Overexpression of the SiLEA5 Gene in Saussurea involucrata Increases the Low-Temperature Tolerance of Transgenic Tomatoes
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Growth Materials
2.2. Bioinformatics Analysis of SiLEA5 Gene
2.3. SiLEA5 Gene Cloning and Plant Expression Vector Construction
2.4. Transformation of Tomato and Identification of Positive Plants
2.5. Treatment of Low-Temperature Stress
2.6. Determination of Semilethal Temperature (LT50)
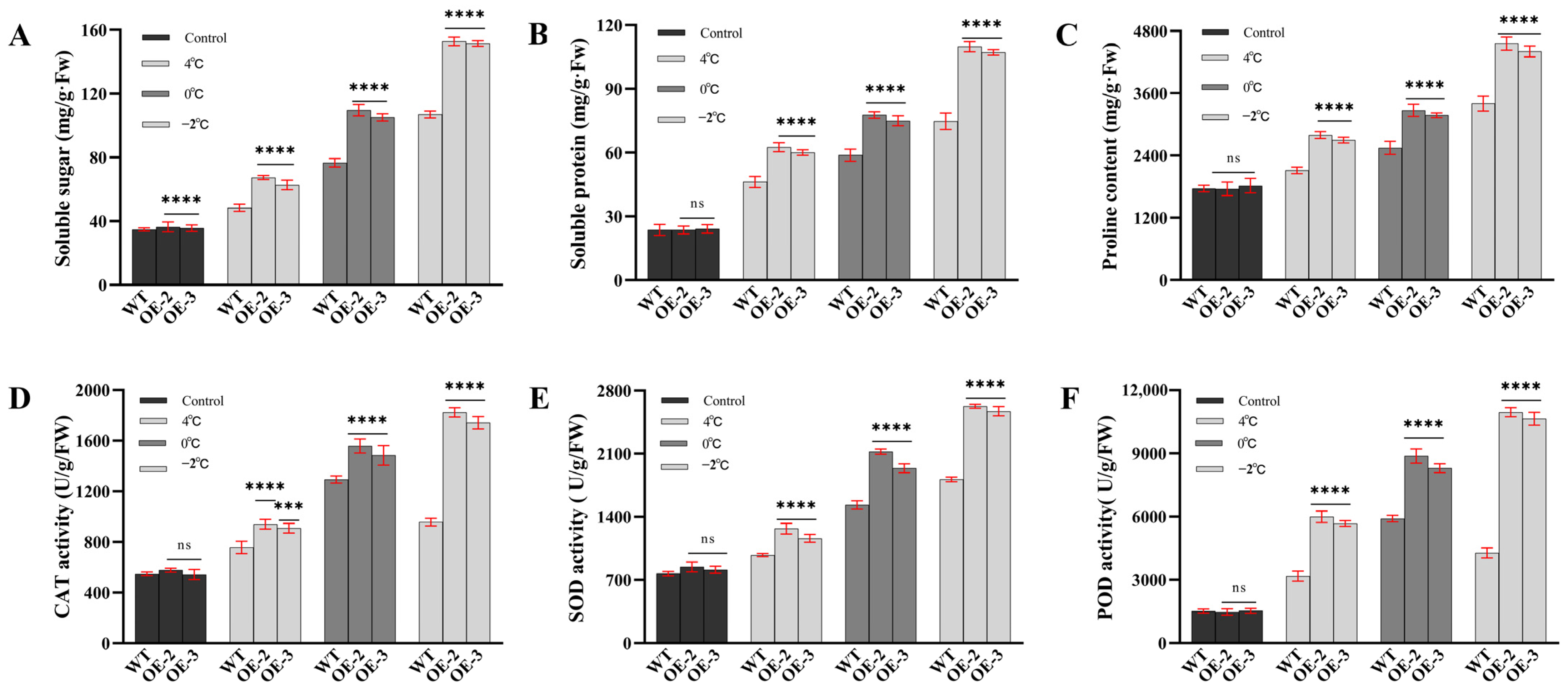
2.7. Physiological Determination of Stress Resistance
2.8. Analysis of Agronomic Characters and Yield
3. Results
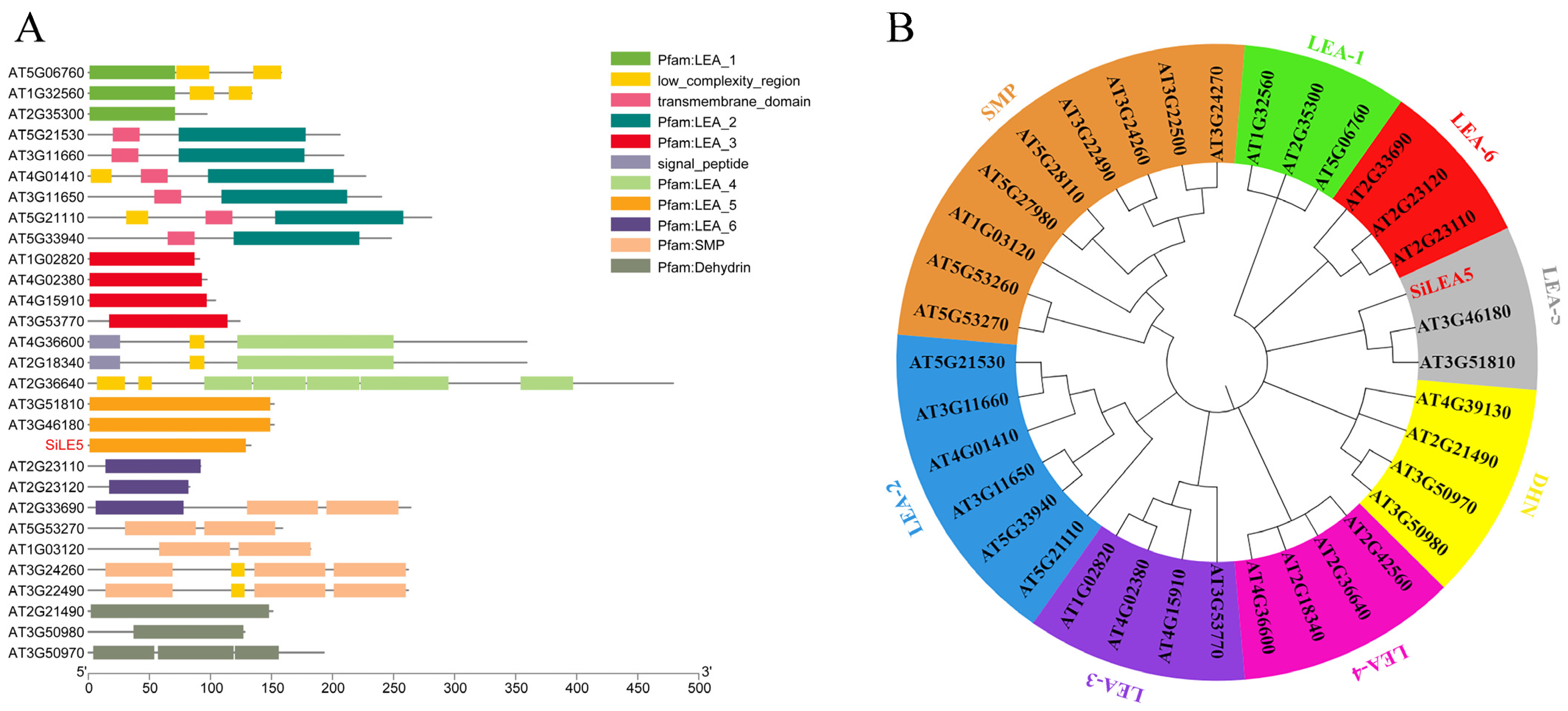
3.1. Bioinformatics Analysis of the SiLEA5 Gene
3.2. Quantitative Real-Time PCR
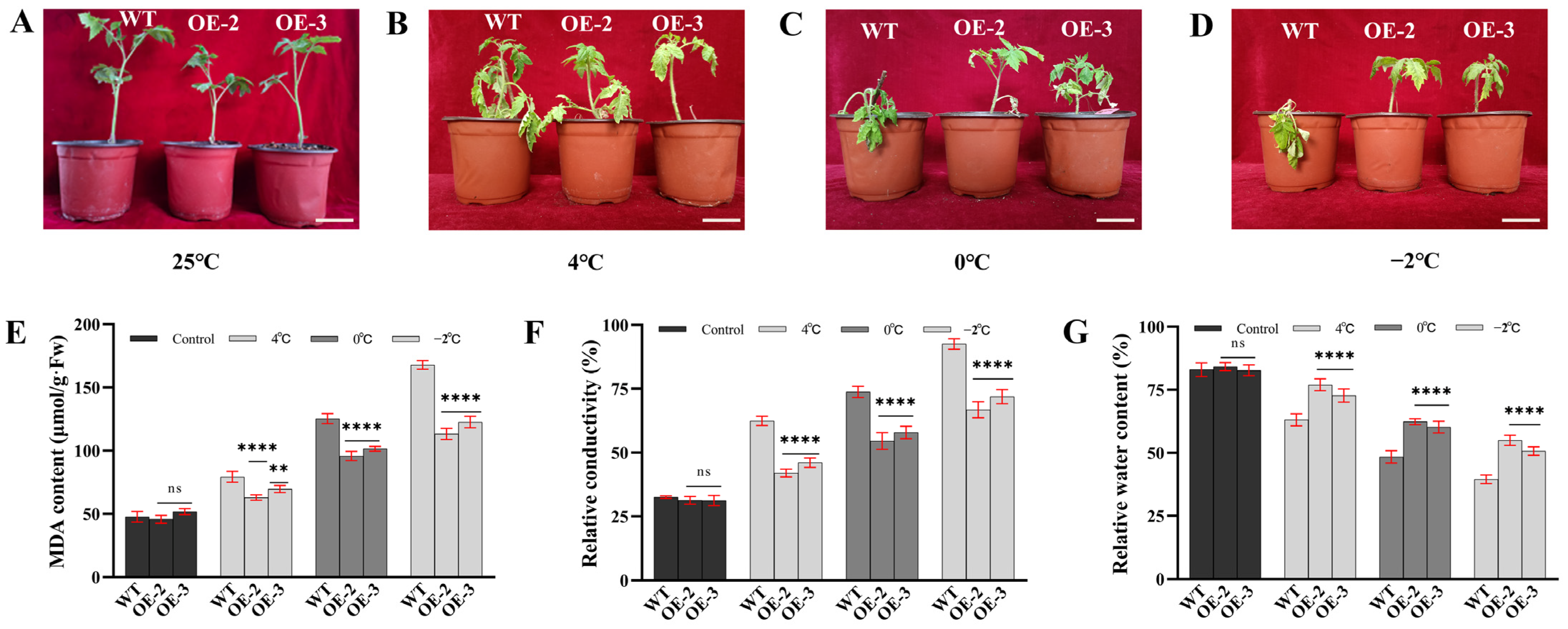
3.3. SiLEA5 Gene Increased the Low-Temperature Tolerance of Transgenic Tomato
3.4. Analysis of Semilethal Temperature (LT50)
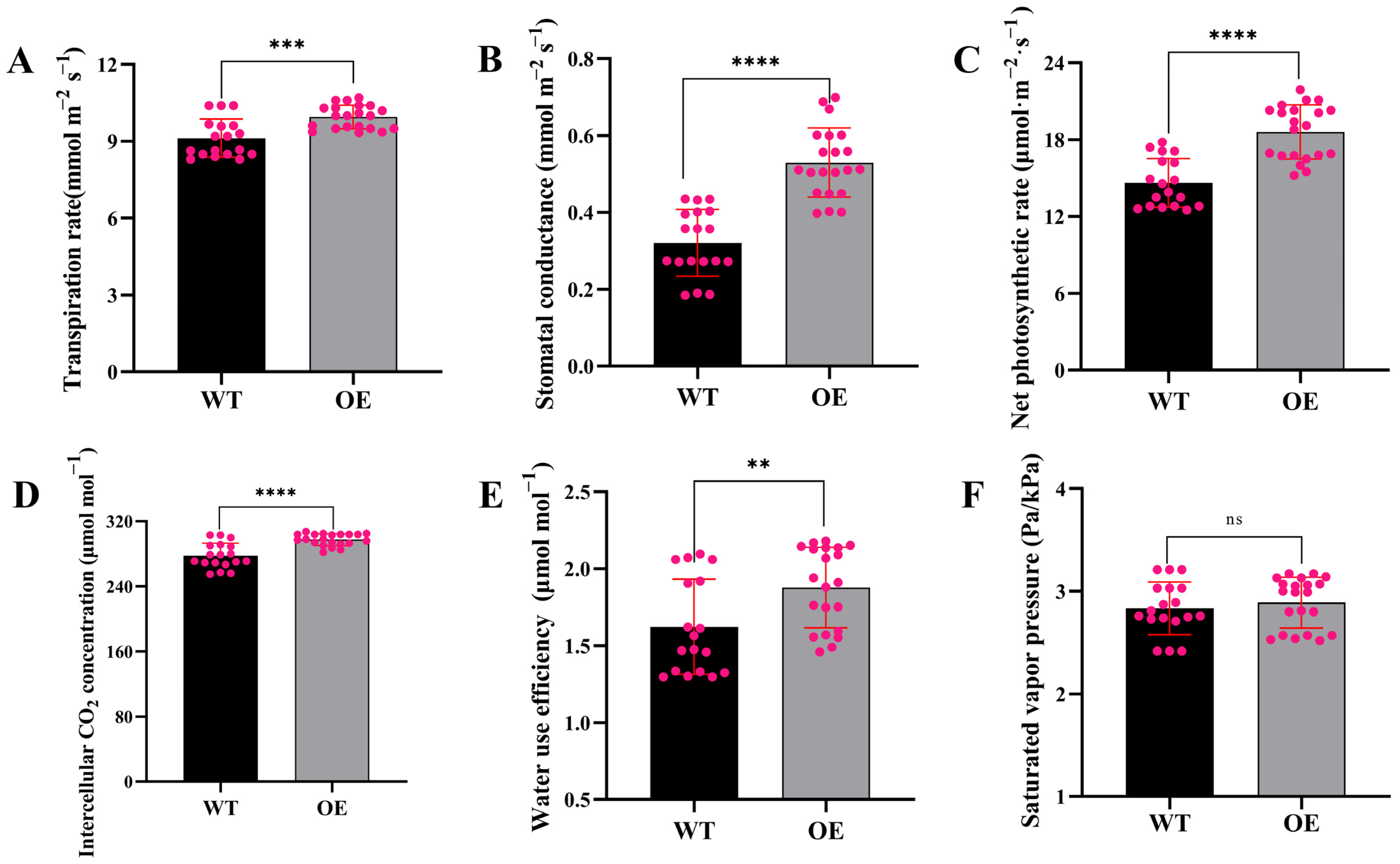
3.5. Transgenic Tomatoes with SiLEA5 Gene Overexpression Have Greater Photosynthetic Efficiency
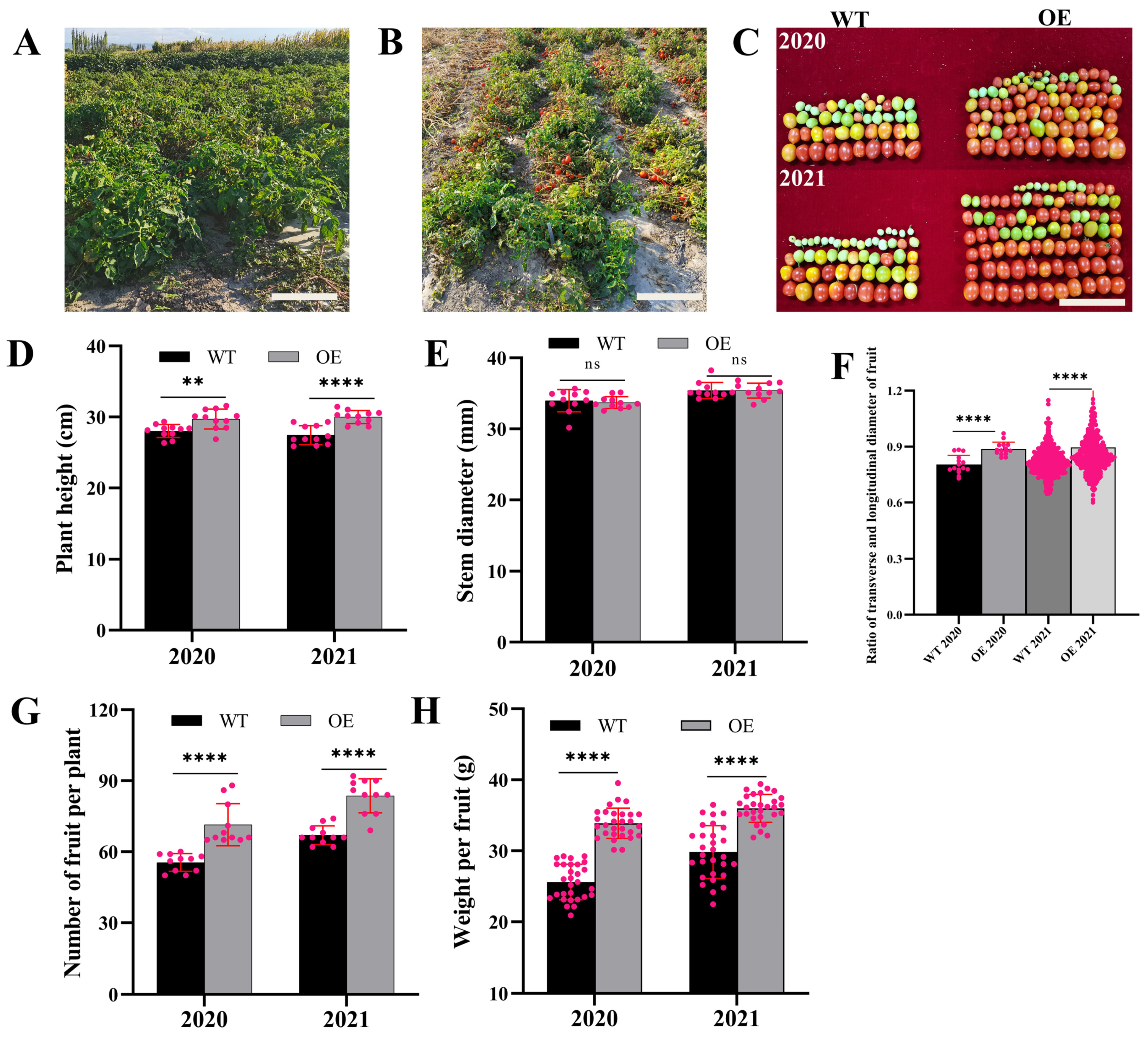
3.6. Analysis of Yields and Agronomic Characteristics
4. Discussion and Conclusions
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Shahzad, A.; Ullah, S.; Dar, A.A.; Sardar, M.F.; Mehmood, T.; Tufail, M.A.; Shakoor, A.; Haris, M. Nexus on climate change: Agriculture and possible solution to cope future climate change stresses. Environ. Sci. Pollut. Res. Int. 2021, 28, 14211–14232. [Google Scholar] [CrossRef] [PubMed]
- Shao, J.; Li, G.; Li, Y.; Zhou, X. Intraspecific responses of plant productivity and crop yield to experimental warming: A global synthesis. Sci. Total Environ. 2022, 840, 156685. [Google Scholar] [CrossRef] [PubMed]
- Filho, W.L.; Setti, A.F.F.; Azeiteiro, U.M.; Lokupitiya, E.; Donkor, F.K.; Etim, N.N.; Matandirotya, N.; Olooto, F.M.; Sharifi, A.; Nagy, G.J.; et al. An overview of the interactions between food production and climate change. Sci. Total Environ. 2022, 838, 156438. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Ruiz, H.; Szurek, B.; Van den Ackerveken, G. Stop helping pathogens: Engineering plant susceptibility genes for durable resistance. Curr. Opin. Biotechnol. 2021, 70, 187–195. [Google Scholar] [CrossRef]
- Sami, A.; Xue, Z.; Tazein, S.; Arshad, A.; He Zhu, Z.; Ping Chen, Y.; Hong, Y.; Tian Zhu, X.; Jin Zhou, K. CRISPR-Cas9-based genetic engineering for crop improvement under drought stress. Bioengineered 2021, 12, 5814–5829. [Google Scholar] [CrossRef]
- Nowicka, B.; Kruk, J. Genetic engineering as a method for the improvement of photosynthesis. Postepy. Biochem. 2018, 64, 13–20. [Google Scholar] [CrossRef]
- Xia, W.; Liu, X.; Xin, H.; Wu, X.; Liu, R.; Li, J.; Zhu, J. Saussurea involucrata PIP2;7 improves photosynthesis and drought resistance by decreasing the stomatal density and increasing intracellular osmotic pressure. Environ. Exp. Bot. 2021, 191, 10406. [Google Scholar] [CrossRef]
- Zandalinas, S.I.; Fichman, Y.; Devireddy, A.R.; Sengupta, S.; Azad, R.K.; Mittler, R. Systemic signaling during abiotic stress combination in plants. Proc. Natl. Acad. Sci. USA 2020, 117, 13810–13820. [Google Scholar] [CrossRef]
- Battaglia, M.; Covarrubias, A.A. Late Embryogenesis Abundant (LEA) proteins in legumes. Front. Plant Sci. 2013, 4, 190. [Google Scholar] [CrossRef]
- Xu, M.; Tong, Q.; Wang, Y.; Wang, Z.; Xu, G.; Elias, G.K.; Li, S.; Liang, Z. Transcriptomic Analysis of the Grapevine LEA Gene Family in Response to Osmotic and Cold Stress Reveals a Key Role for VamDHN3. Plant Cell Physiol. 2020, 61, 775–786. [Google Scholar] [CrossRef]
- Wu, X.; Gong, F.; Yang, L.; Hu, X.; Tai, F.; Wang, W. Proteomic analysis reveals differential accumulation of small heat shock proteins and late embryogenesis abundant proteins between ABA-deficient mutant vp5 seeds and wild-type Vp5 seeds in maize. Front. Plant Sci. 2014, 5, 801. [Google Scholar] [CrossRef] [PubMed]
- Zamora-Briseño, J.A.; de Jiménez, E.S. A LEA 4 protein up-regulated by ABA is involved in drought response in maize roots. Mol. Biol. Rep. 2016, 43, 221–228. [Google Scholar] [CrossRef] [PubMed]
- Abdul Aziz, M.; Sabeem, M.; Mullath, S.K.; Brini, F.; Masmoudi, K. Plant Group II LEA Proteins: Intrinsically Disordered Structure for Multiple Functions in Response to Environmental Stresses. Biomolecules 2021, 11, 1662. [Google Scholar] [CrossRef] [PubMed]
- Hong-Bo, S.; Zong-Suo, L.; Ming-An, S. LEA proteins in higher plants: Structure, function, gene expression and regulation. Colloids Surf. B Biointerfaces 2005, 45, 131–135. [Google Scholar] [CrossRef]
- Mowla, S.B.; Cuypers, A.; Driscoll, S.P.; Kiddle, G.; Thomson, J.; Foyer, C.H.; Theodoulou, F.L. Yeast complementation reveals a role for an Arabidopsis thaliana late embryogenesis abundant (LEA)-like protein in oxidative stress tolerance. Plant J. 2006, 48, 743–756. [Google Scholar] [CrossRef]
- Guo, X.; Zhang, L.; Wang, X.; Zhang, M.; Xi, Y.; Wang, A.; Zhu, J. Overexpression of Saussurea involucrata dehydrin gene SiDHN promotes cold and drought tolerance in transgenic tomato plants. PLoS ONE 2019, 14, e0225090. [Google Scholar] [CrossRef]
- Magwanga, R.O.; Lu, P.; Kirungu, J.N.; Dong, Q.; Hu, Y.; Zhou, Z.; Cai, X.; Wang, X.; Hou, Y.; Wang, K.; et al. Cotton Late Embryogenesis Abundant (LEA2) Genes Promote Root Growth and Confer Drought Stress Tolerance in Transgenic Arabidopsis thaliana. G3 (Bethesda) 2018, 8, 2781–2803. [Google Scholar] [CrossRef]
- Zeng, X.; Ling, H.; Yang, J.; Li, Y.; Guo, S. LEA proteins from Gastrodia elata enhance tolerance to low temperature stress in Escherichia coli. Gene 2018, 646, 136–142. [Google Scholar] [CrossRef]
- Gong, G.; Huang, J.; Yang, Y.; Qi, B.; Han, G.; Zheng, Y.; He, H.; Chan, K.; Tsim, K.W.; Dong, T.T. Saussureae Involucratae Herba (Snow Lotus): Review of Chemical Compositions and Pharmacological Properties. Front. Pharmacol. 2019, 10, 1549. [Google Scholar] [CrossRef]
- Kuo, C.L.; Agrawal, D.C.; Chang, H.C.; Chiu, Y.T.; Huang, C.P.; Chen, Y.L.; Huang, S.H.; Tsay, H.S. In vitro culture and production of syringin and rutin in Saussurea involucrata (Kar. et Kir.)—An endangered medicinal plant. Bot. Stud. 2015, 56, 12. [Google Scholar] [CrossRef]
- Wang, J.; Hu, H.; Wang, W.; Wei, Q.; Hu, T.; Bao, C. Genome-Wide Identification and Functional Characterization of the Heat Shock Factor Family in Eggplant (Solanum melongena L.) under Abiotic Stress Conditions. Plants 2020, 9, 915. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Liu, H.; Xia, W.; Mu, J.; Feng, Y.; Liu, R.; Yan, P.; Wang, A.; Lin, Z.; Guo, Y.; et al. De Novo Transcriptome Sequencing and the Hypothetical Cold Response Mode of Saussurea involucrata in Extreme Cold Environments. Int. J. Mol. Sci. 2017, 18, 1155. [Google Scholar] [CrossRef] [PubMed]
- Chung, S.W.; Rho, H.; Lim, C.K.; Jeon, M.K.; Kim, S.; Jang, Y.J.; An, H.J. Photosynthetic response and antioxidative activity of ’Hass’ avocado cultivar treated with short-term low temperature. Sci. Rep. 2022, 12, 11593. [Google Scholar] [CrossRef] [PubMed]
- Dong, S.; Jiang, Y.; Dong, Y.; Wang, L.; Wang, W.; Ma, Z.; Yan, C.; Ma, C.; Liu, L. A study on soybean responses to drought stress and rehydration. Saudi J. Biol. Sci. 2019, 26, 2006–2017. [Google Scholar] [CrossRef]
- Dai, L.; Feng, Z.; Pan, X.; Xu, Y.; Li, P.; Lefohn, A.S.; Harmens, H.; Kobayashi, K. Increase of apoplastic ascorbate induced by ozone is insufficient to remove the negative effects in tobacco, soybean and poplar. Environ. Pollut. 2019, 245, 380–388. [Google Scholar] [CrossRef]
- Cheng, L.; Li, X.; Huang, X.; Ma, T.; Liang, Y.; Ma, X.; Peng, X.; Jia, J.; Chen, S.; Chen, Y.; et al. Overexpression of sheepgrass R1-MYB transcription factor LcMYB1 confers salt tolerance in transgenic Arabidopsis. Plant Physiol. Biochem. 2013, 70, 252–260. [Google Scholar] [CrossRef]
- Batool, T.; Ali, S.; Seleiman, M.F.; Naveed, N.H.; Ali, A.; Ahmed, K.; Abid, M.; Rizwan, M.; Shahid, M.R.; Alotaibi, M.; et al. Plant growth promoting rhizobacteria alleviates drought stress in potato in response to suppressive oxidative stress and antioxidant enzymes activities. Sci. Rep. 2020, 10, 16975. [Google Scholar] [CrossRef]
- Yoshida, T.; Yamaguchi-Shinozaki, K. Metabolic engineering: Towards water deficiency adapted crop plants. J. Plant Physiol. 2021, 258–259, 153375. [Google Scholar] [CrossRef]
- Xia, X.; Cheng, X.; Li, R.; Yao, J.; Li, Z.; Cheng, Y. Advances in application of genome editing in tomato and recent development of genome editing technology. Theor. Appl. Genet. 2021, 134, 2727–2747. [Google Scholar] [CrossRef]
- Hefferon, K. Plant biotechnology patents: Applications in agriculture and medicine. Recent. Pat. Biotechnol. 2010, 4, 136–152. [Google Scholar] [CrossRef]
- Kumar, V.; Khare, T.; Sharma, M.; Wani, S.H. Engineering Crops for the Future: A Phosphoproteomics Approach. Curr. Protein. Pept. Sci. 2018, 19, 413–426. [Google Scholar] [CrossRef] [PubMed]
- Geng, W.; Wang, Y.; Zhang, J.; Liu, Z.; Chen, X.; Qin, L.; Yang, L.; Tang, H. Genome-wide identification and expression analyses of late embryogenesis abundant (LEA) gene family in tobacco (Nicotiana tabacum L.) reveal their function in abiotic stress responses. Gene 2022, 836, 146665. [Google Scholar] [CrossRef]
- Li, Q.; Wang, M.; Fang, L. BASIC PENTACYSTEINE2 negatively regulates osmotic stress tolerance by modulating LEA4-5 expression in Arabidopsis thaliana. Plant Physiol. Biochem. 2021, 168, 373–380. [Google Scholar] [CrossRef]
- Mareri, L.; Parrotta, L.; Cai, G. Environmental Stress and Plants. Int. J. Mol. Sci. 2022, 23, 5416. [Google Scholar] [CrossRef] [PubMed]
- Dure, L., 3rd; Greenway, S.C.; Galau, G.A. Developmental biochemistry of cottonseed embryogenesis and germination: Changing messenger ribonucleic acid populations as shown by in vitro and in vivo protein synthesis. Biochemistry 1981, 20, 4162–4168. [Google Scholar] [CrossRef] [PubMed]
- Popova, A.V.; Rausch, S.; Hundertmark, M.; Gibon, Y.; Hincha, D.K. The intrinsically disordered protein LEA7 from Arabidopsis thaliana protects the isolated enzyme lactate dehydrogenase and enzymes in a soluble leaf proteome during freezing and drying. Biochim. Biophys. Acta. 2015, 1854, 1517–1525. [Google Scholar] [CrossRef] [PubMed]
- He, C.; Liu, X.; Teixeira da Silva, J.A.; Wang, H.; Peng, T.; Zhang, M.; Si, C.; Yu, Z.; Tan, J.; Zhang, J.; et al. Characterization of LEA genes in Dendrobium officinale and one Gene in induction of callus. J. Plant. Physiol. 2021, 258–259, 153356. [Google Scholar] [CrossRef]
- Shi, K.; Gao, Z.; Shi, T.Q.; Song, P.; Ren, L.J.; Huang, H.; Ji, X.J. Reactive Oxygen Species-Mediated Cellular Stress Response and Lipid Accumulation in Oleaginous Microorganisms: The State of the Art and Future Perspectives. Front. Microbiol. 2017, 8, 793. [Google Scholar] [CrossRef]
- Bobrovskikh, A.; Zubairova, U.; Kolodkin, A.; Doroshkov, A. Subcellular compartmentalization of the plant antioxidant system: An integrated overview. Peer J. 2020, 8, e9451. [Google Scholar] [CrossRef]
- Bouzroud, S.; Gasparini, K.; Hu, G.; Barbosa, M.A.M.; Rosa, B.L.; Fahr, M.; Bendaou, N.; Bouzayen, M.; Zsögön, A.; Smouni, A.; et al. Down Regulation and Loss of Auxin Response Factor 4 Function Using CRISPR/Cas9 Alters Plant Growth, Stomatal Function and Improves Tomato Tolerance to Salinity and Osmotic Stress. Genes 2020, 11, 272. [Google Scholar] [CrossRef]
- Furlan, A.L.; Bianucci, E.; Giordano, W.; Castro, S.; Becker, D.F. Proline metabolic dynamics and implications in drought tolerance of peanut plants. Plant Physiol. Biochem. 2020, 151, 566–578. [Google Scholar] [CrossRef] [PubMed]
- Lin, Z.; Wang, Y.L.; Cheng, L.S.; Zhou, L.L.; Xu, Q.T.; Liu, D.C.; Deng, X.Y.; Mei, F.Z.; Zhou, Z.Q. Mutual regulation of ROS accumulation and cell autophagy in wheat roots under hypoxia stress. Plant Physiol. Biochem. 2021, 158, 91–102. [Google Scholar] [CrossRef]
- Naliwajski, M.; Skłodowska, M. The Relationship between the Antioxidant System and Proline Metabolism in the Leaves of Cucumber Plants Acclimated to Salt Stress. Cells 2021, 10, 609. [Google Scholar] [CrossRef] [PubMed]
- He, H.; He, L.F. Regulation of gaseous signaling molecules on proline metabolism in plants. Plant Cell Rep. 2018, 37, 387–392. [Google Scholar] [CrossRef]
- Nguyen, Q.H.; Vu, L.T.K.; Nguyen, L.T.N.; Pham, N.T.T.; Nguyen, Y.T.H.; Le, S.V.; Chu, M.H. Overexpression of the GmDREB6 gene enhances proline accumulation and salt tolerance in genetically modified soybean plants. Sci. Rep. 2019, 9, 19663. [Google Scholar] [CrossRef]
- Ban, Q.; Liu, G.; Wang, Y. A DREB gene from Limonium bicolor mediates molecular and physiological responses to copper stress in transgenic tobacco. J. Plant Physiol. 2011, 168, 449–458. [Google Scholar] [CrossRef]
- Wang, W.; Gao, T.; Chen, J.; Yang, J.; Huang, H.; Yu, Y. The late embryogenesis abundant gene family in tea plant (Camellia sinensis): Genome-wide characterization and expression analysis in response to cold and dehydration stress. Plant Physiol. Biochem. 2019, 135, 277–286. [Google Scholar] [CrossRef]
- Shibuya, T.; Itai, R.; Maeda, M.; Kitashiba, H.; Isuzugawa, K.; Kato, K.; Kanayama, Y. Characterization of PcLEA14, a Group 5 Late Embryogenesis Abundant Protein Gene from Pear (Pyrus communis). Plants 2020, 9, 1138. [Google Scholar] [CrossRef]
- Mittler, R.; Vanderauwera, S.; Gollery, M.; Van Breusegem, F. Reactive oxygen gene network of plants. Trends Plant Sci. 2004, 9, 490–498. [Google Scholar] [CrossRef]
- Vessal, S.; Siddique, K.H.; Atkins, C.A. Comparative proteomic analysis of genotypic variation in germination and early seedling growth of chickpea under suboptimal soil-water conditions. J. Proteome Res. 2012, 11, 4289–4307. [Google Scholar] [CrossRef]
- Khurana, P.; Vishnudasan, D.; Chhibbar, A.K. Genetic approaches towards overcoming water deficit in plants-special emphasis on LEAs. Physiol. Mol. Biol. Plants 2008, 14, 277–298. [Google Scholar] [CrossRef]
- Lv, A.; Wen, W.; Fan, N.; Su, L.; Zhou, P.; An, Y. Dehydrin MsDHN1 improves aluminum tolerance of alfalfa (Medicago sativa L.) by affecting oxalate exudation from root tips. Plant J. 2021, 108, 441–458. [Google Scholar] [CrossRef]
- Du, B.; Zhao, W.; An, Y.; Li, Y.; Zhang, X.; Song, L.; Guo, C. Overexpression of an alfalfa glutathione S-transferase gene improved the saline-alkali tolerance of transgenic tobacco. Biol. Open 2019, 8, bio043505. [Google Scholar] [CrossRef]
- Rubio, S.; Noriega, X.; Pérez, F.J. Abscisic acid (ABA) and low temperatures synergistically increase the expression of CBF/DREB1 transcription factors and cold-hardiness in grapevine dormant buds. Ann. Bot. 2019, 123, 681–689. [Google Scholar] [CrossRef]
- Li, M.; Duan, X.; Gao, G.; Liu, T.; Qi, H. Running title: ABA pathway meets CBF pathway at CmADC. Hortic. Res. 2022, 9, uhac002. [Google Scholar] [CrossRef]
- Xie, Z.; Lin, W.; Yu, G.; Cheng, Q.; Xu, B.; Huang, B. Improved cold tolerance in switchgrass by a novel CCCH-type zinc finger transcription factor gene, PvC3H72, associated with ICE1-CBF-COR regulon and ABA-responsive genes. Biotechnol. Biofuels 2019, 12, 224. [Google Scholar] [CrossRef]
- Vyse, K.; Schaarschmidt, S.; Erban, A.; Kopka, J.; Zuther, E. Specific CBF transcription factors and cold-responsive genes fine-tune the early triggering response after acquisition of cold priming and memory. Physiol. Plant 2022, 174, e13740. [Google Scholar] [CrossRef]
- Morran, S.; Eini, O.; Pyvovarenko, T.; Parent, B.; Singh, R.; Ismagul, A.; Eliby, S.; Shirley, N.; Langridge, P.; Lopato, S. Improvement of stress tolerance of wheat and barley by modulation of expression of DREB/CBF factors. Plant Biotechnol. J. 2011, 9, 230–249. [Google Scholar] [CrossRef]
- Feng, H.L.; Ma, N.N.; Meng, X.; Zhang, S.; Wang, J.R.; Chai, S.; Meng, Q.W. A novel tomato MYC-type ICE1-like transcription factor, SlICE1a, confers cold, osmotic and salt tolerance in transgenic tobacco. Plant Physiol. Biochem. 2013, 73, 309–320. [Google Scholar] [CrossRef]
- Ali, N.; Hadi, F. CBF/DREB transcription factor genes play role in cadmium tolerance and phytoaccumulation in Ricinus communis under molybdenum treatments. Chemosphere 2018, 208, 425–432. [Google Scholar] [CrossRef]
- Cao, S.; Wang, Y.; Li, X.; Gao, F.; Feng, J.; Zhou, Y. Characterization of the AP2/ERF Transcription Factor Family and Expression Profiling of DREB Subfamily under Cold and Osmotic Stresses in Ammopiptanthus nanus. Plants 2020, 9, 455. [Google Scholar] [CrossRef]
- Hassan, S.; Berk, K.; Aronsson, H. Evolution and identification of DREB transcription factors in the wheat genome: Modeling, docking and simulation of DREB proteins associated with salt stress. J. Biomol. Struct. Dyn. 2021, 40, 7191–7204. [Google Scholar] [CrossRef]
- Wang, R.; Yin, J.; Li, S.; Zhang, W.; Zhang, J.; Dong, Z.; Li, X. Crop Cultivation Science, 2nd ed.; Higher Education Press: Beijing, China, 2015; p. 15. ISBN 978-7-04-041881-1. [Google Scholar]
- Nuccio, M.L.; Potter, L.; Stiegelmeyer, S.M.; Curley, J.; Cohn, J.; Wittich, P.E.; Tan, X.; Davis, J.; Ni, J.; Trullinger, J.; et al. Strategies and tools to improve crop productivity by targeting photosynthesis. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2017, 372, 1730. [Google Scholar] [CrossRef]
- Zhang, Y.; Kaiser, E.; Marcelis, L.F.M.; Yang, Q.; Li, T. Salt stress and fluctuating light have separate effects on photosynthetic acclimation, but interactively affect biomass. Plant Cell Environ. 2020, 43, 2192–2206. [Google Scholar] [CrossRef]
- Chen, X.; Ma, J.; Wang, X.; Lu, K.; Liu, Y.; Zhang, L.; Peng, J.; Chen, L.; Yang, M.; Li, Y.; et al. Functional modulation of an aquaporin to intensify photosynthesis and abrogate bacterial virulence in rice. Plant J. 2021, 108, 330–346. [Google Scholar] [CrossRef]
- Karpinska, B.; Razak, N.; Shaw, D.S.; Plumb, W.; Van De Slijke, E.; Stephens, J.; De Jaeger, G.; Murcha, M.W.; Foyer, C.H. Late Embryogenesis Abundant (LEA)5 Regulates Translation in Mitochondria and Chloroplasts to Enhance Growth and Stress Tolerance. Front. Plant Sci. 2022, 13, 875799. [Google Scholar] [CrossRef]
- Nowicka, B.; Ciura, J.; Szymańska, R.; Kruk, J. Improving photosynthesis, plant productivity and abiotic stress tolerance-current trends and future perspectives. J. Plant Physiol. 2018, 231, 415–433. [Google Scholar] [CrossRef]
- Juszczak, I.; Bartels, D. LEA gene expression, RNA stability and pigment accumulation in three closely related Linderniaceae species differing in desiccation tolerance. Plant Sci. 2017, 255, 59–71. [Google Scholar] [CrossRef]
- Snowdon, R.J.; Wittkop, B.; Chen, T.W.; Stahl, A. Crop adaptation to climate change as a consequence of long-term breeding. Theor. Appl. Genet. 2021, 134, 1613–1623. [Google Scholar] [CrossRef]
- García-Estrada, R.S.; Diaz-Lara, A.; Aguilar-Molina, V.H.; Tovar-Pedraza, J.M. Viruses of Economic Impact on Tomato Crops in Mexico: From Diagnosis to Management-A Review. Viruses 2022, 14, 1251. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, X.; Xia, W.; Zhang, X.; Li, A.; Qin, J.; Sun, H.; Li, J.; Zhu, J. Overexpression of the SiLEA5 Gene in Saussurea involucrata Increases the Low-Temperature Tolerance of Transgenic Tomatoes. Horticulturae 2022, 8, 1023. https://doi.org/10.3390/horticulturae8111023
Liu X, Xia W, Zhang X, Li A, Qin J, Sun H, Li J, Zhu J. Overexpression of the SiLEA5 Gene in Saussurea involucrata Increases the Low-Temperature Tolerance of Transgenic Tomatoes. Horticulturae. 2022; 8(11):1023. https://doi.org/10.3390/horticulturae8111023
Chicago/Turabian StyleLiu, Xiaoyan, Wenwen Xia, Xiaoli Zhang, Aowei Li, Jiawang Qin, Huili Sun, Jin Li, and Jianbo Zhu. 2022. "Overexpression of the SiLEA5 Gene in Saussurea involucrata Increases the Low-Temperature Tolerance of Transgenic Tomatoes" Horticulturae 8, no. 11: 1023. https://doi.org/10.3390/horticulturae8111023
APA StyleLiu, X., Xia, W., Zhang, X., Li, A., Qin, J., Sun, H., Li, J., & Zhu, J. (2022). Overexpression of the SiLEA5 Gene in Saussurea involucrata Increases the Low-Temperature Tolerance of Transgenic Tomatoes. Horticulturae, 8(11), 1023. https://doi.org/10.3390/horticulturae8111023





