Identification and Expression Analysis of Zinc Finger A20/AN1 Stress-Associated Genes SmSAP Responding to Abiotic Stress in Eggplant
Abstract
:1. Introduction
2. Materials and Methods
2.1. Identification of SAP Family Members in the Eggplant
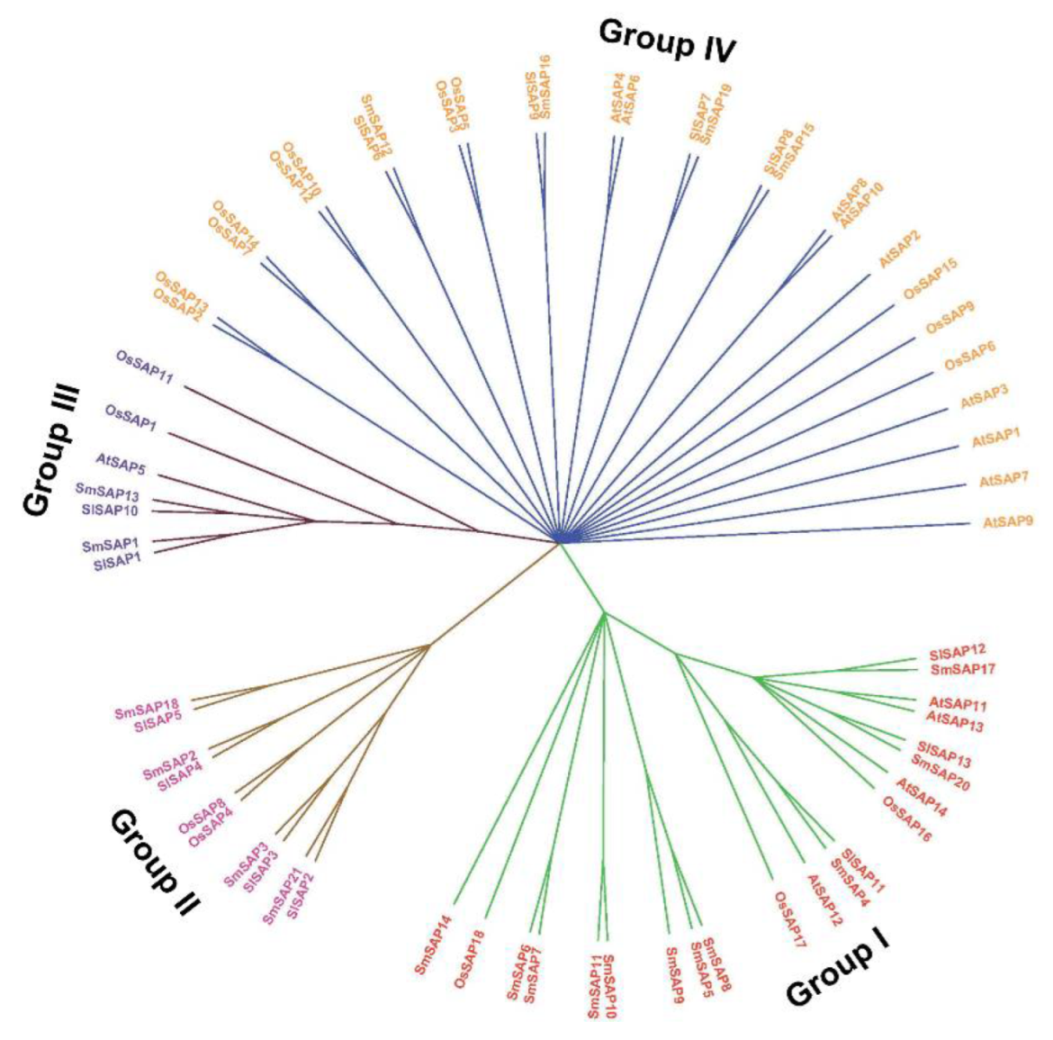
2.2. Phylogenetic Analysis of SAP Family Members in the Eggplant and Other Species
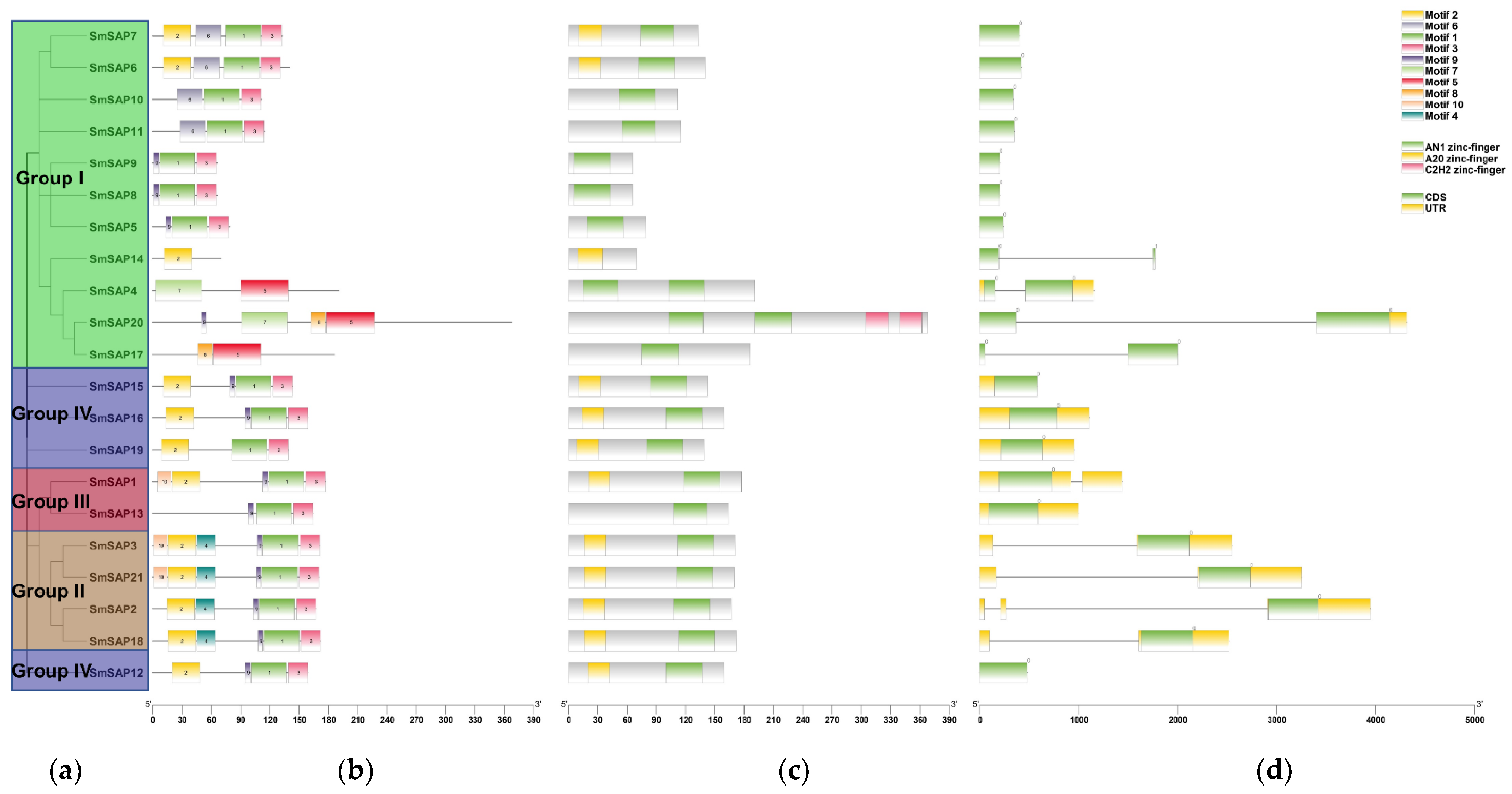
2.3. Analysis of Motifs, Domains and Gene Structure of SAP Family Members in Eggplant
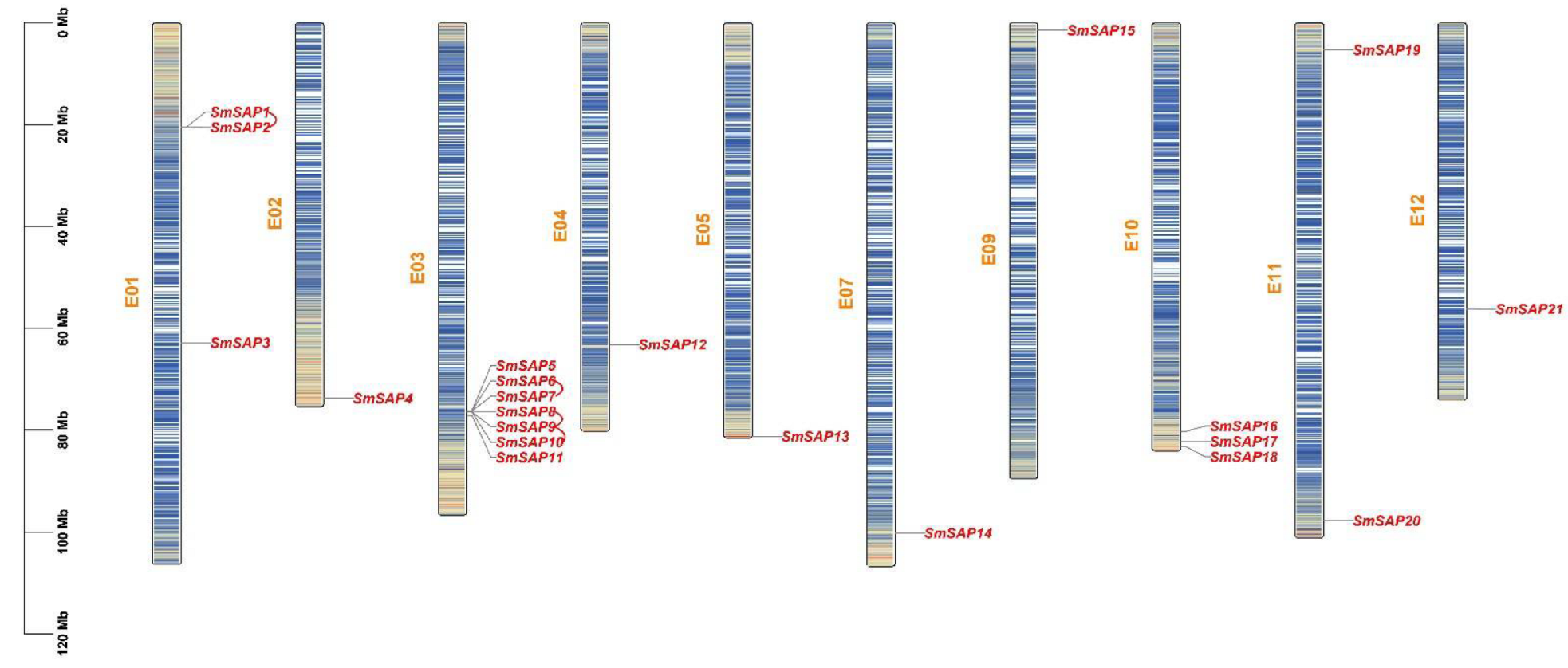
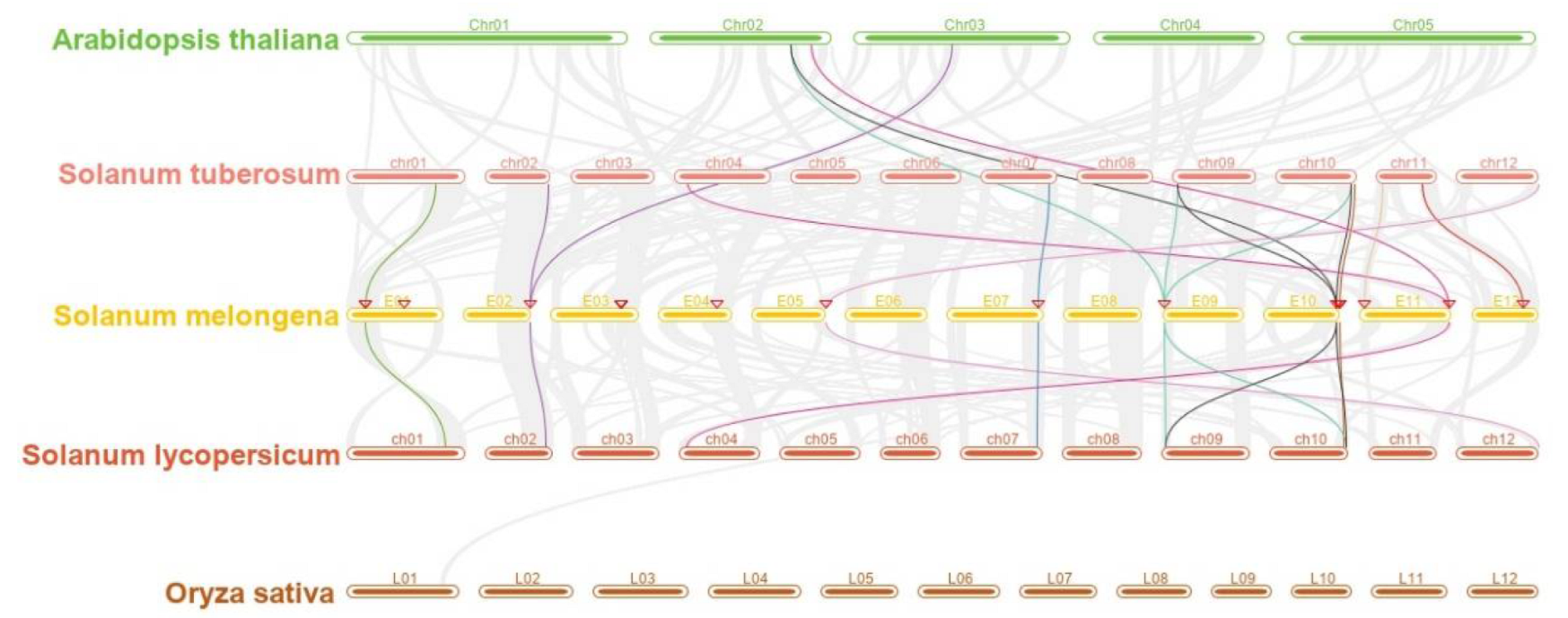
2.4. Analysis of Location on Chromosomes, Gene Duplication, and Collinearity of SAP Family Genes in Eggplant
2.5. Identification of Putative Cis-Elements in the Promoters of SmSAP Genes
2.6. Plant Material, Environmental Conditions and Abiotic Stress Treatments
2.7. RNA Extraction, and Expression Analysis
2.8. Statistical Analysis
3. Results
3.1. Identification of SAP Gene Family Members in Eggplant
3.2. Phylogenetic Analysis of SAP Genes
3.3. Analysis of Motifs, Domains, Exons and Introns of SAP Family Members in Eggplant
3.4. Location on Chromosomes, Gene Duplication, and Collinearity Analysis of SAP Family Genes in Eggplant
3.5. Cis-Elements Analysis in the Promoters of SmSAP Genes
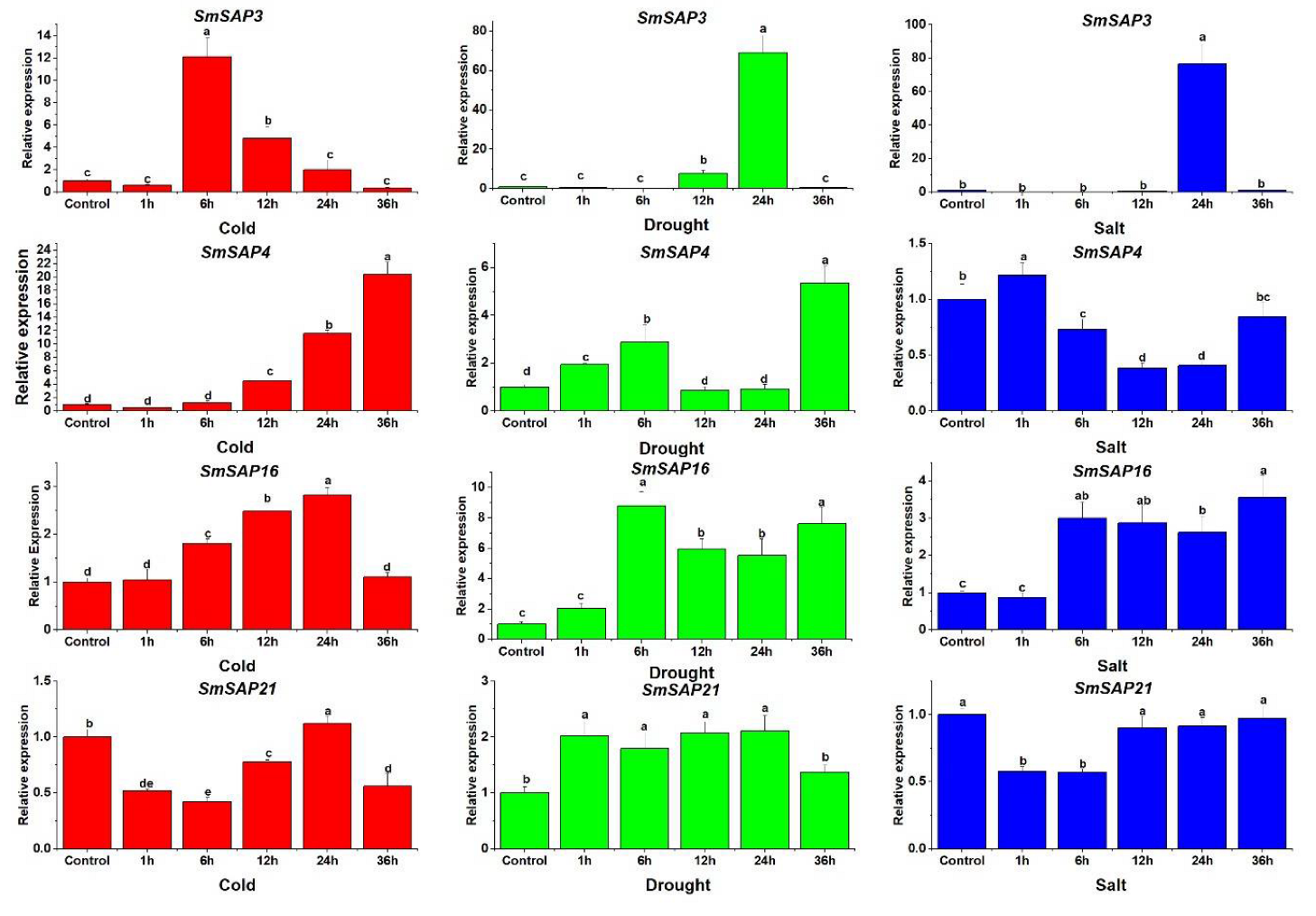
3.6. Expression Analysis of Four Selected SmSAP Genes under Abiotic Stress Treatments
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhang, X.Z.; Zheng, W.J.; Cao, X.Y.; Cui, X.Y.; Zhao, S.P. Genomic analysis of stress associated proteins in soybean and the role of GmSAP16 in abiotic stress responses in Arabidopsis and soybean. Front. Plant Sci. 2019, 10, 1453. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Giri, J.; Dansana, P.; Kothari, K.; Sharma, G.; Vij, S.; Tyagi, A. SAPs as novel regulators of abiotic stress response in plants. BioEssays 2013, 35, 639–648. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Zeng, L.; Chen, R.; Wang, Y.; Song, J. Genome-wide identification and characterization of stress-associated protein (SAP) gene family encoding A20/AN1 zinc-finger proteins in Medicago truncatula. Arch. Biol. Sci. 2018, 70, 28. [Google Scholar] [CrossRef]
- Vij, S.; Tyagi, A.K. Genome-wide analysis of the stress associated protein (SAP) gene family containing A20/AN1 zinc-finger(s) in rice and their phylogenetic relationship with Arabidopsis. Mol. Genet. Genom. 2006, 276, 565–575. [Google Scholar] [CrossRef]
- Solanke, A.U.; Sharma, M.K.; Tyagi, A.K.; Sharma, A.K. Characterization and phylogenetic analysis of environmental stress-responsive SAP gene family encoding A20/AN1 zinc finger proteins in tomato. Mol. Genet. Genom. 2009, 282, 153–164. [Google Scholar] [CrossRef]
- Gao, W.; Long, L.; Tian, X.; Jin, J.; Liu, H.; Zhang, H.; Xu, F.; Song, C. Genome-wide identification and expression analysis of stress-associated proteins (SAPs) containing A20/AN1 zinc finger in cotton. Mol. Genet. Genom. 2016, 291, 2199–2213. [Google Scholar] [CrossRef]
- Baidyussen, A.; Aldammas, M.; Kurishbayev, A.; Myrzabaeva, M.; Zhubatkanov, A.; Sereda, G.; Porkhun, R.; Sereda, S.; Jatayev, S.; Langridge, P.; et al. Identification, gene expression and genetic polymorphism of zinc finger A20/AN1 stress-associated genes, HvSAP, in salt stressed barley from Kazakhstan. BMC Plant Biol. 2020, 20, 156. [Google Scholar] [CrossRef]
- Mukhopadhyay, A.; Vij, S.; Tyagi, A.K. Overexpression of a zinc-finger protein gene from rice confers tolerance to cold, dehydration, and salt stress in transgenic tobacco. Proc. Natl. Acad. Sci. USA 2004, 101, 6309–6314. [Google Scholar] [CrossRef] [Green Version]
- He, X.A.; Xie, S.A.; Xie, P.A.; Yao, M.A.; Liu, W.A.; Qin, L.A.; Liu, Z.; Zheng, M.; Liu, H.D.; Guan, M. Genome-wide identification of stress-associated proteins (SAP) with A20/AN1 zinc finger domains associated with abiotic stresses responses in Brassica napus. Environ. Exp. Bot. 2019, 165, 108–119. [Google Scholar] [CrossRef]
- Lai, W.; Zhou, Y.; Pan, R.; Liao, L.; Liu, S. Identification and expression analysis of stress-associated proteins (SAPs) containing A20/AN1 zinc finger in cucumber. Plants 2020, 9, 400. [Google Scholar] [CrossRef] [Green Version]
- Dixit, A.R.; Parkash, D.O.; Abidur, R. A novel stress-associated protein ‘AtSAP10’ from Arabidopsis thaliana confers tolerance to nickel, manganese, zinc, and high temperature stress. PLoS ONE 2011, 6, 38–44. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharma, G.; Giri, J.; Tyagi, A.K. Rice OsiSAP7 negatively regulates ABA stress signalling and imparts sensitivity to water-deficit stress in Arabidopsis. Plant Sci. 2015, 237, 80–92. [Google Scholar] [CrossRef] [PubMed]
- Ben, S.R.; Safi, H.; Ben, H.A.; Brini, F.; Ben, R.W. Functional domain analysis of LmSAP protein reveals the crucial role of the zinc-finger A20 domain in abiotic stress tolerance. Protoplasma 2019, 256, 1333–1344. [Google Scholar]
- Dansana, P.K.; Kothari, K.S.; Vij, S.; Tyagi, A.K. OsiSAP1 overexpression improves water-deficit stress tolerance in transgenic rice by affecting expression of endogenous stress-related genes. Plant Cell Rep. 2014, 33, 1425–1440. [Google Scholar] [CrossRef]
- Kothari, K.S.; Dansana, P.K.; Jitender, G.; Tyagi, A.K. Rice stress associated protein 1 (OsSAP1) interacts with aminotransferase (OsAMTR1) and pathogenesis-related 1a protein (OsSCP) and regulates abiotic stress responses. Front. Plant Sci. 2016, 7, 1057. [Google Scholar] [CrossRef] [Green Version]
- Kanneganti, V.; Gupta, A.K. Overexpression of OsiSAP8, a member of stress associated protein (SAP) gene family of rice confers tolerance to salt, drought and cold stress in transgenic tobacco and rice. Plant Mol. Biol. 2008, 66, 445–462. [Google Scholar] [CrossRef]
- Kang, M.; Fokar, M.; Abdelmageed, H.; Allen, R.D. Arabidopsis SAP5 functions as a positive regulator of stress responses and exhibits E3 ubiquitin ligase activity. Plant Mol. Biol. 2011, 75, 451–466. [Google Scholar] [CrossRef]
- Zhang, N.; Yin, Y.; Liu, X.; Tong, S.; Xing, J.; Zhang, Y.; Pudake, R.N.; Izquierdo, E.M.; Peng, H.; Xin, M.; et al. The E3 Ligase TaSAP5 alters drought stress responses by promoting the degradation of DRIP proteins. Plant Physiol. 2017, 175, 1878–1892. [Google Scholar] [CrossRef] [Green Version]
- Dixit, A.; Tomar, P.; Vaine, E.; Abdullah, H.; Hazen, S.; Dhankher, O.P. A stress-associated protein, AtSAP13, from Arabidopsis thaliana provides tolerance to multiple abiotic stresses. Plant Cell Environ. 2018, 41, 1171–1185. [Google Scholar] [CrossRef]
- Liu, S.; Wang, J.; Jiang, S.; Wang, H.; Gao, Y.; Zhang, H.; Li, D.; Song, F. Tomato SlSAP3, a member of the stress-associated protein family, is a positive regulator of immunity against Pseudomonas syringae pv. tomato DC3000. Mol. Plant Pathol. 2019, 20, 815–830. [Google Scholar] [CrossRef] [Green Version]
- Wei, Q.; Wang, J.; Wang, W.; Hu, T.; Hu, H.; Bao, C. A high-quality chromosome-level genome assembly reveals genetics for important traits in eggplant. Hortic. Res. 2020, 7, 153. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; von Haeseler, A.; Lanfear, R. IQ-TREE 2: New Models and Efficient Methods for Phylogenetic Inference in the Genomic Era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Z.; Fu, M.; Li, H.; Chen, Y.; Liu, R. Distributed under creative commons CC-BY 4.0 systematic analysis of NAC transcription factors in Gossypium barbadense uncovers their roles in response to Verticillium wilt. PeerJ 2019, 2019, e7995. [Google Scholar] [CrossRef] [Green Version]
- Gantasala, N.P.; Papolu, P.K.; Thakur, P.K.; Kamaraju, D.; Sreevathsa, R.; Rao, U. Selection and validation of reference genes for quantitative gene expression studies by real-time PCR in eggplant (Solanum melongena L). BMC Res. Notes 2013, 6, 312. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Dong, Q.; Duan, D.; Zhao, S.; Xu, B.; Luo, J.; Wang, Q.; Huang, D.; Liu, C.; Li, C.; Gong, X.; et al. Genome-wide analysis and cloning of the apple stress-associated protein gene family reveals MdSAP15, which confers tolerance to drought and osmotic stresses in transgenic Arabidopsis. Int. J. Mol. Sci. 2018, 19, 2478. [Google Scholar] [CrossRef] [Green Version]
- Beyaert, R.; Heyninck, K.; Huffel, S.V. A20 and A20-binding proteins as cellular inhibitors of nuclear factor-kappa B-dependent gene expression and apoptosis. Biochem. Pharmacol. 2000, 60, 1143–1151. [Google Scholar] [CrossRef]
- Stroher, E.; Wang, X.J.; Roloff, N.; Klein, P.; Husemann, A.; Dietz, K.J. Redox-dependent regulation of the stress-induced zinc-finger protein SAP12 in Arabidopsis thaliana. Mol. Plant 2009, 2, 357–367. [Google Scholar] [CrossRef]
- Vij, S.; Tyagi, A.K. A20/AN1 zinc-finger domain-containing proteins in plants and animals represent common elements in stress response. Funct. Integr. Genom. 2008, 8, 301–307. [Google Scholar] [CrossRef]
- Rogozin, I.B.; Sverdlov, A.V.; Babenko, V.N.; Koonin, E.V. Analysis of evolution of exon-intron structure of eukaryotic genes. Brief. Bioinform. 2005, 6, 118–134. [Google Scholar] [CrossRef] [Green Version]
- Rose, A.B. Intron-Mediated Regulation of Gene Expression; Springer: Berlin/Heidelberg, Germany, 2008. [Google Scholar]
- Grzybowska, E.A. Human intronless genes: Functional groups, associated diseases, evolution, and mRNA processing in absence of splicing. Biochem. Biophys. Res. Commun. 2012, 424, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Magadum, S.; Banerjee, U.; Murugan, P.; Gangapur, D.; Ravikesavan, R. Gene duplication as a major force in evolution. J. Genet. 2013, 92, 155–161. [Google Scholar] [CrossRef] [PubMed]
- Dong, Q.; Zhao, S.; Duan, D.; Tian, Y.; Wang, Y.; Mao, K.; Zhou, Z.; Ma, F. Structural and functional analyses of genes encoding VQ proteins in apple. Plant Sci. 2018, 272, 208–219. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wan, F.; Xu, Y.; Wang, S.; Gao, J.; Lu, D.; Zhou, C.; Liao, Y.; Ma, Y.; Zheng, Y. Identification and Expression Analysis of Zinc Finger A20/AN1 Stress-Associated Genes SmSAP Responding to Abiotic Stress in Eggplant. Horticulturae 2022, 8, 108. https://doi.org/10.3390/horticulturae8020108
Wan F, Xu Y, Wang S, Gao J, Lu D, Zhou C, Liao Y, Ma Y, Zheng Y. Identification and Expression Analysis of Zinc Finger A20/AN1 Stress-Associated Genes SmSAP Responding to Abiotic Stress in Eggplant. Horticulturae. 2022; 8(2):108. https://doi.org/10.3390/horticulturae8020108
Chicago/Turabian StyleWan, Faxiang, Yuhu Xu, Sulong Wang, Jun Gao, Dan Lu, Chenghong Zhou, Yanqing Liao, Yanyan Ma, and Yu Zheng. 2022. "Identification and Expression Analysis of Zinc Finger A20/AN1 Stress-Associated Genes SmSAP Responding to Abiotic Stress in Eggplant" Horticulturae 8, no. 2: 108. https://doi.org/10.3390/horticulturae8020108
APA StyleWan, F., Xu, Y., Wang, S., Gao, J., Lu, D., Zhou, C., Liao, Y., Ma, Y., & Zheng, Y. (2022). Identification and Expression Analysis of Zinc Finger A20/AN1 Stress-Associated Genes SmSAP Responding to Abiotic Stress in Eggplant. Horticulturae, 8(2), 108. https://doi.org/10.3390/horticulturae8020108





