Abstract
Tomato yellow leaf curl virus (TYLCV) is a serious pathogen transmitted by the whitefly (Bemisia tabaci). Due to the quick spread of the virus, which is assisted by its vector, tomato yield and quality have suffered a crushing blow. Resistance to TYLCV has been intensively investigated in transmission, yet the mechanism of anti-TYLCV remains elusive. Herein, we conducted transcriptome profiling with a TYLCV-resistant cultivar (CLN2777A) and a susceptible line (Moneymaker) to identify the potential mechanism of resistance to TYLCV. Compared to the susceptible line, CLN2777A maintained a lower level of lipid peroxidation (LPO) after TYLCV infection. Through RNA-seq, over 1000 differentially expressed genes related to the metabolic process, cellular process, response to stimulus, biological regulation, and signaling were identified, indicating that the defense response was activated after the virus attack. Further analysis showed that TYLCV infection could induce the expression of the genes involved in salicylic and jasmonic acid biosynthesis and the signal transduction of phytohormones, which illustrated that phytohormones were essential for tomatoes to defend against TYLCV. These findings provide greater insight into the effective source of resistance for TYLCV control, indicating a potential molecular tool for the design of TYLCV-resistant tomatoes.
1. Introduction
Tomato yellow leaf curl virus (TYLCV), belonging to the genus Begomovirus (family Geminiviridae), is a devastating pathogen and can lead to great crop loss, especially in tropical and subtropical regions with high heat and humidity [1]. The migration of infected whitefly (Bemisia tabaci) is considered the main route of transmission. However, planting or grafting seedlings with TYLCV could also spread the virus [2,3,4,5]. In addition to vegetables such as tomatoes, peppers, and cucumbers, TYLCV can infect various crops, including tobacco, cassava, and cotton, and can cause great damage to their production [6]. As the main host of TYLCV, the typical symptom in tomatoes induced by the virus is arrested development with upward curling of the leaves and chlorosis, which results in a reduced yield and poor quality [7,8]. It is hard to completely eliminate TYLCV or control an outbreak of it as its vector, Bemisia tabaci, has a strong reproduction capability and migration rate. Hence, characterization of the TYLCV resistance gene to select and breed cultivars against the virus may be an ideal strategy to defend against the pathogen [2,3,9,10].
Previous studies have identified six TYLCV-resistant genes designated Ty-1 [11], Ty-2 [12,13], Ty-3 [14], Ty-4 [15], ty-5 [16,17], and Ty-6 [18,19]. Ty-1/Ty-3 encoding the RNA-dependent RNA polymerases (RDRs) induce TYLCV resistance through RNA interference [20]. Compared to susceptible cultivars, tomatoes harboring the Ty-1 gene generate more siRNA, and the genome of the virus shows the hypermethylation of cytosine in vivo, repressing the virus replication [21]. However, with the promotion of tomatoes carrying Ty-1/Ty-3, the recombinant TYLCV occurs in this cultivar. Thus, new variants of Ty-1 against the virus have been explored [22,23]. Through mapping-based cloning, TYNBS1, a nucleotide-binding domain and leucine-rich repeat-containing (NB-LRR) gene, is proved to contribute to Ty-2’s Begomovirus resistance; however, the mechanism is still elusive [24]. Moreover, the locus of the minor resistance gene Ty-4, the recessive gene ty-5, and the incomplete dominant gene Ty-6 remains unclear [15,16,17,18,19]. With the development of sequencing technology, such as whole genome sequencing [25], as well as RNA-seq, more genes have been isolated for TYLCV resistance [26,27,28]. Comparative analysis between the resistant line CLN2777A (R) and the susceptible line TMXA48-4-0 (S) has indicated that the WRKY transcription factor, protein kinase, and protein receptor kinase can respond to virus infection [29]. Apart from this, strand-specific RNA sequencing (ssRNA-seq) has shown that lncRNAs also play a potential role in TYLCV resistance in tomatoes [30]. Proteomics and metabolomics analysis has revealed that the expression level of antioxidative proteins maintains a higher level in TYLCV-resistant varieties compared to the sensitive variety invaded by the virus [31].
Plant hormones are a key part of plant–pathogen interactions. Exogenous application of salicylic acid (SA) can inhibit virus accumulation and improve the resistance of susceptible cultivars. Furthermore, the blocking of SA signaling results in more detectable TYLCV in tomatoes [32,33]. Jasmonic acid (JA) can regulate the biosynthesis of phytoalexin to avoid the virus transmitting from the vector to the plant [34]. The virulence factor βC1 of TYLCV directly interacts with MYC2, the fundamental transcription factor in JA signaling, and compromises its transcription activation, resulting in the suppression of terpene biosynthesis and subverting plant resistance [35]. To date, researchers have mainly focused on gene expression and function with samples after a long period of infection in TYLCV and tomato interaction [36,37,38]. To investigate the differential expression genes induced by TYLCV in a short time, we carried out RNA-seq with Moneymaker (MK; a susceptible line) and CLN2777A (T2; a resistant line) infecting them with virus after 24 and 48 h. Meanwhile, the activity of reactive oxygen species (ROS)-related enzymes and plant hormones was detected to uncover the mechanism of TYLCV resistance in tomatoes.
2. Materials and Methods
2.1. Plant Materials and TYLCV Inoculation
Moneymaker (MK; susceptible to TYLCV infection) and CLN2777A (T2; harboring the Ty-2 gene, resistant to TYLCV infection) were used for TYLCV inoculation. The germplasms of MK and T2 were obtained from Jiangsu Academy of Agricultural Sciences. The seeds were germinated at 25 °C, and then put in plastic boxes containing nutritional soil (soil/perlite/vermiculite = 2:1:1, v/v/v). Seedlings of the two-leaf stage were transferred into plastic pots. The temperature and relative humidity for all of the experiments were set at 25 °C and 70%–80% with a 16/8 h day/night photoperiod. For TYLCV inoculation, we used an Agrobacterium tumefaciens vector constructed by Zhang et al. [39], which contained the effective infection fragment of TYLCV. The method of inoculation of TYLCV was performed as described by Zhang et al. [39] with slight modifications. The first leaf of four-leaf stage plants was injected with 1 mL of A. tumefaciens (OD600 = 0.6) harboring TYLCV or not (control (CK)). After inoculating with TYLCV for 24 and 48 h, the leaves of plants were harvested for ROS-related enzyme activity determination, RNA sequencing, phytohormones assay, and gene expression detection. Thirty seedlings were planted for each treatment of each tomato line, and the leaves of six random plants were collected at designated time points; then, all samples were frozen in liquid nitrogen and immediately stored at −80 °C.
2.2. Growth Parameter Measurement
After 26 d of TYLCV inoculation, plant height and fresh and dry weight (shoots and roots) were measured. The fresh weight was measured on a scale (±0.01 g) with an ambient temperature of 25 °C. Then, the samples were dried at 80 °C in an oven until the weight no longer changed, and the dry weight was measured on a scale (±0.01 g). For each measurement, one biological replicate containing six seedlings and three biological replicates was performed.
2.3. Enzyme Activity Assays
For detection of the enzyme activities, tomato leaves (0.2 g) were ground to homogenate in 1 mL of ice-cold phosphate buffer (pH = 7.8, 50 mM), and then centrifuged at 12,000× g for 20 min at 4 °C. SOD and POD activity was measured according to Macadam [40]. The activity of ascorbate oxidase (AAO) was detected by the method from Nakano [41]. Three biological replicates were performed for each measurement.
2.4. Library Construction and Illumina Sequencing
For library preparation, RNA of leaf samples collected at 0, 24, and 48 h post-TYLCV inoculation (hpi) were isolated, respectively, and the preparation strategy can be found in previous study [42]. First, total RNA was extracted using a Trizol reagent kit (Invitrogen, Carlsbad, CA, USA), and the extracted RNA concentration and quality were assessed by Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA, USA). Next, the eukaryotic mRNA was enriched by Oligo (dT)-attached magnetic beads, while the ribosomal RNA was removed by Ribo-zeroTM rRNA removal kit (Epicentre, Madison, WI, USA). Then the enriched mRNA was fragmented into short fragments using fragmentation buffer and reverse-transcribed into double-strand cDNA with random primers. The libraries were constructed using TruSeq RNA sample Preparation kit (Illumina, San Diego, CA, USA) following the manufacturer’s recommendations, and then sequenced on Illumina HiSeq 2500 platform with 2 × 150 bp PE mode by Gene Denovo Biotechnology Co. (Guangzhou, China).
2.5. Read Mapping, Statistical Analysis, and Functional Annotation
Useless sequences such as sequencing adapters, rRNA reads, and short fragment reads were filtered out. Then, the clean reads were mapped to the Solanum lycopersicum V4.0 reference genome (https://solgenomics.net/organism/Solanum_lycopersicoides/genome) on 22 April 2021 by HISAT2 software [43] (version 2.2.4, Johns Hopkins University, Baltimore, MD, USA, http://daehwankimlab.github.io/hisat2/, 22 April 2021) with “-rna-strandness RF” and other parameters set as a default. After this, the FPKM (fragments per kilobase of transcript per million mapped reads) values for known gene models were identified by StringTie software (version 1.3.1, Johns Hopkins University, Baltimore, MD, USA, http://ccb.jhu.edu/software/stringtie/index.shtml on 22 April 2021) according to Pertea et al. [44]. The threshold of the p-value was determined using the false discovery rate (FDR). In this study, differentially expressed genes (DEGs) were identified using edgeR [45], with the rules FDR ≤ 0.05 and fold change ≥2. The GO annotation of DEGs was analyzed by Blast2go software (http://www.blast2go.org) on 26 April 2021. KEGG (Kyoto Encyclopedia of Genes and Genomes) pathway enrichment analysis was performed by a KEGG enrichment tool of the platform OmicShare (http://www.omicshare.com/tools/Home/Soft/pathwaygsea) on 28 April 2021.
2.6. Phytohormone Content Assays
The phytohormone contents were detected as previously described [46], and the detection was performed at Genepioneer Biotechnologies Co. Ltd, Nanjing., China. Briefly, 0.2 g of the leaf sample was ground and extracted with a mixture buffer (2-propanol/H2O/concentrated HCl, 2:1:0.002, v/v/v). The sample solution was loaded into the reverse-phase C18 Gemini HPLC column for HPLC–ESI–MS/MS analysis.
2.7. Validation of DEGs with Quantitative RT-PCR
Ten genes involved in the TYLCV resistance of tomatoes were selected to conduct quantitative RT-PCR validation. Primers were designed and their specificity for quantitative RT-PCR were evaluated using NCBI primer-blast (Table S1). First-strand cDNA synthesis, PCR amplifications, and calculations of the relative gene expression level were performed as previously described [47]. The gene α-Tubulin (Solyc04g077020.2) was used as the internal reference. Three independent biological replicates and two technical repeats per biological replicate were performed for each chosen gene.
3. Results
3.1. TYLCV Leads to Tomato Development Arrest
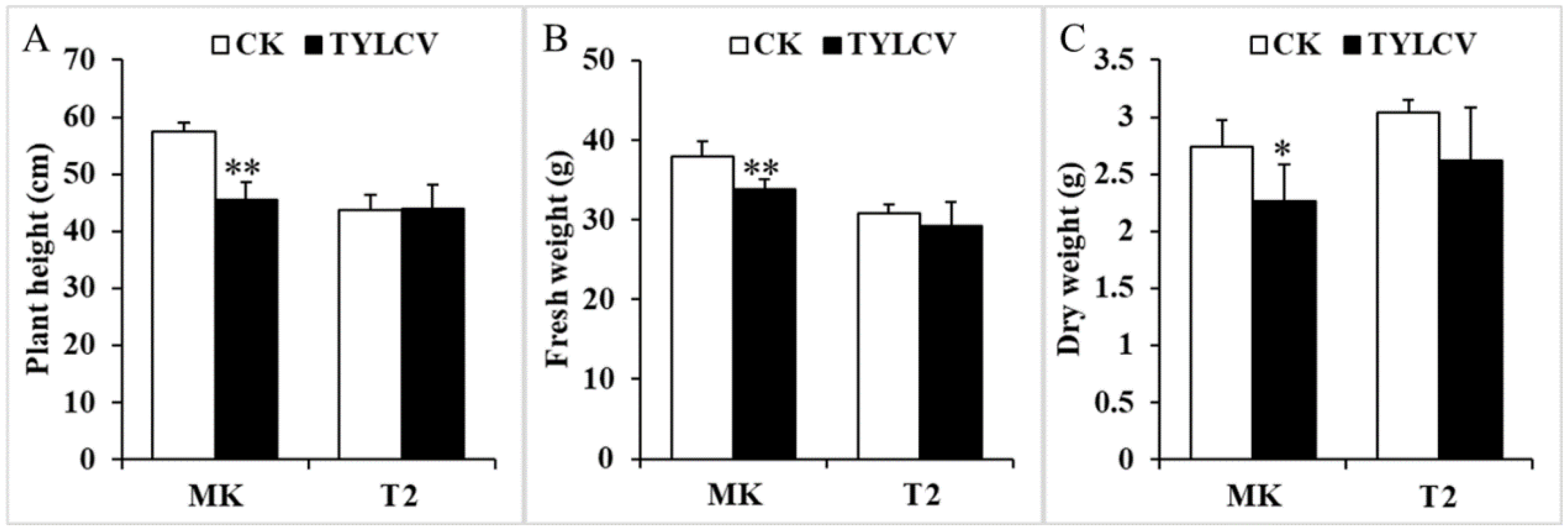
To further investigate the influence of the virus on tomato growth, the TYLCV-susceptible line MK and the resistant line T2 were infected with the virus. The biomass, including plant weight and fresh and dry weight, was measured after 26 d of infection. The plant height of MK was reduced by 20%, while no significant difference was noticed in T2 invaded by TYLCV (Figure 1A). Similarly, both the fresh and dry weights of MK infected by TYLCV were significantly decreased (Figure 1B,C). In contrast, the decrease in the biomass of T2 showed no significant difference (Figure 1B,C). These data suggest that TYLCV can repress tomato growth and reduce biomass.

Figure 1.
Plant height and biomass differences between the MK (S) and T2 (R) genotypes under TYLCV treatment. (A–C) the plant height (A), fresh weight (B), and dry weight (C) of MK and T2 infected by TYLCV for 26 d. The data represent the mean ± SD (n = 3). Data were analyzed by an ANOVA in the SPSS software. * (p < 0.05) and ** (p < 0.01) indicate statistically significant differences.
3.2. TYLCV Leads to Lipid Peroxidation Stress at the Initial Time of Infection
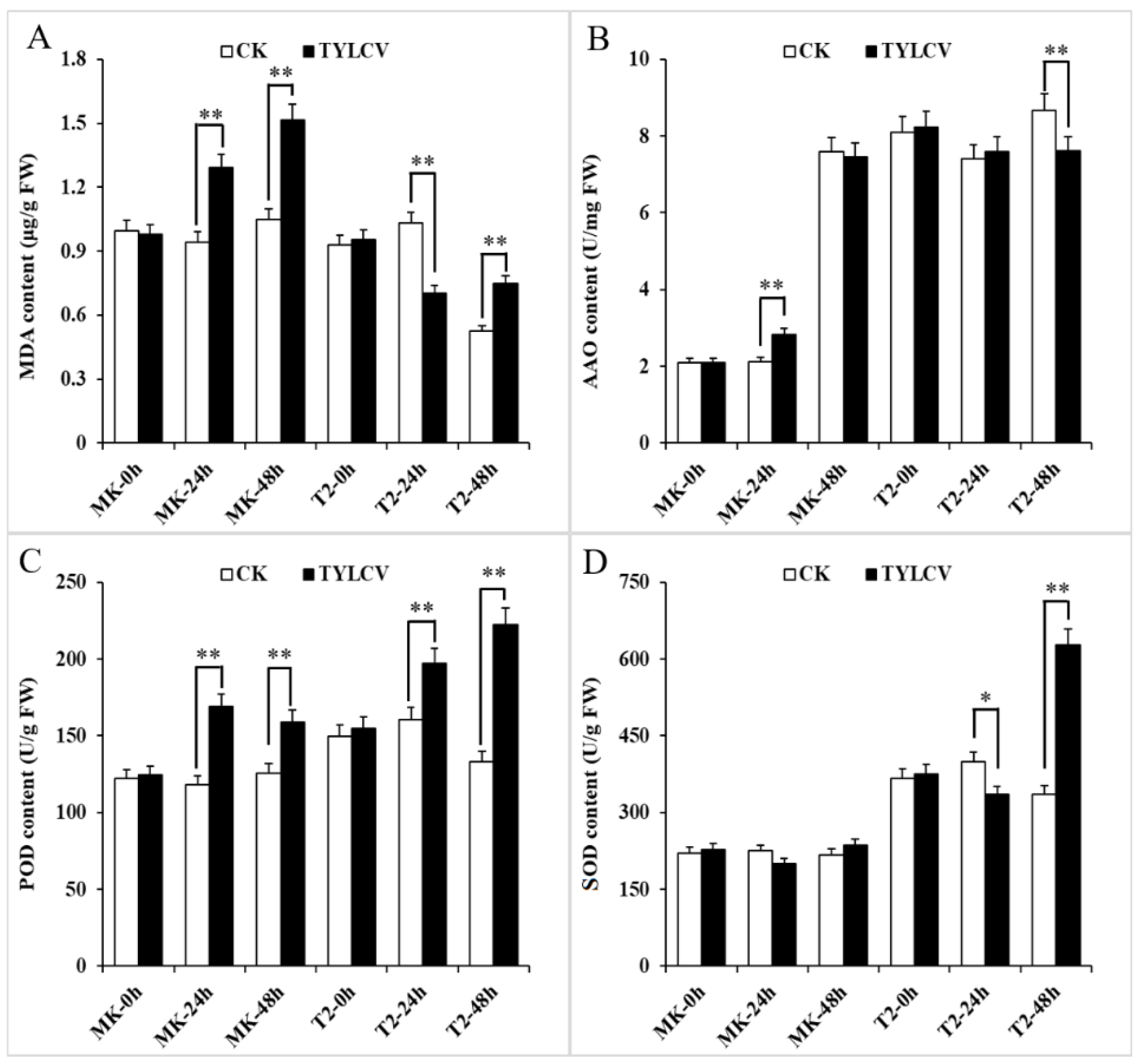
The accumulation of ROS is a stress reaction of a plant to disease. The amount of ROS could be elevated in tomatoes, breaking the state of its production and degradation balance under virus treatment for 2–14 d [33]. However, few explorations have focused on the initial time of symptom appearance in plants attacked by TYLCV. We checked the content of MDA along with the ROS-scavenging enzymes in response to TYLCV infection at early times (within 48 h). The results showed that more MDA accumulated from 24 h in the TYLCV-sensitive cultivar MK (S), while the process was delayed in T2 (R) after virus infection (Figure 2A). Furthermore, the MDA content in TYLCV-attacked T2 maintained half of that in MK, which suggested that the virus-resistant lines could alleviate TYLCV-induced lipid peroxidation. All of the ROS-scavenging enzymes had higher activity in T2 than those in MK with the virus treatment at the same time point. Remarkably, the activity of POD and SOD of T2 at 48 h increased by 60% and 90%, respectively, which was 30% and 150% higher than that in MK (Figure 2B–D).
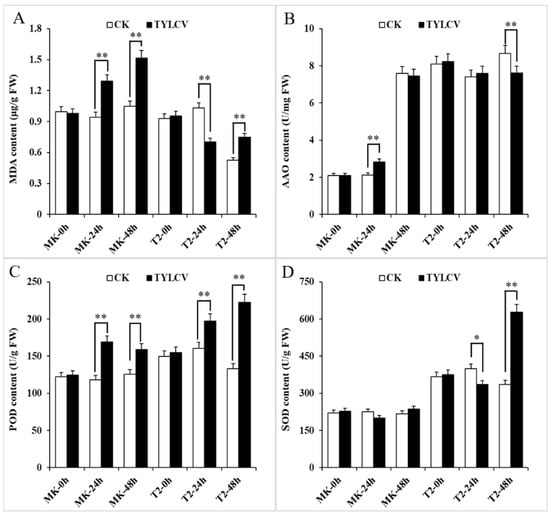
Figure 2.
The MDA content and activity of reactive oxygen species (ROS)-scavenging enzymes in TYLCV-infected plant leaves. (A) The MDA content in treated plants. (B–D) The activities of ascorbate oxidase (AAO), peroxidase (POD), and superoxide dismutase (SOD) in treated plants, respectively. The data represent the mean ± SD (n = 3). Data were analyzed by an ANOVA in the SPSS software. * (p < 0.05) and ** (p < 0.01) indicate statistically significant differences.
3.3. Comparative Transcriptome of Tomatoes in Response to TYLCV Infection
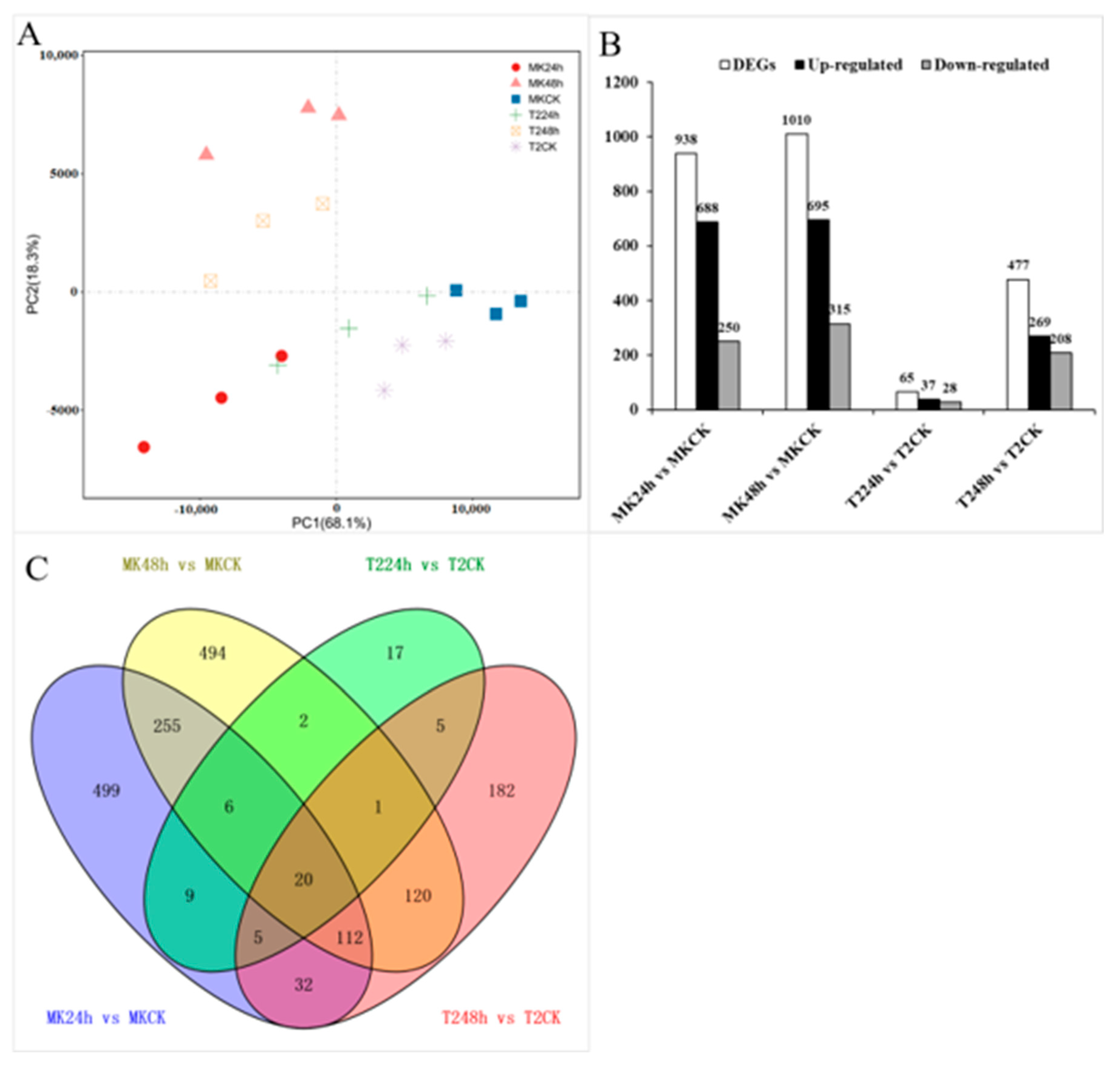
To investigate the gene expression pattern at the initial time of virus inoculation, the leaves of MK and T2, before (0 h) and after (24 and 48 h) TYLCV inoculation, were extracted for RNA sequencing. The raw data are summarized in Table S2 and the gene expression level (estimated by FPKM values) is shown in Figure S1. To assess the data quality, Pearson’s correlation coefficient between biological replicates was observed (R2 > 0.91 for all samples (Figure S2), which indicated that the biological replicates were reliable. Additionally, PCA was performed (Figure 3A) and the results revealed that the 18 samples could be assigned to different groups, with PC1 and PC2 capturing most of the variance in the data. In addition, the biological replicates were projected closely in the space, which indicated a good correlation between replicates (Figure 3A).

Figure 3.
Transcriptomes of plants under TYLCV infection. (A) Principal component analysis (PCA) of the RNA sequencing data. The 18 samples (three biological replicates of CK (0), 24, and 48 h) were projected on the first and second principal components (PCs). PC1 and PC2 account for > 86% of the total variance in the data. (B) Numbers of differentially expressed genes (DEGs). (C) Venn diagram depicting the number of all DEGs expressed in the leaves of MK and T2 under TYLCV infection and control conditions.
The genes with the parameter of false discovery rate (FDR) below 0.05 and absolute fold change ≥ 2 were considered DEGs. To determine the gene expression changes resulting from TYLCV infection, the DEGs between the control samples (CK; 0 h) and the infected samples (24 and 48 h) were identified according to the criterion. A total of 938, 1010, 65, and 477 DEGs were found after 24/48 h of TYLCV infection in comparison to CK in MK and T2, respectively. Of these DEGs, 688, 695, 37, and 269 genes were up-regulated and 250, 315, 28, and 208 genes were down-regulated, respectively (Figure 3B and Table S3). Interestingly, the up-regulated DEGs occupied 70%, which suggested that a batch of TYLCV-responsive genes were induced in the initial stage. Moreover, there were 20 DEGs common to two infection stages in MK and T2, compared to the uninfected at 0 h (Figure 3C), which indicated that these 20 DEGs might be the hub genes in response to TYLCV infection.
3.4. Functional Annotation of DEGs
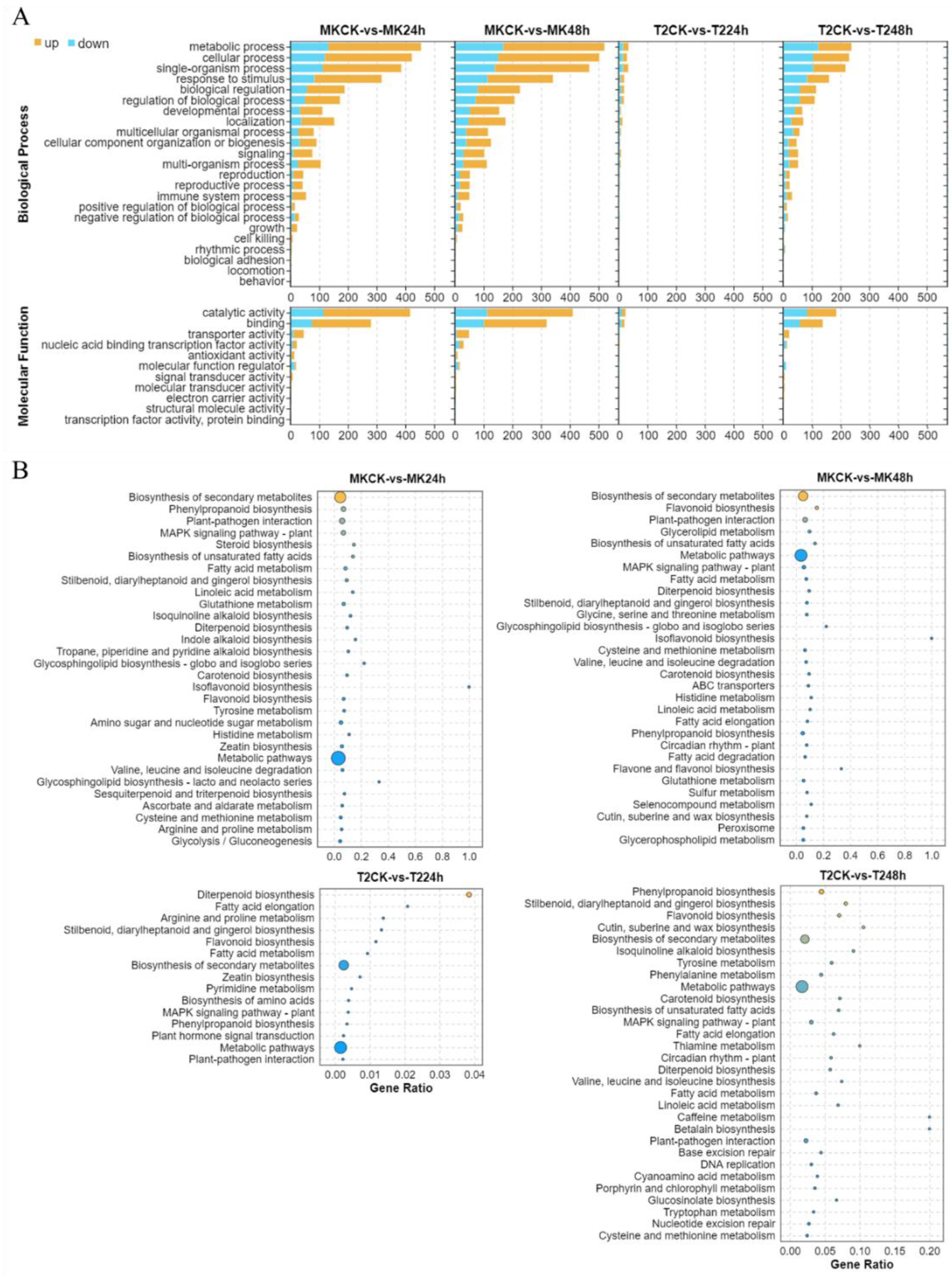
To gain an insight into the functions of the DEGs, GO and KEGG pathway enrichment analysis was applied. The graphical results revealed that a large number of DEGs were enriched in GO terms, such as the metabolic process, cellular process, response to stimulus, biological regulation, and signaling (Figure 4). Notably, the number of DEGs involved in all the categories was much greater in the S line MK than that in the R line T2, which indicated that TYLCV might severely damage the biological processes in the S line. In addition, a few DEGs were found to participate in the immune system process, cell killing, and the rhythmic process. As for the molecular function class, several DEGs were classified into metabolic pathways, biosynthesis of secondary metabolites, plant–pathogen interaction, and MAPK signaling pathway. On top of this, a set of DEGs were involved in the phytohormone signaling pathway, e.g., zeatin biosynthesis (ZT), sesquiterpenoid and triterpenoid biosynthesis (SA), linolenic acid metabolism (JA), carotenoid biosynthesis (ABA), tryptophan metabolism (IAA), and plant hormone signal transduction. Hence, we further explored the DEGs involved in the metabolism of phytohormones and their signal pathways systematically.

Figure 4.
GO and KEGG categories of DEGs in MK and T2 under TYLCV infection. GO terms (A) and KEGG pathways (B) involved in TYLCV resistance. The X-axis in (A) indicates the DEG numbers in each GO term; the X-axis in (B) indicates the percentage of DEG numbers vs. the background gene numbers in each KEGG pathway. Detailed information is shown in Table S4.
3.5. Response of Plant Hormones to TYLCV in Tomatoes
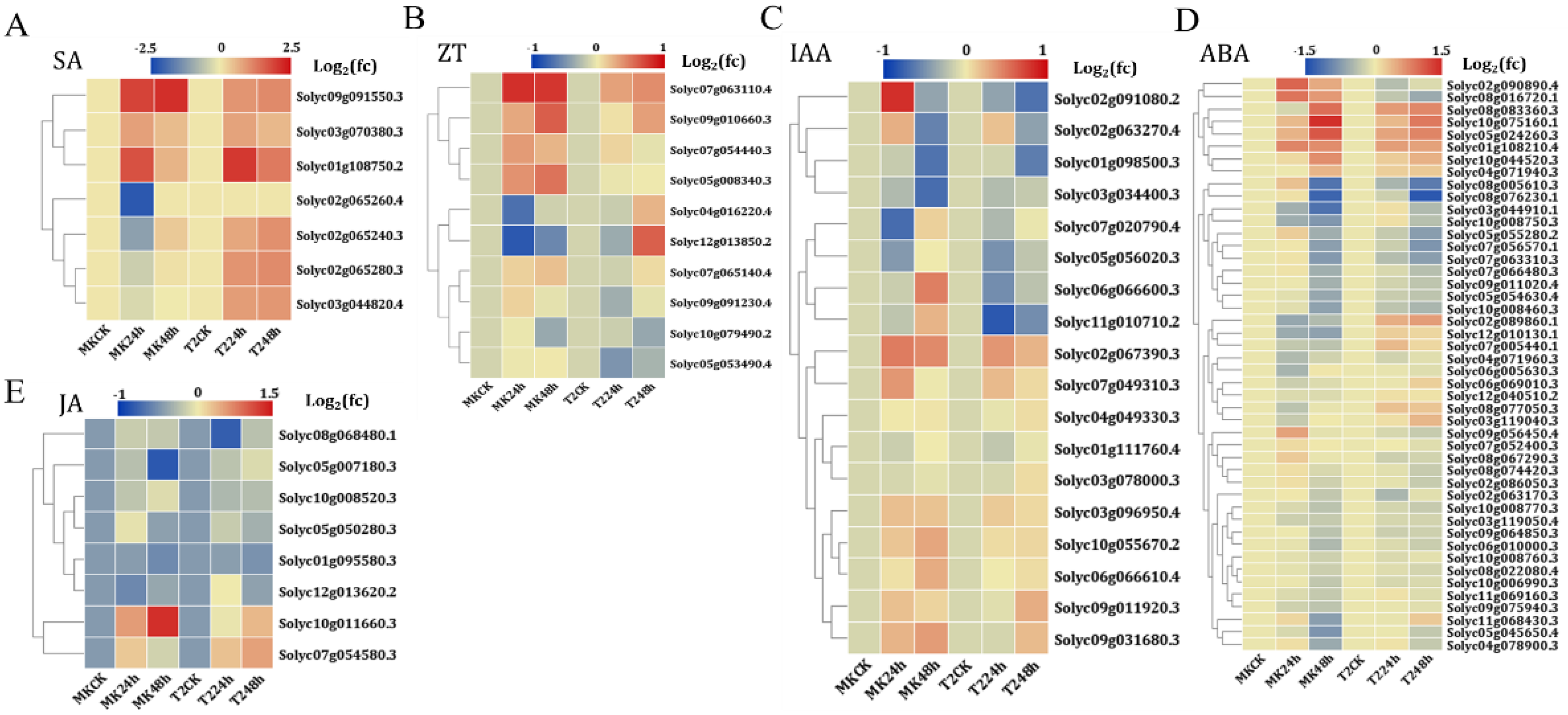
Among the genes involved in the SA biosynthesis and signal pathway, SA carboxyl methyltransferase (SAMT, Solyc09g091550.3) and a set of SA-binding protein 2 homologs (Solyc02g065280.3, Solyc02g065240.3, Solyc09g091550.3, Solyc03g044820.4, Solyc03g070380.3, Solyc02g065260.4, and Solyc01g108750.2) were up-regulated in the R line T2 after 24/48 h of TYLCV infection, while Solyc02g065240.3 and Solyc02g065260.4 showed a down-regulated expression pattern in the S line MK (Figure 5 and Table S5). For the genes involved in the zeatin biosynthetic and signal pathway, beta-glucuronosyltransferase GlcAT14 homologs (Solyc09g010660.3, Solyc07g054440.3, Solyc07g063110.4, and Solyc05g008340.3) were up-regulated after 24/48 h of TYLCV infection in both the S and R lines, whereas Solyc04g016220.4 (zeatin O-xylosyltransferase-like), Solyc10g079490.2, Solyc07g065140.4 (beta-(1,2)-xylosyltransferase), Solyc05g053490.4, Solyc09g091230.4, and Solyc12g013850.2 showed the opposite expression pattern in the S and R lines in response to TYLCV infection (Figure 5 and Table S5). In the IAA-mediated signaling pathway, there were nine down-regulated genes and 10 up-regulated genes after TYLCV inoculation (Figure 5 and Table S5). In the case of the JA signaling pathway, six and two genes were down-regulated and up-regulated, respectively. The genes involved in the ABA signaling pathway are detailed in Table S5. Taken together, a KEGG map of all of the plant hormone signal transductions is displayed, in which the DEGs induced by TYLCV infection are highlighted (Figure S3). These results indicated that TYLCV universally reprogramed the expression of the genes related to the biosynthesis and signaling transduction of plant hormones.

Figure 5.
Plant hormone-related genes were differently expressed in TYLCV infection. Gene expression patterns by heatmap representations in the SA (A), ZT (B), IAA (C), ABA (D), JA (E) biosynthetic, and signal pathway. fc is the fold change of gene expression after 24/48 h of TYLCV infection.
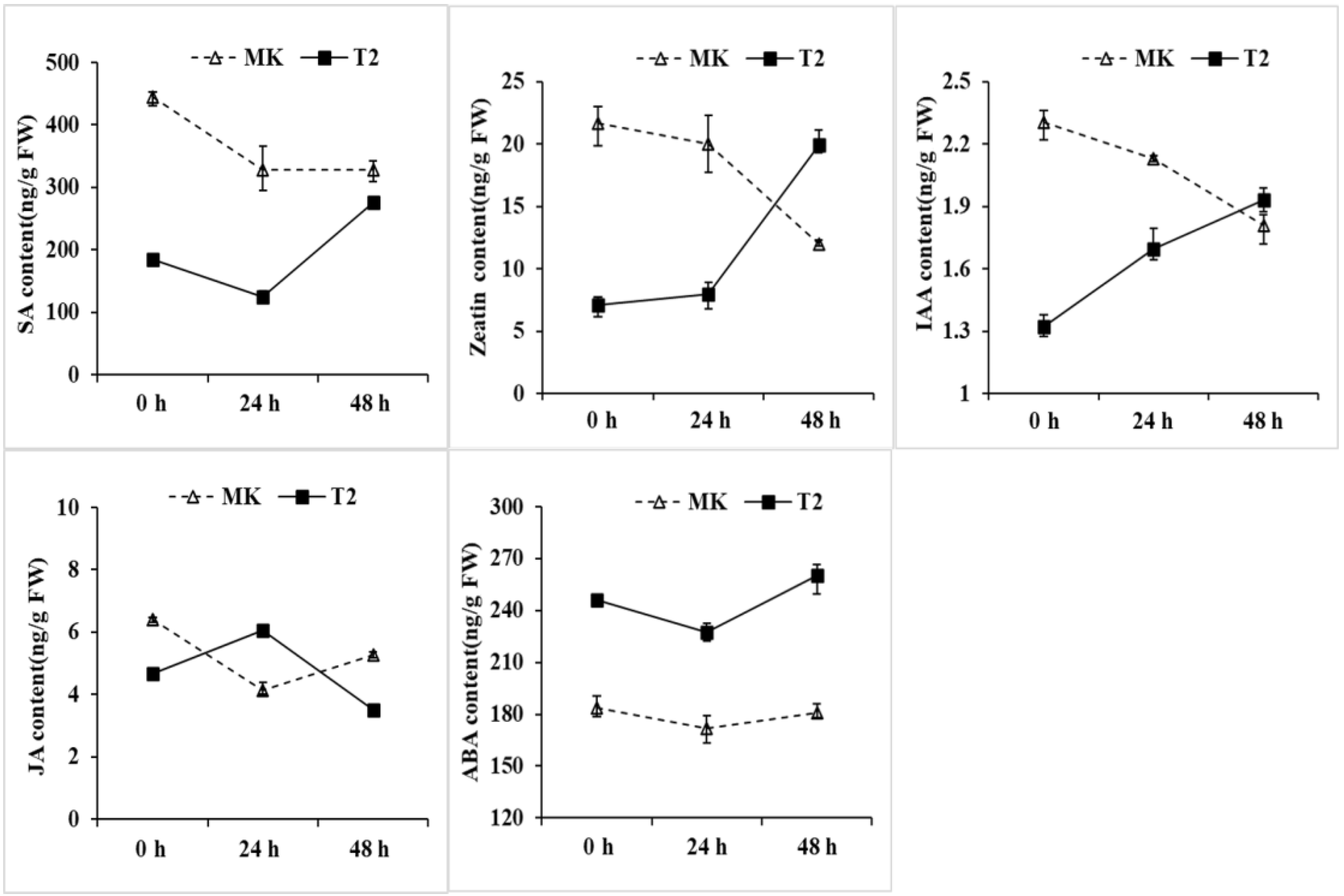
To further explore the roles of plant hormones in the crosstalk between TYLCV and tomatoes, the contents of several plant hormones were measured after 24/48 h of TYLCV infection (Figure 6). The contents of zeatin and indole‐3‐acetic acid (IAA) in TYLCV‐resistant cultivar (T2) were sharply increased after 48 h of TYLCV inoculation, while it is opposite in susceptible line (MK) (Figure 6). SA is essential to plants for acquiring resistance to many pathogens, whose content was up-regulated in the R line T2 after 48 h of TYLCV infection. However, SA was reduced in the S line MK (Figure 6). Interestingly, in the first 48 h of infection of TYLCV, the contents of JA and ABA were also obviously affected. These results indicate that tomato plants of T2 probably regulate the biosynthesis of SA to defend against pathogen attack, making sure that plants produce more ZT and IAA for growth.
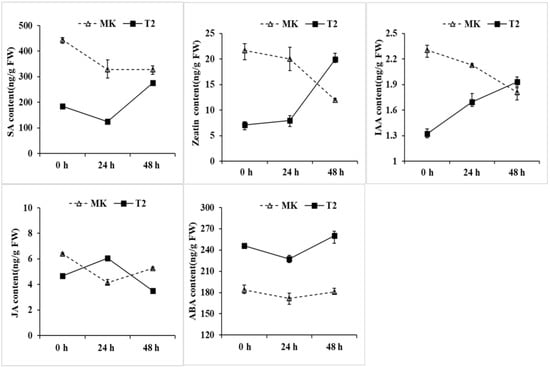
Figure 6.
The contents of SA, ZT, IAA, JA, and ABA in the plant leaves after TYLCV infection. The values are the means of three biological replicates ± SD. Vertical bars represent the standard error of the mean of three biological replicates.
3.6. Quantitative RT-PCR Validation of the RNA-Seq Expression
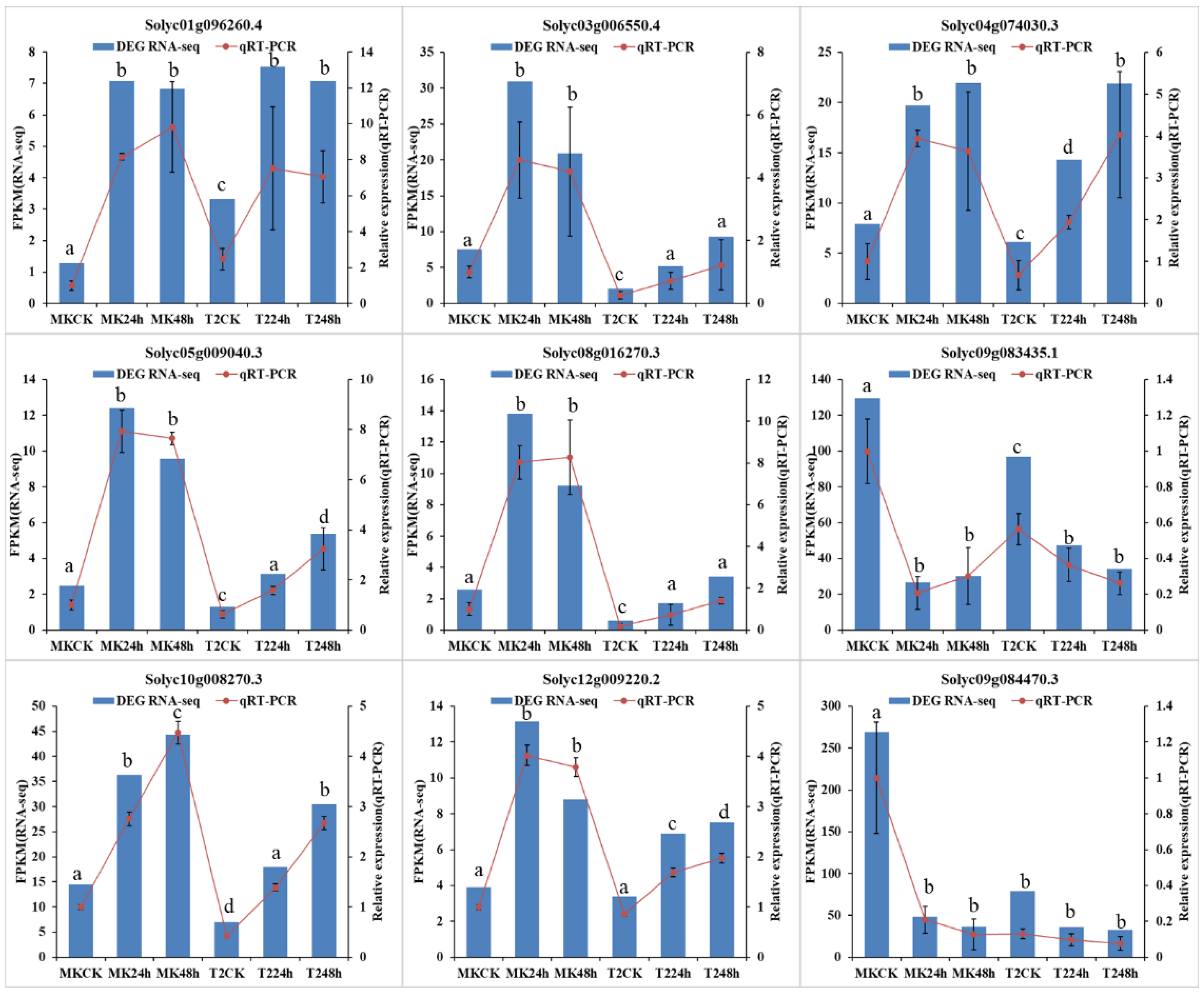
To validate the reliability of the RNA-seq data, we randomly selected nine DEGs to perform qRT-PCR. These genes included jasmonate ZIM-domain protein 1 (Solyc12g009220.2), LRR receptor-like serine/threonine-protein kinase GSO1 isoform X1 (Solyc08g016270.3), and transcription factor bHLH120-like (Solyc10g008270.3) (see Table S1 for more details). The results showed a good correlation between the FPKM values and the qRT-PCR results (Figure 7), which suggested that the gene expression analysis by RNA-seq was reliable.
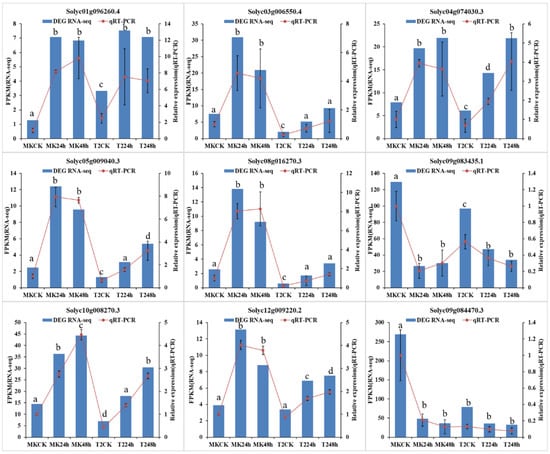
Figure 7.
Verification of nine randomly selected DEGs by qRT-PCR. Comparison of RNA-seq data (blue bar) with qRT-PCR data (red line). The normalized expression level (FPKM) of RNA-seq is indicated on the Y-axis to the left, and the relative qRT-PCR expression level is shown on the Y-axis to the right. α-Tubulin (Solyc04g077020.2) was used as the internal control. Both methods show similar gene expression trends. Three biological replicates were performed. Data were analyzed by an ANOVA in the SPSS software, and a, b, c, and d indicate statistically significant differences (p < 0.05) for the designated time point.
4. Discussion
Over the past decade, a great deal has been learned about viral vector–host interactions. The dramatic rise in the populations of Bemisia tabaci worldwide and the subsequent virus epidemics have led to increased research aimed at controlling the economic losses resulting from TYLCV. To block transmissible whitefly-borne geminiviruses, the host selection behavior of the vector has been studied [5,48]. Moreover, from an ecological perspective, the application of chemical insecticides and the improvement in cultural methods could protect tomatoes from attack by whitefly carrying the TYLCV [7,8,9]. To disturb the virus transferring to plants, the features of crops should be another key point to explore [5,8]. Generally, visual cues, such as plant color, and olfactory cues, such as plant volatiles, are the first barriers to prevent crops from whitefly [5]. Furthermore, the inbred lines or cultivars harboring the TYLCV-resistant locus or genes have been utilized for breeding and production. Among the germplasm, six genes have been identified, including Ty-1 to Ty-6 [11,12,13,14,15,16,17,18,19]. Ty-1 and its allelic gene Ty-3, which have been widely used in tomato breeding for a long time and have been studied several times concerning their siRNA [20], and now some new variants of TYLCV have emerged to break through resistance to this type of gene [23]. The understanding of Ty-4, ty-5, and Ty-6 is still limited. Moreover, Ty-2, which was identified as TYNBS1, plays an increasingly important role in the breeding of resistant varieties, but its specific mechanism of anti-TYLCV remains unclear.
Data from plant physiology and molecular analyses demonstrated that TYLCV can repress tomato growth from the initial time of TYLCV inoculation through modulating the ROS balance and plant hormone biosynthesis and signaling. As reported, TYLCV causes great damage to susceptible cultivars [1], but the process could be delayed when tomatoes possess the Ty-2 gene (Figure 1). Thus, Ty-2 has been widely introduced to tomato cultivars to combat TYLCV. Through mapping-based cloning, Ty-2 was identified as the NB-LRR family gene TYNBS1, providing the genetic and molecular basis for further studies [24]. Under biotic and abiotic stresses, ROS, superoxide, hydrogen peroxide, and nitric oxide were over accumulated in plants [49]. This process was considered to be harmful by damaging the protein structure, enhancing membrane lipid peroxidation, and producing MDA, leading to cell cytotoxicity [50]. To overcome ROS toxicity, antioxidant enzymes, such as SOD, POD, CAT, APX, and PPO, can clear ROS [51]. As for plant–pathogen defense, ROS may also orchestrate a plant’s hypersensitive disease resistance response [51]. Combined with proteomics and metabolomics analysis with TYLCV R/S lines, the expression level of reactive oxygen species (ROS) compounds and antioxidative proteins remains high in tomatoes invaded by the virus [31]. In our study, the MDA content combined with ROS-eliminating enzymes suggested that Ty-2 could maintain the normal physiological condition for more time. Given that Ty-1 suppresses virus infection in tomatoes via the induction of RNA silencing [21], recent studies have shown that siRNAs are also involved in ROS signals to regulate plant immunity [52]. Hence, Ty-2 may have crosstalk with Ty-1 in the regulation of the host defense pathway. Coordinating the two genes could be a potential strategy for tomato TYLCV-resistant breeding.
Plant hormones play a crucial role in crop growth and virus defense. SA involves the production of ROS and the accumulation of pathogenesis-related protein, leading to characteristic hypersensitive responses (HRs), which restrict viruses locally [53]. In SA-signaling-active mutants such as cpr1 and cim10 [54] or the application of exogenous SA [33], the accumulation of TYLCV can be reduced, which can enhance the virus resistance. Our results showed that both SA biosynthesis and signal genes were dramatically induced by TYLCV at the time of infection (Figure 5A), which illustrates that SA acts as the principal defender. JA is another key phytohormone that tackles viruses and vectors. A previous study showed that the C2 protein from TYLCV decreases the sensitivity toward JA and suppresses JA-mediated defenses [55]. Interestingly, our results argue that JA-related genes can still be up-regulated after TYLCV attack, presenting the drastic arms race between plants and the pathogen. Since plant resistance is usually promoted at the cost of development, growth hormones are also involved in plant immunity. When attacked by geminiviruses, brassinosteroid can induce an ROS burst to resist the virus in an SA-independent manner [56]. Similarly, cytokinin plays a role in ROS induction and synergistic association with SA to participate in the plant immunity [57]. Apart from these essential antivirus hormones, others—including zeatin and IAA—might have also been involved in TYLCV resistance in our results (Figure 5).
RNA-seq is a powerful tool for identifying the key genes and pathways involved in particular biological processes [58]. Data from RNA-seq with tomatoes infected with TYLCV within 48 h showed that the virus has a big impact on the catalytic activity and metabolic process (Figure 4). This is consistent with the results that both biosynthetic genes and the contents of phytohormones, including SA, IAA, and zeatin, were up-regulated (Figure 5 and Figure 6). Moreover, the increase in the activity of ROS-scavenging enzymes revealed that they should be downstream of phytohormones to maintain the ROS balance participating in TYLCV resistance as per a previous report [53,57]. Meanwhile, JA, the major defense phytohormone, which could enhance the production of defense compounds against insect herbivores and pathogens [34], was also induced in the initial stage of TYLCV inoculation (24 h) (Figure 6). This indicates that the contents of phytoalexins such as terpene regulated by JA [35] may be affected by the virus just after infection. For further studies, the metabolome—especially phytoalexin—should be detected to explain TYLCV resistance.
5. Conclusions
TYLCV is a common pathogen causing damage to tomato production. We used the susceptible line MK and the resistant line T2 to perform TYLCV inoculation. The infection of the virus promoted the accumulation of MDA, resulting in lipid peroxidation stress at the initial time of treatment. The activity of the ROS-scavenging enzymes was higher in T2 (R), suggesting that the ROS balance was important in the virus resistance. RNA-seq was carried out to detect the antivirus-related genes or pathways. Combined with qRT-PCR and the measurement of the phytohormones, both the biosynthesis and signal of SA, IAA, and zeatin were involved in the tomato defense against TYLCV. These insights will likely facilitate the genetic modification of tomatoes for desirable resistance traits and possibly many other species of plants infected by TYLCV.
Supplementary Materials
The following are available online at www.mdpi.com/article/10.3390/horticulturae8020143/s1, Figure S1: Distribution of gene expression (FPKM) level. Figure S2: Pearson correlation analysis of the FPKM values of leaf samples in tomato under TYLCV infection. Figure S3: The Kyoto encyclopedia of genes and genomes (KEGG) map of the signal transduction of different plant hormones. Table S1: Genes and primers for qRT-PCR. Table S2: Summary of aligned and mapped reads. Table S3: Differentially expressed genes in Moneymaker (MK) and CLN2777A (T2). Table S4: GO and KEGG pathway annotation of DEGs. Table S5: Expressed genes for phytohormones biosynthesis and signaling pathway.
Author Contributions
Conceptualization, L.S.; formal analysis, Y.W.; funding acquisition, L.S., T.Z. and Y.W.; project administration, L.Z.; resources, T.Z.; supervision, T.Z.; validation, L.S.; writing—original draft, L.S.; writing—review and editing, Y.W. and T.Z. All authors have read and agreed to the published version of the manuscript.
Funding
This work was funded by Natural Science Foundation of Youth in Jiangsu Province (BK20180306), National Natural Science Foundation of China (31902025), National Natural Science Foundation of China (31972424), the Independent Innovation of Agricultural Sciences in Jiangsu Province (CX (20)3102), the Seed Industry Revitalization Project of Jiangsu Province (JBGS [2021] 066).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The transcriptome data used in this study have been submitted to the NCBI database under the accession number PRJNA787385. These data have not been released due to unfinished research content but could be available upon request.
Acknowledgments
We thank Yu Wang (College of Horticulture, Nanjing Agricultural University) for his help in the draft and revision process.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Varsani, A.; Navas-Castillo, J.; Moriones, E.; Hernandez-Zepeda, C.; Idris, A.; Brown, J.K.; Murilo Zerbini, F.; Martin, D.P. Establishment of three new genera in the family Geminiviridae: Becurtovirus, Eragrovirus and Turncurtovirus. Arch. Virol. 2014, 159, 2193–2203. [Google Scholar] [CrossRef] [PubMed]
- Kheyr-Pour, A.; Bendahmane, M.; Matzeit, V.; Accotto, G.P.; Crespi, S.; Gronenborn, B. Tomato yellow leaf curl virus from Sardinia is a whitefly-transmitted monopartite geminivirus. Nucleic Acids Res. 1991, 19, 6763–6769. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Navot, N.; Pichersky, E.; Zeidan, M.; Zamir, D.; Czosnek, H. Tomato yellow leaf curl virus: A whitefly-transmitted geminivirus with a single genomic component. Virology 1991, 185, 151–161. [Google Scholar] [CrossRef]
- Moriones, E.; Navas-Castillo, J. Tomato yellow leaf curl virus, an emerging virus complex causing epidemics worldwide. Virus Res. 2000, 71, 123–134. [Google Scholar] [CrossRef]
- Johnston, N.; Martini, X. The influence of visual and olfactory cues in host selection for Bemisia tabaci Biotype B in the presence or absence of Tomato yellow leaf curl virus. Insects 2020, 11, 115. [Google Scholar] [CrossRef] [Green Version]
- Vidavski, F.; Czosnek, H.; Gazit, S.; Levy, D.; Lapidot, M. Pyramiding of genes conferring resistance to Tomato yellow leaf curl virus from different wild tomato species. Plant Breed. 2008, 127, 625–631. [Google Scholar] [CrossRef]
- Cohen, S.; Antignus, Y. Tomato yellow leaf curl virus, a whitefly-borne Geminivirus of tomatoes. In Advances in Disease Vector Research; Harris, K.F., Ed.; Springer: New York, NY, USA, 1994; pp. 259–288. [Google Scholar]
- Hanssen, I.M.; Lapidot, M.; Thomma, B.P. Emerging viral diseases of tomato crops. Mol. Plant Microbe Interact. 2010, 23, 539–548. [Google Scholar] [CrossRef] [Green Version]
- Bass, C.; Denholm, I.; Williamson, M.S.; Nauen, R. The global status of insect resistance to neonicotinoid insecticides. Pestic. Biochem. Physiol. 2015, 121, 78–87. [Google Scholar] [CrossRef] [Green Version]
- Julián, O.; Herráiz, J.; Corella, S.; Di-Lolli, I.; Soler, S.; Díez, M.; Pérez-De-Castro, A. Initial development of a set of introgression lines from Solanum peruvianum PI 126944 into tomato: Exploitation of resistance to viruses. Euphytica 2013, 193, 183–196. [Google Scholar] [CrossRef]
- Zamir, D.; Ekstein-Michelson, I.; Zakay, Y.; Navot, N.; Zeidan, M.; Sarfatti, M.; Eshed, Y.; Harel, E.; Pleban, T.; van-Oss, H.; et al. Mapping and introgression of a Tomato yellow leaf curl virus tolerance gene, TY-1. Theor. Appl. Genet. 1994, 88, 141–146. [Google Scholar] [CrossRef]
- Hanson, P.M.; Green, S.K.; Kuo, G. Ty-2, a gene on chromosome 11 conditioning geminivirus resistance in tomato. Tomato Genet. Coop Rep. 2000, 56, 17–18. [Google Scholar]
- Yang, X.; Caro, M.; Hutton, S.F.; Scott, J.W.; Guo, Y.; Wang, X.; Rashid, M.H.; Szinay, D.; de Jong, H.; Visser, R.G.; et al. Fine mapping of the Tomato yellow leaf curl virus resistance gene Ty-2 on chromosome 11 of tomato. Mol. Breed. 2014, 34, 749–760. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ji, Y.; Schuster, D.J.; Scott, J.W. Ty3, a begomovirus resistance locus near the Tomato yellow leaf curl virus resistance locus Ty1 on chromosome 6 of tomato. Mol. Breed. 2007, 20, 271–284. [Google Scholar] [CrossRef]
- Ji, Y.; Scott, J.W.; Schuster, D.J.; Maxwell, D.P. Molecular Mapping of Ty-4, a new Tomato yellow leaf curl virus resistance locus on chromosome 3 of tomato. J. Am. Soc. Hortic. Sci. 2009, 134, 281–288. [Google Scholar] [CrossRef] [Green Version]
- Anbinder, I.; Reuveni, M.; Azari, R.; Paran, I.; Nahon, S.; Shlomo, H.; Chen, L.; Lapidot, M.; Levin, I. Molecular dissection of Tomato leaf curl virus resistance in tomato line TY172 derived from Solanum peruvianum. Theor. Appl. Genet. 2009, 119, 519–530. [Google Scholar] [CrossRef]
- Hutton, S.F.; Scott, J.W.; Schuster, D.J. Recessive resistance to Tomato yellow leaf curl virus from the tomato cultivar Tyking is located in the same region as Ty-5 on Chromosome 4. HortScience 2012, 47, 324–327. [Google Scholar] [CrossRef] [Green Version]
- Kadirvel, P.; De la Peña, R.; Schafleitner, R.; Huang, S.; Geethanjali, S.; Kenyon, L.; Tsai, W.; Hanson, P. Mapping of QTLs in tomato line FLA456 associated with resistance to a virus causing tomato yellow leaf curl disease. Euphytica 2013, 190, 297–308. [Google Scholar] [CrossRef]
- Upinder, G.; John, W.S.; Reza, S.; Eben, O.; Cees, S.; David, M.F.; Sung-Chur, S.; Hugh, S.; Samuel, F.H. Ty-6, a major begomovirus resistance gene on chromosome 10, is effective against Tomato yellow leaf curl virus and Tomato mottle virus. Theor. Appl. Genet. 2019, 132, 1543–1554. [Google Scholar]
- Verlaan, M.G.; Hutton, S.F.; Ibrahem, R.M.; Kormelink, R.; Visser, R.G.; Scott, J.W.; Edwards, J.D.; Bai, Y. The tomato yellow leaf curl virus resistance genes Ty-1 and Ty-3 are allelic and code for DFDGD-class RNA-dependent RNA polymerases. PLoS Genet. 2013, 9, e1003399. [Google Scholar] [CrossRef] [Green Version]
- Butterbach, P.; Verlaan, M.G.; Dullemans, A.; Lohuis, D.; Visser, R.G.; Bai, Y.; Kormelink, R. Tomato yellow leaf curl virus resistance by Ty-1 involves increased cytosine methylation of viral genomes and is compromised by Cucumber mosaic virus infection. Proc. Natl. Acad. Sci. USA 2014, 111, 12942–12947. [Google Scholar] [CrossRef] [Green Version]
- Belabess, Z.; Urbino, C.; Granier, M.; Tahiri, A.; Blenzar, A.; Peterschmitt, M. The typical RB76 recombination breakpoint of the invasive recombinant Tomato yellow leaf curl virus of Morocco can be generated experimentally but is not positively selected in tomato. Virus Res. 2018, 243, 44–51. [Google Scholar] [CrossRef]
- Panno, S.; Caruso, A.G.; Davino, S. The nucleotide sequence of a recombinant Tomato yellow leaf curl virus strain frequently detected in Sicily isolated from tomato plants carrying the Ty-1 resistance gene. Arch. Virol. 2018, 163, 795–797. [Google Scholar] [CrossRef] [PubMed]
- Hirotaka, Y.; Jun, O.; Atsushi, S.; Akio, O.; Tsukasa, N.; Koji, M.; Hiroyuki, F. An NB-LRR gene, TYNBS1, is responsible for resistance mediated by the Ty-2 Begomovirus resistance locus of tomato. Theor. Appl. Genet. 2018, 131, 1345–1362. [Google Scholar] [CrossRef] [PubMed]
- Tomato Genome Consortium. The tomato genome sequence provides insights into fleshy fruit evolution. Nature 2012, 485, 635–641. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gupta, S.; Shi, X.; Lindquist, I.E.; Devitt, N.; Mudge, J.; Rashotte, A.M. Transcriptome profiling of cytokinin and auxin regulation in tomato root. J. Exp. Bot. 2013, 64, 695–704. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, X.; Gupta, S.; Lindquist, I.E.; Cameron, C.T.; Mudge, J.; Rashotte, A.M. Transcriptome analysis of cytokinin response in tomato leaves. PLoS ONE 2013, 8, e55090. [Google Scholar]
- Westermann, A.J.; Gorski, S.A.; Vogel, J. Dual RNA-seq of pathogen and host. Nat. Rev. Microbiol. 2012, 10, 618–630. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Lv, Y.; Zhao, T.; Li, N.; Yang, Y.; Yu, W.; He, X.; Liu, T.; Zhang, B. Comparative transcriptome profiling of a resistant vs. susceptible tomato (Solanum lycopersicum) cultivar in response to infection by Tomato yellow leaf curl virus. PLoS ONE 2013, 8, e80816. [Google Scholar] [CrossRef]
- Wang, J.; Yu, W.; Yang, Y.; Li, X.; Chen, T.; Liu, T.; Ma, N.; Yang, X.; Liu, R.; Zhang, B. Genome-wide analysis of tomato long non-coding RNAs and identification as endogenous target mimic for microRNA in response to TYLCV infection. Sci. Rep. 2015, 5, 16946. [Google Scholar] [CrossRef] [Green Version]
- Moshe, A.; Pfannstiel, J.; Yariv, B.; Kolot, M.; Sobol, I.; Czosnek, H.; Gorovits, R. Stress responses to Tomato yellow leaf curl virus (TYLCV) infection of resistant and susceptible tomato plants are different. Metabolomics S1 2012, 6, 2153-0769. [Google Scholar]
- Sade, D.; Sade, N.; Shriki, O.; Lerner, S.; Gebremedhin, A.; Karavani, A.; Brotman, Y.; Osorio, S.; Fernie, A.R.; Willmitzer, L. Water balance, hormone homeostasis, and sugar signaling are all involved in tomato resistance to Tomato yellow leaf curl virus. Plant Physiol. 2014, 165, 1684–1697. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, T.; Huang, Y.; Xu, Z.S.; Wang, F.; Xiong, A.S. Salicylic acid-induced differential resistance to the Tomato yellow leaf curl virus among resistant and susceptible tomato cultivars. BMC Plant Biol. 2019, 19, 173. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Wang, D.D.; Fang, X.; Chen, X.Y.; Mao, Y.B. Plant specialized metabolism regulated by jasmonate signaling. Plant Cell Physiol. 2019, 60, 2638–2647. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Weldegergis, B.T.; Li, J.; Jung, C.; Qu, J.; Sun, Y.; Qian, H.; Tee, C.; van Loon, J.J.; Dicke, M.; et al. Virulence factors of geminivirus interact with MYC2 to subvert plant resistance and promote vector performance. Plant Cell 2014, 26, 4991–5008. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Yang, Y.; Jin, L.; Ling, X.; Liu, T.; Chen, T.; Ji, Y.; Yu, W.; Zhang, B. Re-analysis of long non-coding RNAs and prediction of circRNAs reveal their novel roles in susceptible tomato following TYLCV infection. BMC Plant Biol. 2018, 18, 104. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Li, T.; Xu, Z.S.; Wang, F.; Xiong, A.S. Six NAC transcription factors involved in response to TYLCV infection in resistant and susceptible tomato cultivars. Plant Physiol. Biochem. 2017, 120, 61–74. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Zhang, X.-Y.; Huang, Y.; Xu, Z.-S.; Wang, F.; Xiong, A.-S. An R2R3-MYB transcription factor, SlMYB28, involved in the regulation of TYLCV infection in tomato. Sci. Hortic. 2018, 237, 192–200. [Google Scholar] [CrossRef]
- Zhang, H.; Gong, H.; Zhou, X. Molecular characterization and pathogenicity of Tomato yellow leaf curl virus in China. Virus Genes 2009, 39, 249–255. [Google Scholar] [CrossRef]
- Macadam, J.W.; Nelson, C.J.; Sharp, R.E. Peroxidase activity in the leaf elongation zone of tall fescue: I. spatial distribution of ionically bound peroxidase activity in genotypes differing in length of the elongation zone. Plant Physiol. 1992, 99, 872–878. [Google Scholar] [CrossRef] [Green Version]
- Nakano, Y.; Asada, K. Hydrogen peroxide is scavenged by ascorbate-specific peroxidase in spinach chloroplasts. Plant Cell Physiol. 1980, 22, 867–880. [Google Scholar]
- Eybishtz, A.; Peretz, Y.; Sade, D.; Akad, F.; Czosnek, H. Silencing of a single gene in tomato plants resistant to Tomato yellow leaf curl virus renders them susceptible to the virus. Plant Mol. Biol. 2009, 71, 157–171. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pertea, M.; Kim, D.; Pertea, G.M.; Leek, J.T.; Salzberg, S.L. Transcript-level expression analysis of RNA-seq experiments with hisat, stringtie and ballgown. Nat. Protoc. 2016, 11, 1650–1667. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgerR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pan, X.; Welti, R.; Wang, X. Quantitative analysis of major plant hormones in crude plant extracts by high-performance liquid chromatography-mass spectrometry. Nat. Protoc. 2010, 5, 986–992. [Google Scholar] [CrossRef]
- Shi, Y.; Li, Y.; Guo, Y.; Borrego, E.J.; Wei, Z.; Ren, H.; Ma, Z.; Yan, Y. A rapid pipeline for pollen-and anther-specific gene discovery based on transcriptome profiling analysis of maize tissues. Int. J. Mol. Sci. 2021, 22, 6877. [Google Scholar] [CrossRef]
- Garzo, E.; Moreno, A.; Plaza, M.; Fereres, A. Feeding behavior and virus-transmission ability of insect vectors exposed to systemic insecticides. Plants 2020, 9, 895. [Google Scholar] [CrossRef]
- Castro, B.; Citterico, M.; Kimura, S.; Stevens, D.M.; Wrzaczek, M.; Coaker, G. Stress-induced reactive oxygen species compartmentalization, perception and signalling. Nat. Plants 2021, 7, 403–412. [Google Scholar] [CrossRef]
- Morales, M.; Munne-Bosch, S. Malondialdehyde: Facts and artifacts. Plant Physiol. 2019, 180, 1246–1250. [Google Scholar] [CrossRef] [Green Version]
- Waszczak, C.; Carmody, M.; Kangasjarvi, J. Reactive oxygen species in plant signaling. Annu. Rev. Plant Biol. 2018, 69, 209–236. [Google Scholar] [CrossRef] [Green Version]
- Liu, P.; Zhang, X.; Zhang, F.; Xu, M.; Ye, Z.; Wang, K.; Liu, S.; Han, X.; Cheng, Y.; Zhong, K.; et al. A virus-derived siRNA activates plant immunity by interfering with ROS scavenging. Mol. Plant 2021, 14, 1088–1103. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, D.; Chakraborty, S. Molecular interplay between phytohormones and geminiviruses: A saga of a never-ending arms race. J. Exp. Bot. 2021, 72, 2903–2917. [Google Scholar] [CrossRef] [PubMed]
- Ascencio-Ibanez, J.T.; Sozzani, R.; Lee, T.J.; Chu, T.M.; Wolfinger, R.D.; Cella, R.; Hanley-Bowdoin, L. Global analysis of Arabidopsis gene expression uncovers a complex array of changes impacting pathogen response and cell cycle during geminivirus infection. Plant Physiol. 2008, 148, 436–454. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rosas-Diaz, T.; Macho, A.P.; Beuzon, C.R.; Lozano-Duran, R.; Bejarano, E.R. The C2 protein from the geminivirus Tomato yellow leaf curl Sardinia virus decreases sensitivity to jasmonates and suppresses jasmonate-mediated defences. Plants 2016, 5, 8. [Google Scholar] [CrossRef] [PubMed]
- Deng, X.G.; Zhu, T.; Peng, X.J.; Xi, D.H.; Guo, H.; Yin, Y.; Zhang, D.W.; Lin, H.H. Role of brassinosteroid signaling in modulating Tobacco mosaic virus resistance in Nicotiana benthamiana. Sci. Rep. 2016, 6, 20579. [Google Scholar] [CrossRef]
- Choi, J.; Choi, D.; Lee, S.; Ryu, C.M.; Hwang, I. Cytokinins and plant immunity: Old foes or new friends? Trends Plant Sci. 2011, 16, 388–394. [Google Scholar] [CrossRef]
- Wang, Z.; Gerstein, M.; Snyder, M. RNA-Seq: A revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009, 10, 57–63. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).