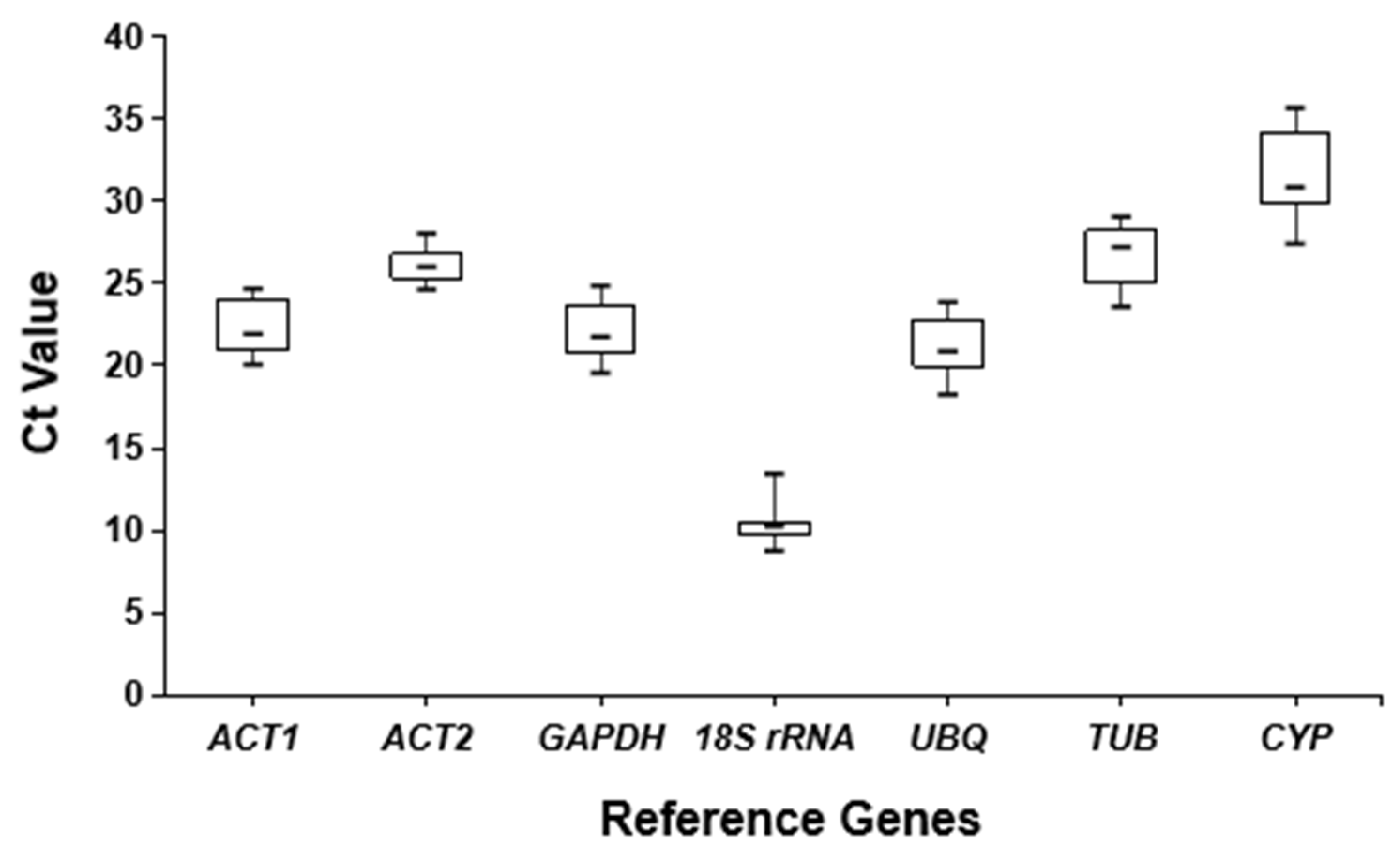
3.1. Amplification Efficiency of Candidate Reference Genes
3.2. Expression Analysis of Candidate Reference Genes
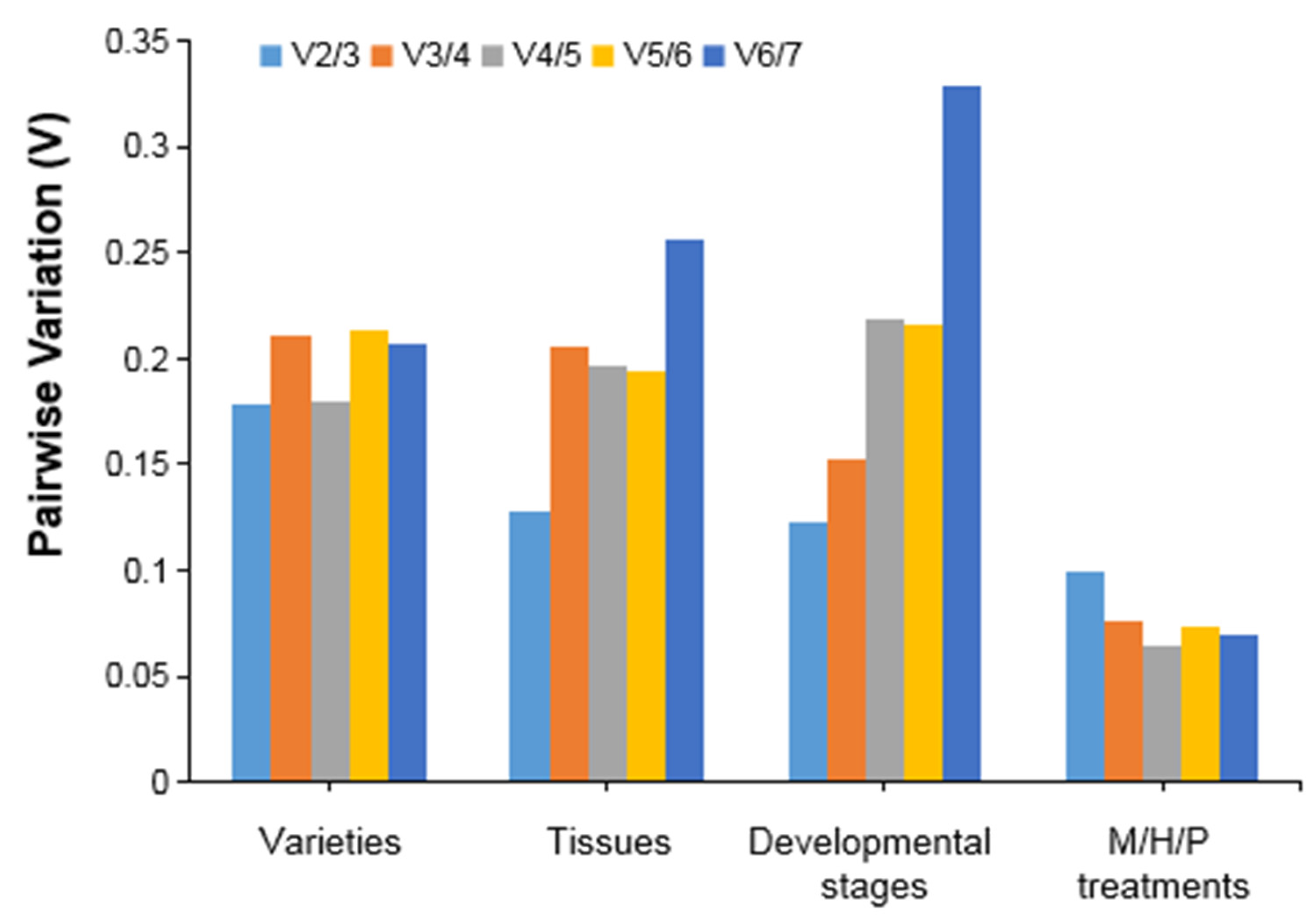
3.3. Expression Stability Analysis of Candidate Reference Genes
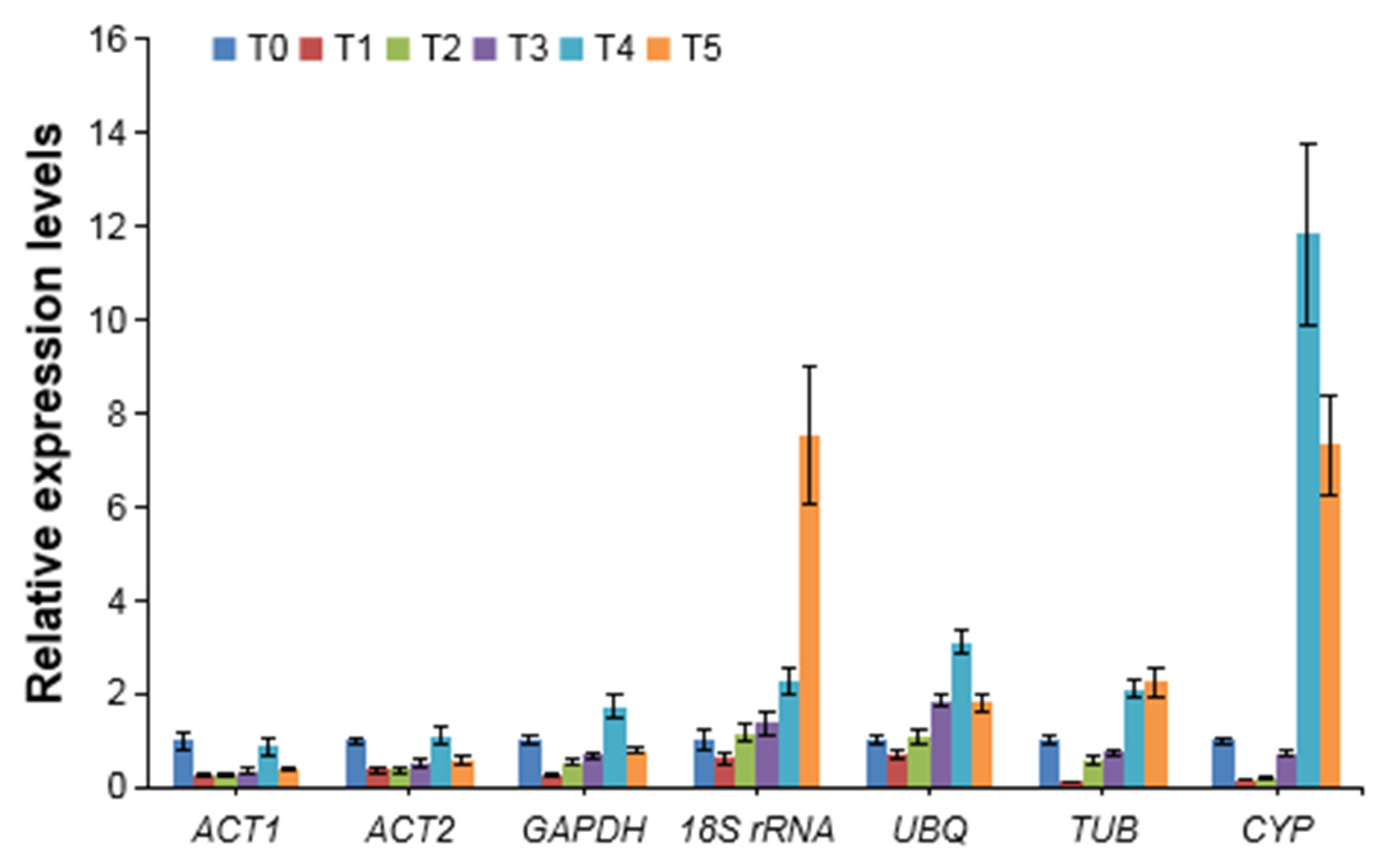
3.4. Validation of Selected Reference Genes by AcPSY Expression Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Higuchi, R.; Fockler, C.; Dollinger, G.; Watson, R.J.B.T. Kinetic PCR Analysis: Real-time Monitoring of DNA Amplification Reactions. Biotechnology 1993, 11, 1026–1030. [Google Scholar] [CrossRef] [PubMed]
- Gachon, C.; Mingam, A.; Charrier, B. Real-time PCR: What relevance to plant studies? J. Exp. Bot. 2004, 55, 1445–1454. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huggett, J.; Dheda, K.; Bustin, S.; Zumla, A.J.G. Real-time RT-PCR normalisation; strategies and considerations. Genes Immun. 2005, 6, 279–284. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Exposito-Rodriguez, M.; Borges, A.A.; Borges-Perez, A.; Perez, J.A. Selection of internal control genes for quantitative real-time RT-PCR studies during tomato development process. BMC Plant Biol. 2008, 8, 131. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bustin, S.A. Quantification of mRNA using real-time reverse transcription PCR (RT-PCR): Trends and problems. J. Mol. Endocrinol. 2002, 29, 23–39. [Google Scholar] [CrossRef]
- Thellin, O.; ElMoualij, B.; Heinen, E.; Zorzi, W. A decade of improvements in quantification of gene expression and internal standard selection. Biotechnol. Adv. 2009, 27, 323–333. [Google Scholar] [CrossRef]
- Radonić, A.; Thulke, S.; Mackay, I.M.; Landt, O.; Siegert, W.; Nitsche, A. Guideline to reference gene selection for quantitative real-time PCR. Biochem. Biophys. Res. Commun. 2004, 313, 856–862. [Google Scholar] [CrossRef]
- Dheda, K.; Huggett, J.F.; Bustin, S.A.; Johnson, M.A.; Rook, G.; Zumla, A.J.B. Validation of housekeeping genes for normalizing RNA expression in real-time PCR. Biotechniques 2004, 37, 112–119. [Google Scholar] [CrossRef] [Green Version]
- Hu, R.; Fan, C.; Li, H.; Zhang, Q.; Fu, Y.-F. Evaluation of putative reference genes for gene expression normalization in soybean by quantitative real-time RT-PCR. BMC Mol. Biol. 2009, 10, 93. [Google Scholar] [CrossRef] [Green Version]
- Dheda, K.; Huggett, J.F.; Chang, J.S.; Kim, L.U.; Bustin, S.A.; Johnson, M.A.; Rook, G.A.W.; Zumla, A. The implications of using an inappropriate reference gene for real-time reverse transcription PCR data normalization. Anal. Biochem. 2005, 344, 141–143. [Google Scholar] [CrossRef]
- Podevin, N.; Krauss, A.; Henry, I.; Swennen, R.; Remy, S. Selection and validation of reference genes for quantitative RT-PCR expression studies of the non-model crop Musa. Mol. Breed. 2012, 30, 1237–1252. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ye, Y.; Lu, Y.; Wang, G.; Liu, Y.; Zhang, Y.; Tang, H. Stable reference gene selection for qRT-PCR normalization in strawberry (Fragaria × ananassa) leaves under different stress and light-quality conditions. Horticulturae 2021, 7, 452. [Google Scholar] [CrossRef]
- Ampomah-Dwamena, C.; Thrimawithana, A.H.; Dejnoprat, S.; Lewis, D.; Espley, R.V.; Allan, A.C.J.N.P. A kiwifruit (Actinidia deliciosa) R2R3-MYB transcription factor modulates chlorophyll and carotenoid accumulation. New Phytol. 2019, 221, 309–325. [Google Scholar] [CrossRef] [Green Version]
- Ferradas, Y.; Rey, L.; Martinez, O.; Rey, M.; Victoria Gonzalez, M. Identification and validation of reference genes for accurate normalization of real-time quantitative PCR data in kiwifruit. Plant Physiol. Biochem. 2016, 102, 27–36. [Google Scholar] [CrossRef] [PubMed]
- Pattyn, F.; Speleman, F.; De Paepe, A.; Vandesompele, J. RTPrimerDB: The real-time PCR primer and probe database. Nucleic Acids Res. 2003, 31, 122–123. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andersen, C.L.; Jensen, J.L.; Orntoft, T.F. Normalization of real-time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004, 64, 5245–5250. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pfaffl, M.W.; Tichopad, A.; Prgomet, C.; Neuvians, T.P. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper--Excel-based tool using pair-wise correlations. Biotechnol. Lett. 2004, 26, 509–515. [Google Scholar] [CrossRef]
- Liang, D.; Shen, Y.; Ni, Z.; Wang, Q.; Lei, Z.; Xu, N.; Deng, Q.; Lin, L.; Wang, J.; Lv, X.; et al. Exogenous melatonin application delays senescence of kiwifruit leaves by regulating the antioxidant capacity and biosynthesis of flavonoids. Front. Plant Sci. 2018, 9, 426. [Google Scholar] [CrossRef]
- Khanlou, K.M.; Van Bockstaele, E. A critique of widely used normalization software tools and an alternative method to identify reliable reference genes in red clover (Trifolium pratense L.). Planta 2012, 236, 1381–1393. [Google Scholar] [CrossRef]
- Taylor, S.; Wakem, M.; Dijkman, G.; Alsarraj, M.; Nguyen, M. A practical approach to RT-qPCR-Publishing data that conform to the MIQE guidelines. Methods 2010, 50, S1–S5. [Google Scholar] [CrossRef]
- Dang, Q.; Sha, H.; Nie, J.; Wang, Y.; Yuan, Y.; Jia, D. An apple (Malus domestica) AP2/ERF transcription factor modulates carotenoid accumulation. Hortic. Res. 2021, 8, 223. [Google Scholar] [CrossRef] [PubMed]
- Ye, X.; Zhang, F.; Tao, Y.; Song, S.; Fang, J. Reference gene selection for quantitative real-time PCR normalization in different cherry genotypes, developmental stages and organs. Sci. Hortic. 2015, 181, 182–188. [Google Scholar] [CrossRef]
- Zhu, L.; Yang, C.; You, Y.; Liang, W.; Wang, N.; Ma, F.; Li, C. Validation of reference genes for qRT-PCR analysis in peel and flesh of six apple cultivars (Malus domestica) at diverse stages of fruit development. Sci. Hortic. 2019, 244, 165–171. [Google Scholar] [CrossRef]
- Kumar, G.; Singh, A.K. Reference gene validation for qRT-PCR based gene expression studies in different developmental stages and under biotic stress in apple. Sci. Hortic. 2015, 197, 597–606. [Google Scholar] [CrossRef]
- Liu, D.; Huang, X.; Lin, Y.; Wang, X.; Yan, Z.; Wang, Q.; Ding, J.; Gu, T.; Li, Y. Identification of reference genes for transcript normalization in various tissue types and seedlings subjected to different abiotic stresses of woodland strawberry Fragaria vesca. Sci. Hortic. 2020, 261, e108840. [Google Scholar] [CrossRef]
- Doddaraju, P.; Kumar, P.; Dashyal, M.S.; Girigowda, M. Identification of suitable reference genes for expression studies in pomegranate under different biotic and abiotic stress conditions. Mol. Biol. Rep. 2021, 48, 3935–3943. [Google Scholar] [CrossRef] [PubMed]
- Jose, S.; Abbey, J.; Jaakola, L.; Percival, D. Selection and validation of reliable reference genes for gene expression studies from Monilinia vaccinii-corymbosi infected wild blueberry phenotypes. Sci. Rep. 2020, 10, 11688. [Google Scholar] [CrossRef]
- Jin, Y.; Liu, F.; Huang, W.; Sun, Q.; Huang, X. Identification of reliable reference genes for qRT-PCR in the ephemeral plant Arabidopsis pumila based on full-length transcriptome data. Sci. Rep. 2019, 9, 84408. [Google Scholar] [CrossRef]
- Liang, C.; Hao, J.; Meng, Y.; Luo, L.; Li, J. Identifying optimal reference genes for the normalization of microRNA expression in cucumber under viral stress. PLoS ONE 2018, 13, e0194436. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Han, J.; Hu, Y.; Yang, J. Selection of reference genes for quantitative real-time PCR during flower development in tree peony (Paeonia suffruticosa Andr.). Front. Plant Sci. 2016, 7, 516. [Google Scholar] [CrossRef] [Green Version]
- Andrade, L.M.; Dos Santos Brito, M.; Favero Peixoto Junior, R.; Marchiori, P.E.R.; Nobile, P.M.; Martins, A.P.B.; Ribeiro, R.V.; Creste, S. Reference genes for normalization of qPCR assays in sugarcane plants under water deficit. Plant Methods 2017, 13, 28. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, C.; Wu, J.; Hua, Q.; Tel-Zur, N.; Xie, F.; Zhang, Z.; Chen, J.; Zhang, R.; Hu, G.; Zhao, J.; et al. Identification of reliable reference genes for quantitative real-time PCR normalization in pitaya. Plant Methods 2019, 15, 70. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, H.; Hu, B.; Zhao, L.; Shi, D.; She, Z.; Huang, X.; Priyadarshani, S.V.G.N.; Niu, X.; Qin, Y. Differential expression analysis of reference genes in pineapple (Ananas comosus L.) during reproductive development and response to abiotic stress, hormonal stimuli. Trop. Plant Biol. 2019, 12, 67–77. [Google Scholar] [CrossRef]
- You, S.; Cao, K.; Chen, C.; Li, Y.; Wu, J.; Zhu, G.; Fang, W.; Wang, X.; Wang, L. Selection and validation reference genes for qRT-PCR normalization in different cultivars during fruit ripening and softening of peach (Prunus persica). Sci. Rep. 2021, 11, 7302. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Huang, J.; Li, X. Identification of appropriate reference genes for RT-qPCR analysis in Ziziphus jujuba Mill. Sci. Hortic. 2015, 197, 166–169. [Google Scholar] [CrossRef]
- Chen, L.; Zhong, H.; Kuang, J.; Li, J.; Lu, W.; Chen, J. Validation of reference genes for RT-qPCR studies of gene expression in banana fruit under different experimental conditions. Planta 2011, 234, 377–390. [Google Scholar] [CrossRef] [PubMed]
- Wan, H.; Zhao, Z.; Qian, C.; Sui, Y.; Malik, A.A.; Chen, J. Selection of appropriate reference genes for gene expression studies by quantitative real-time polymerase chain reaction in cucumber. Anal. Biochem. 2010, 399, 257–261. [Google Scholar] [CrossRef]
- Xia, H.; Wang, X.; Zhou, Y.; Su, W.; Jiang, L.; Deng, H.; Li, M.; Zhuang, Q.; Xie, Y.; Liang, D. Biochemical and molecular factors governing flesh-color development in two yellow-fleshed kiwifruit cultivars. Sci. Hortic. 2021, 280, 109929. [Google Scholar] [CrossRef]
- Chen, J.; Zhou, J.; Hong, Y.; Li, Z.; Cheng, X.; Zheng, A.; Zhang, Y.; Song, J.; Xie, G.; Chen, C.; et al. Genome-wide identification of ubiquitin proteasome subunits as superior reference genes for transcript normalization during receptacle development in strawberry cultivars. BMC Genom. 2021, 22, 88. [Google Scholar] [CrossRef]
- Kou, X.; Zhang, L.; Yang, S.; Li, G.; Ye, J. Selection and validation of reference genes for quantitative RT-PCR analysis in peach fruit under different experimental conditions. Sci. Hortic. 2017, 225, 195–203. [Google Scholar] [CrossRef]
- Galimba, K.; Tosetti, R.; Loerich, K.; Michael, L.; Pabhakar, S.; Dove, C.; Dardick, C.; Callahan, A. Identification of early fruit development reference genes in plum. PLoS ONE 2020, 15, e0230920. [Google Scholar] [CrossRef] [PubMed]
| Gene | Primer Sequence (5′→3′) | Tm (°C) |
|---|---|---|
| ACT1 | GCAGGAATCCATGAGACTACC GTCTGCGATACCAGGGAACAT | 58 |
| ACT2 | TGCATGAGCGATCAAGTTTCAAG TGTCCCATGTCTGGTTGATGACT | 57 |
| 18S rRNA | CTGTGAAACTGCGAATGGCTC TTCCAGAAGTCGGGGTTTGT | 56.5 |
| UBQ | CCACCACGGAGACGGAGCAC TGCAGATCTTCGTGAAAACC | 58 |
| GAPDH | ACACTCCATCACTGCGACA CACCTTGCCAACAGCCTTA | 56.5 |
| TUB | TGAGCACTAAAGAGGTGGATGA TGGGATGTCACACACACTGG | 56.5 |
| CYP | TGATGGCACTGGAGGAGAATC ACTGAGACCCGTTTGTGTTAGG | 58 |
| Gene | R2 | Amplification Efficiency (%) |
|---|---|---|
| ACT1 | 0.999 | 94.57 |
| ACT2 | 0.999 | 91.01 |
| GAPDH | 0.999 | 92.21 |
| 18S rRNA | 0.998 | 87.38 |
| UBQ | 0.989 | 104.07 |
| TUB | 0.989 | 106.91 |
| CYP | 0.981 | 103.66 |
| Ranking | Varieties | Tissues | Developmental Stages | M/H/P Treatments | ||||
|---|---|---|---|---|---|---|---|---|
| Gene | Stability | Gene | Stability | Gene | Stability | Gene | Stability | |
| 1 | GAPDH | 0.377 | ACT1 | 0.407 | ACT2 | 0.322 | GAPDH | 0.242 |
| 2 | 18S rRNA | 0.377 | GAPDH | 0.407 | GAPDH | 0.322 | UBQ | 0.242 |
| 3 | CYP | 0.498 | UBQ | 0.424 | ACT1 | 0.372 | ACT1 | 0.294 |
| 4 | ACT1 | 0.693 | ACT2 | 0.638 | UBQ | 0.503 | TUB | 0.318 |
| 5 | ACT2 | 0.810 | 18S rRNA | 0.804 | 18S rRNA | 0.752 | ACT2 | 0.341 |
| 6 | TUB | 0.987 | TUB | 0.951 | TUB | 0.966 | CYP | 0.390 |
| 7 | UBQ | 1.141 | CYP | 1.221 | CYP | 1.366 | 18S rRNA | 0.432 |
| Ranking | Varieties | Tissues | Developmental Stages | M/H/P Treatments | ||||
|---|---|---|---|---|---|---|---|---|
| Gene | Stability | Gene | Stability | Gene | Stability | Gene | Stability | |
| 1 | GAPDH | 0.110 | GAPDH | 0.240 | GAPDH | 0.490 | ACT1 | 0.120 |
| 2 | 18S rRNA | 0.360 | UBQ | 0.270 | ACT1 | 0.650 | TUB | 0.210 |
| 3 | CYP | 0.480 | ACT1 | 0.340 | UBQ | 0.660 | UBQ | 0.230 |
| 4 | ACT1 | 0.530 | ACT2 | 0.640 | ACT2 | 0.800 | GAPDH | 0.230 |
| 5 | ACT2 | 0.810 | 18S rRNA | 0.950 | TUB | 0.830 | ACT2 | 0.320 |
| 6 | UBQ | 1.410 | TUB | 1.240 | 18S rRNA | 0.930 | CYP | 0.460 |
| 7 | TUB | 1.440 | CYP | 1.770 | CYP | 2.280 | 18S rRNA | 0.470 |
| Ranking | Varieties | Tissues | Developmental Stages | M/H/P Treatments | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene | SD | CV | Gene | SD | CV | Gene | SD | CV | Gene | SD | CV | |
| 1 | CYP | 0.427 | 1.414 | ACT2 | 0.422 | 1.645 | ACT2 | 0.253 | 0.996 | ACT1 | 0.184 | 0.759 |
| 2 | GAPDH | 0.535 | 2.503 | UBQ | 0.614 | 3.025 | ACT1 | 0.259 | 1.260 | 18S rRNA | 0.202 | 1.962 |
| 3 | 18S rRNA | 0.683 | 6.871 | GAPDH | 0.719 | 3.426 | GAPDH | 0.459 | 2.248 | UBQ | 0.259 | 1.128 |
| 4 | ACT1 | 0.758 | 3.541 | 18S rRNA | 0.867 | 8.504 | UBQ | 0.647 | 3.223 | GAPDH | 0.323 | 1.349 |
| 5 | ACT2 | 0.773 | 2.990 | ACT1 | 0.955 | 4.482 | 18S rRNA | 0.899 | 8.536 | TUB | 0.335 | 1.173 |
| 6 | UBQ | 1.470 | 7.057 | TUB | 1.296 | 4.970 | TUB | 1.168 | 4.494 | ACT2 | 0.411 | 1.522 |
| 7 | TUB | 1.171 | 4.520 | CYP | 2.002 | 6.496 | CYP | 2.375 | 7.603 | CYP | 0.458 | 1.328 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, Y.; Xia, H.; Liu, X.; Lin, Z.; Guo, Y.; Deng, H.; Wang, J.; Lin, L.; Deng, Q.; Lv, X.; et al. Identification of Suitable Reference Genes for qRT-PCR Normalization in Kiwifruit. Horticulturae 2022, 8, 170. https://doi.org/10.3390/horticulturae8020170
Zhou Y, Xia H, Liu X, Lin Z, Guo Y, Deng H, Wang J, Lin L, Deng Q, Lv X, et al. Identification of Suitable Reference Genes for qRT-PCR Normalization in Kiwifruit. Horticulturae. 2022; 8(2):170. https://doi.org/10.3390/horticulturae8020170
Chicago/Turabian StyleZhou, Yuanjie, Hui Xia, Xinling Liu, Zhiyi Lin, Yuqi Guo, Honghong Deng, Jin Wang, Lijin Lin, Qunxian Deng, Xiulan Lv, and et al. 2022. "Identification of Suitable Reference Genes for qRT-PCR Normalization in Kiwifruit" Horticulturae 8, no. 2: 170. https://doi.org/10.3390/horticulturae8020170
APA StyleZhou, Y., Xia, H., Liu, X., Lin, Z., Guo, Y., Deng, H., Wang, J., Lin, L., Deng, Q., Lv, X., Xu, K., & Liang, D. (2022). Identification of Suitable Reference Genes for qRT-PCR Normalization in Kiwifruit. Horticulturae, 8(2), 170. https://doi.org/10.3390/horticulturae8020170






