Abstract
‘Huangguan’ pear (Pyrus bretschneideri Rehd. cv. Huangguan) fruit is sensitive to chilling injury (CI), which exhibits peel browning spots (PBS) during cold storage. Dehydrin (DHN) is considered to be related to cold tolerance in plants, but its function in postharvest pear fruit during storage remains unclear. In this study, six PbDHNs (PbDHN1–6) genes were identified and characterized, and the PbDHN proteins were sorted into YnKn, SKn and YnSKn according to the major conserved motifs related to the number and location of K-segments, S-segments, and Y-segments. In addition, there were five cold-responsive related cis-acting elements in the promoter region of the PbDHNs. The analysis of fruit quality suggested that compared with a storage temperature at 20 °C, a storage temperature of 0 °C results in CI in ‘Huangguan’ pear fruit, while a storage temperature of 10 °C and low temperature conditioning (LTC) alleviates the CI. Moreover, gene expression results indicated that the six PbDHNs were markedly enhanced at low temperatures, especially at 0 °C. The transcripts of PbDHN1, PbDHN4, PbDHN5 and PbDHN6 were also increased in the fruit stored at 10 °C, but they were lower than that at 0 °C except PbDHN5. Compared with low temperature storage at 0 °C, LTC treatment significantly depressed the expression of PbDHN1, PbDHN2, PbDHN3, PbDHN4, and PbDHN6, while enhanced the mRNA amount of PbDHN5. In conclusion, PbDHN1, PbDHN4, PbDHN5, and PbDHN6 were closely related to the CI, and LTC lowered the CI by down-regulating the expression of PbDHN1, PbDHN4, and PbDHN6 and by up-regulating PbDHN5 in ‘Huangguan’ pear fruit.
1. Introduction
‘Huangguan’ (Pyrus bretschneideri Rehd. cv. Huangguan) is one of the most important cultivars of pear in China, and it is favored by consumers because of the regular shape and excellent quality. However, ‘Huangguan’ pear fruit is prone to PBS, which is a CI phenomenon that develops during cold storage [1,2,3], which results in the loss of fruit commodity value.
Dehydrin (DHN) is a class of hydrophilic, thermostable stress-responsive proteins that is widely present in plants [4]. DHN is characterized depending on the presence of three major conserved motifs (K-, Y-, and S-segments). The K-segment has a conserved lysine-rich 15-amino acid motif (EKKGIMDKIKEKLPG), which is usually located at the C-terminal region of DHN. The known DHNs contain at least one K-segment. These K-segments can form a putative amphiphilic α-helix structure that can interact with both membranes and denatured proteins, maintaining the stability of the membranes to protect cell function when the plant has suffered from multiple abiotic stresses, such as osmotic, drought, salinity and low temperature stresses [5,6,7,8,9,10,11,12]. Many DHNs also contain one or more Y-segments (DEYGNP) located at the N-terminus; these segments display amino acid identity at the nucleotide binding site motif of chaperone proteins. The S-segment contains multiple serine residues that can be modified through phosphorylation, which may be involved in ion-binding [13]. According to the number and distribution of the three conserved motifs, the DHNs are divided into five subclasses: Kn, YnKn, KnS, SKn, and YnSKn [14].
The function of DHNs involved in the response to cold stress has been widely studied [3,12,13,14,15,16,17,18,19,20]. In Arabidopsis, the transcripts of the four characterized DHN genes of AtRAB18 (Responsive to ABA 18), AtCOR47 (Cold-regulated 47), AtLTI29 (Low temperature-induced 29), and AtLTI30 (Low temperature-induced 30) accumulate in response to low temperature. Moreover, co-overexpressing AtRAB18 and AtCOR47 or co-overexpressing AtLTI29 and AtLTI30 could significantly improve the cold tolerance of the Arabidopsis seedling [21,22]. Twelve DHN genes (MdDHNs) were identified in apple (Malus domestica), and four DHNs in leaves, MdDHN2, MdDHN4, MdDHN6, and MdDHN8, were found to be strongly induced by cold treatment (4 °C) [23]. The grapevine (Vitis vinifera) DHN genes family comprises four divergent members (VvDHN1–4), although only VvDHN1 and VvDHN2 appeared to respond to cold treatment [24]. Freezing treatment (−3 °C) resulted in the transcripts of all the seven EjDHNs (EjDHN1–7) increasing in loquat (Eriobotrya japonica) fruit [25]. In citrus unshiu, the DHN gene CuCOR19 (Cold-regulated 19) was found to be cold-responsive; furthermore, over-expression of CuCOR19 could prevent lipid peroxidation and subsequently enhance cold tolerance in transgenic tobacco [26]. However, the function of DHNs on the CI development of the postharvest fruit is still little known.
In this study, to clarify the members of DHNs family and their role in the CI development in ‘Huangguan’ pear fruit under low temperature storage, six DHN genes were identified and their expression patterns subsequently analyzed under different storage temperatures.
2. Materials and Methods
2.1. Materials and Treatments
‘Huangguan’ pear fruit with paper-bagging were harvested at commercial maturity (average single fruit weight: 317.20 ± 35.45 g; soluble solid content: 12.16 ± 0.19%; starch content: 1.88 g/kg; firmness: 68.22 ± 3.50 N) from an orchard located in Jinzhou City (115 04′34.80″ E, 38 03′13.50″ N), Hebei Province, and then immediately transported to the laboratory within 2 h. After being removed from the paper bag, the pear fruit with uniform weight and shape without visible defects were selected as materials, and the treatments were carried out as follows: The fruit were divided into four groups. The first, second and third groups were directly stored at 20 ± 1 °C (20 °C), 10 ± 0.5 °C (10 °C), and 0 ± 0.5 °C (0 °C), respectively, while the fourth group was first stored at 10 ± 0.5 °C for 3 days and then transferred to 0 ± 0.5 °C and marked as low temperature conditioning (LTC).
For all treatments, the CI was observed at 0, 3, 5, 10, 15, 20, and 30 days of storage, and then the peel samples were quickly frozen in liquid nitrogen and stored at −80 °C until used. Three replicates were performed for each treatment with 10 fruit per replicate.
2.2. Cloning of PbDHN Genes
Total RNA was extracted from the peel sample by the improved CTAB method [27]. After digestion by DNAse-I, the first strand cDNA was synthesized using the AMV reverse transcription kit (TaKaRa Biomedicals, Dalian, China) according to the manufacturer’s instruction. Primers for cloning of DHNs were designed based on the genomic sequences of Pyrus bretschneideri (https://www.ncbi.nlm.nih.gov/, accessed on 15 September 2019) using the Omega 2.0 software. The primer sequences are listed in Table 1.

Table 1.
Sequence of primers for the PbDNHs cloning.
2.3. Multiple Sequence Alignment, Gene Structure Construction, and Phylogenetic Analysis
The exon-intron structures of the PbDHNs were determined from the alignments of cDNA and pear (Pyrus bretschneideri) genomic sequences using the software GSDS 2.0 (http://gsds.cbi.pku.edu.cn, accessed on 25 October 2019). The protein molecular weight and isoelectric point of the six isolated PbDHN proteins were predicted using the ProtParam program (http://au.expasy.org/tools/protparam. html, accessed on 23 October 2019) based on their amino acid compositions. The deduced amino acid sequences were aligned using the software Clustal X. The DHN proteins sequences of arabidopsis (Arabidopsis thaliana) were obtained from the TAIR website (http://www.arabidopsis.org, accessed on 6 November 2019): AtCOR47 (At1g20400), AtLTI29 (At1g20450), AtHIRD11 (At1g54410), AtERD14 (At1g76180), AtLTI30 (At3g50970), AtXERO1 (At3g50980), AtRAB18 (At5g66400), At2G21490, At4G38410, and At4G39130. DHNs sequences of peach (Prunus persica) were downloaded from the GDR database (http://www.rosaceae.org, accessed on 19 November 2019): PpDHN1 (ppa005514m), PpDHN2 (ppa011637m), PpDHN3 (ppa010326m), PpDHN4 (ppa026861m), PpDHN5 (ppa010975m), and PpDHN6 (ppa009997m). DHNs sequences of apple (Malus domestica), loquat (Eriobotrya japonica), grape (Vitus yeshanensis), Chinese plum (Prunus mume), sorghum (Sorghum bicolor), pepper (Capsicum annuum), and tomato (Solanum lycopersicum) were downloaded from the NCBI database (http://www.ncbi.nlm.nih.gov, accessed on 20 November 2019). Apple: MdDHN1 (JQ649456), MdDHN2 (JQ649457), MdDHN3 (JQ649458), MdDHN4 (JQ649459), MdDHN5 (JQ649460), MdDHN6 (JQ649461), MdDHN7 (JQ649462), MdDHN8 (JQ649463), and MdDHN9 (JQ649464). Loquat: EjDHN1 (FJ472835), EjDHN2 (FJ472836), EjDHN3 (KF277187), EjDHN4 (KF277188), EjDHN5 (KF277189), EjDHN6 (KF277190), and EjDHN7 (KF277191). Grape: VyDHN1 (JF900497), VyDHN2 (JQ408442), VyDHN3 (JQ408443), and VyDHN4 (JQ408444). Chinese plum: PmLEA8 (XM_016796210.1), PmLEA10 (XM_008243512.2), PmLEA19 (XM_008237879.1), PmLEA20 (XM_008238721.1), and PmLEA29 (XM_008222942.1). Sorghum: SbDHN1 (KT865881) and SbDHN2 (KT780443). Pepper: CaDHN1 (JX402924). Tomato: SlDHN (KF585024.1). Based on the result of multiple alignment, a phylogenetic tree was generated using the MEGA 5.0 software (http://www.megasoftware.net/mega.html, accessed on 11 December 2019) using a neighbor-joining method with 1000 bootstrap replicates.
2.4. Prediction of Cis-Acting Elements in Promoter Region
The promoter sequences representing approximately 1500 bp upstream of the translation start codon of PbDHNs were obtained from the NCBI database (https://www.ncbi.nlm.nih.gov/, accessed on 10 January 2020), and the putative cis-acting elements in the upstream acting regions of the PbDHNs were scanned using the PlantCARE database (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/, accessed on 12 January 2020).
2.5. Fruit Quality and Estimation of CI
The fruit firmness was measured using the GY-4 digital fruit hardness meter (TOP Instruments Co., Hangzhou, Zhejiang, China) at two equidistant points on the equatorial region with the skin removed. The soluble solids content (SSC) was measured using a PAL-1 pocket digital refractometer (ATAGO Co., Ltd., Tokyo, Japan). Titratable acidity (TA) was determined using a base buret by titration with 0.01 mol/L NaOH up to pH 8.1, and the TA content was expressed as malic acid (%).
The CI index was evaluated according to the PBS score that was described by a previous report [3]: 0 (no peel browning spots), 1 (area of the peel browning spots < 25%), 2 (area of the peel browning spots ≥ 25% but < 50%), and 3 (area of the peel browning spots ≥ 50%). The CI index was calculated using the following formula:
CI index = Σ (peel browning spots scale × number of fruit at the scale level)/[3 × (total number of fruit)]
2.6. Quantitative RT-PCR (qRT-PCR) Analysis
The First-strand cDNA was synthesized using the PrimeScriptRT Reagent Kit with gDNA Eraser (TaKaRa Biomedicals, Dalian, China) according to the manufacturer’s recommended protocol. The reactions contained 10.0 μL SYBR Green PCR Premix Ex Taq™ (TaKaRa Biomedicals, Dalian, China), 0.5 μL of ROX Reference DyeⅡ, 0.5 μL of forward and reverse primer (Each for 10 μmol/L), 5 ng of cDNA, and 6.0 μL of ddH2O. The qRT-PCR was performed using the ABI 7500 instrument (Applied Biosystems, Foster City, CA, USA), and the cycling parameters were as follows: 10 s at 95 °C, 40 cycles of 95 °C for 5 s, and 60 °C for 34 s. The primers of PbDHN genes for qRT-PCR were designed using the software of OMIGA 2.0, and they are listed in Table 2. The PbActin2 (PbACT2) was used to normalize the amount of gene-specific qRT-PCR products.

Table 2.
Primers of PbDHNs for qRT-PCR.
2.7. Statistical Analysis
The software GraphPad Prism 4 was used to prepare the figures. All values were expressed as mean ± SD. The results were statistically analyzed with ANOVA using SPSS 18 software (SPSS Inc., Chicago, IL, USA). Differences among treatments were assessed through Duncan’s multiple comparison test (p < 0.05).
3. Results
3.1. Identification and Classification of DHN Genes in ‘Huangguan’ Pear Fruit
Based on the RNA-Seq data of ‘Huangguan’ pear fruit (Data unpublished), six candidates PbDHNs with FPKM > 1 were selected for the present study. After being confirmed by RT-PCR cloning, sequencing, and aligned with the NCBI genome database, six PbDHNs with the length of CDS varied from 564 to 1245 bp were identified. The sequences of PbDHN1, PbDHN4, and PbDHN6 were completely consistent with three cold-shock protein CS120-like genes (XM_009347774.2, XM_009347773.1, and XM_009347771.2, respectively), PbDHN2 was identical with a dehydrin DHN2-like (XM_009347776.2), PbDHN3 was identical with a dehydrin DHN3-like (XM_00934778.1), and PbDHN5 was identical with COR47 (XM_009336070.1). In addition, the six PbDHNs were mapped to 2 of 17 pear chromosomes, and PbDHN1, PbDHN2, PbDHN3, PbDHN4, and PbDHN6 were located on Chr 2, while PbDHN5 was present on Chr 15. To verify whether these genes were dehydrins, the protein sequences of the six PbDHNs were aligned online (www.expasy.org/tools/, accessed on 23 October 2019) to predict the basic parameters, and all of the six PbDHNs contained the PF00257 (Dehydrin) conserved domain. The analysis of protein characteristic revealed that six deduced PbDHNs varied from 187 (PbDHN3) to 474 (PbDHN4) in amino acids length, the predicted molecular weights were between 19.77 kDa (PbDHN3) and 50.33 kDa (PbDHN4), and the isoelectric points ranged from 5.25 (PbDHN5) to 7.98 (PbDHN3) (Table 3).

Table 3.
Characteristics analysis of PbDHNs.
3.2. Multiple Sequence Alignment, Structure, and Phylogenetic Analysis of PbDHNs
The alignment was performed using Clustal X, indicating that the amino acid sequence of each dehydrin was quite different except for the domain Y-, K-, and S-segments. PbDHN2, PbDHN3, PbDHN5, and PbDHN6 contained three K-segments, PbDHN1 contained four K-segments, and PbDHN4 contained nine K-segments. With the exception of PbDHN1 that had no S-segment, the other five PbDHNs contained one S-segment. PbDHN1, PbDHN2, PbDHN4, and PbDHN6 included one Y-segment, while PbDHN3 had two Y-segments, and PbDHN5 did not contain Y-segments (Figure 1).

Figure 1.
Alignment of multiple amino acid sequences of PbDHNs in ‘Huangguan’ pear fruit. The sequences in black boxes indicate Y-segment, the sequences in the blue box indicate S-segment, the sequences in red boxes indicate K-segment, and the sequences in the yellow box indicate phosphorylation site.
The six PbDHNs were sorted into three classes by neighbor-joining analysis, including class I (YnSKn, containing PbDHN2, PbDHN3, PbDHN4, and PbDHN6), class II (YnKn, containing PbDHN1), and class III (SKn, containing PbDHN5) (Figure 2A). The intron-exon structure analysis was determined by aligning the cDNA to pear genomic sequences, and it was found that PbDHN1 contained only one exon, while the other five PbDHNs contained two exons and one intron (Figure 2B).
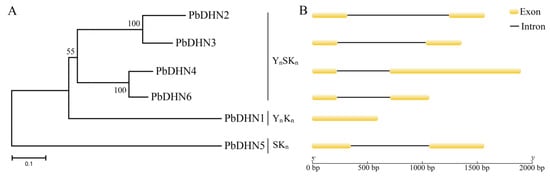
Figure 2.
Phylogenetic relationships (A) and gene structure (B). The phylogenetic tree of PbDHN proteins was performed using MEGA 5.0 software with the neighbor-joining method. The bootstrap test was performed with 1000 replicates. Percentage bootstrap scores of >50% were displayed. Exons were indicated by yellow round-corner rectangles and introns by black lines.
To study the phylogenetic relationships among different plant species, an unrooted phylogenetic tree based on sequence alignments of the DHN protein sequences of pear (Pyrus bretschneideri), apple (Malus domestica), peach (Prunus persica), Chinese plum (Prunus mume), loquat (Eriobotrya japonica), grape (Vitus yeshanensis), arabidopsis (Arabidopsis thaliana), sorghum (Sorghum bicolor), pepper (Capsicum annuum), and tomato (Solanum lycopersicum) was constructed to identify putative orthologs via MEGA5.0. The results revealed that DHN proteins from different species could be roughly divided into four groups: I, II, III, and IV. Group I included PbDHN5, two MdDHNs (MdDHN8 and MdDHN9), two EjDHNs (EjDHN2 and EjDHN5), two VyDHNs (VyDHN2 and VyDHN3), two PpDHNs (PpDHN3 and PpDHN4), two PmDHNs (PmLEA19 and PmLEA29), and three AtDHNs (AtCOR47, AtLTI29, and AtEDR14). Group II included two PbDHNs (PbDHN2 and PbDHN3), two MdDHNs (MdDHN1 and MdDHN7), one EjDHN (EjDHN1), one PpDHN (PpDHN2), and one PmDHN (PmLEA10). Group III included one PbDHN (PbDHN1), two MdDHNs (MdDHN5 and MdDHN6), two EjDHNs (EjDHN4 and EjDHN6), one PpDHN (PpDHN6), and one PmDHN (PmLEA8). Group IV included two PbDHNs (PbDHN4 and PbDHN6), three MdDHNs (MdDHN2, MdDHN3, and MdDHN4), two EjDHNs (EjDHN3 and EjDHN7), two VyDHNs (VyDHN1 and VyDHN4), two PpDHNs (PpDHN1 and PpDHN5), one PmDHN (PmLEA20), and two AtDHNs (AtRAB18 and AtXREO1) (Figure 3). The results of this phylogenetic analysis suggested that the six PbDHNs were closely related to similar such proteins of other plant species.
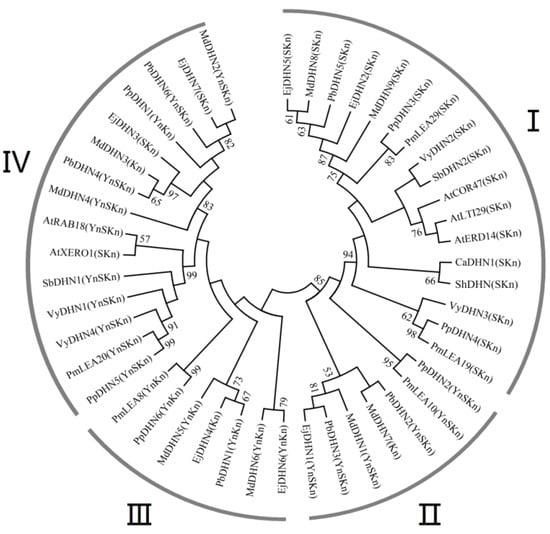
Figure 3.
Phylogenetic tree analysis of DHNs from different plant species. The unrooted phylogenetic tree was constructed using MEGA 5.0 with the neighbor-joining method. The bootstrap test was performed with 1000 replicates. Percentage bootstrap scores of >50% were displayed. vThe results revealed that DHN proteins from different species could be roughly divided into four groups: I, II, III, and IV.
3.3. Comparison of Cis-Acting Elements in the Promoter Region of PbDHNs
To reveal the regulation mechanism of PbDHNs, the promoter region, including 1500 bp of sequence upstream of the start codon of six PbDHNs, were scanned for the putative cis-acting elements using the PlantCARE database. The six PbDHNs mainly contained four sorts of cis-acting elements related to cold stress, including a ABA response element (ABRE/ACGTG), a drought response element (DRE/GCCGAC), MYB (C/TAACCA), and MYC (CAA/TT/GTG), as well as a ethylene response element (ERE/ATTTCATA). There were obvious differences in the abundance and distribution of cis-acting elements among the promoter regions of the six PbDHNs. The promoter region of PbDHN1 had six ABREs, three MYBs, and three MYCs. PbDHN2 contained one ABRE and six MYBs. PbDHN3 contained seven ABREs, one DRE, two EREs, three MYBs, and three MYCs. PbDHN4 contained three ABREs, two DREs, three MYBs, and two MYCs. PbDHN5 contained seven ABREs, two DREs, two EREs, two MYBs, and four MYCs, while PbDHN6 contained five ABREs, two DREs, two EREs, one MYB, and four MYCs (Figure 4).
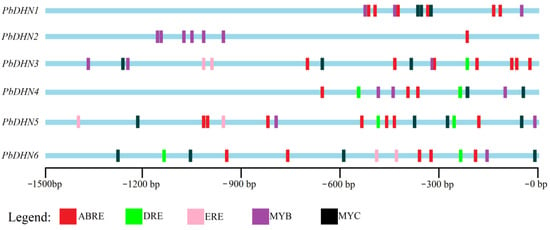
Figure 4.
Location of putative cis-acting elements in the promoter regions of PbDHNs. The promoter sequences were obtained from 1500 bp upstream of the translation start codon (ATG), and the putative cold stress-related cis-acting elements were presented.
3.4. The fruit Quality and CI Index
The firmness, SSC, and TA content were the important factors of fruit quality. After 30 days of storage, the firmness and TA content of pear fruit stored at 20 °C were significantly lower than the fruit stored at 0 °C with LTC treatment, indicating the degree of more advanced ripening when stored at 20 °C. The TA content of pear fruit stored at 10 °C was also lower than the fruit stored at 0 °C with LTC treatment, while the firmness changed slightly, suggesting that the quality of the fruit stored at 10 °C began to decline. There was no obvious difference in firmness, SSC, and TA content between the fruit stored at 0 °C with LTC treatment (Table 4).

Table 4.
Fruit quality of ‘Huangguan’ pear fruit with different treatments after 30 days of storage.
The CI indexes of ‘Huangguan’ pear fruit under different temperature treatments were quite different. Compared with fruit stored at 20 °C (no CI development), the fruit stored at 0 °C appeared CI at day 10, and then the CI index increased sharply, reaching the highest value (0.441) at day 30. Although the fruit stored under 10 °C developed CI at day 10, the CI index was much lower than the fruit stored under 0 °C, and the highest value of CI index was only was 0.053. Compared with the fruit stored under 0 °C, the LTC-treated fruit displayed markedly lower CI index after 10 days of storage, and the highest CI index value was 0.078 (Figure 5).
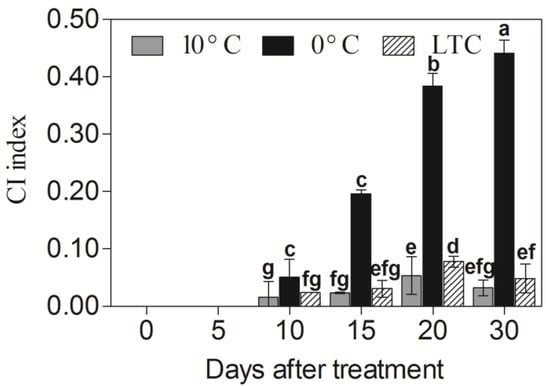
Figure 5.
Estimation of CI in ‘Huangguan’ pear fruit at different temperature storage. Data are means of three replicates, and error bars indicate SD. Different letters (a–g) above the bar chart indicated significant differences (p < 0.05) by Duncan’s multiple comparison test.
3.5. Expression Patterns of PbDHNs under Different Storage Temperatures
The expression profiles of the six PbDHNs were different when the fruit was stored at 0 °C, 10 °C, and 20 °C. When the fruit was stored at 20 °C, the expression profiles of PbDHN1, PbDHN2, and PbDHN5 increased at first, and then decreased (Figure 6A,E,Q). The expression level of PbDHN4 rapidly decreased when the fruit was stored at 20 °C, while the mRNA amounts of PbDHN3 and PbDHN6 changed slightly before day 20 of storage, and displayed higher expression level at day 30 (Figure 6I,M,U). When the fruit was stored at 10 °C, the expression levels of PbDHN1, PbDHN2, PbDHN4, and PbDHN6 rapidly increased at day 3 of storage, and then decreased, while the expression levels PbDHN3 and PbDHN5 gradually increased till 20 days of storage (Figure 6B,F,J,N,R,V). In addition, the mRNA amounts of PbDHN1, PbDHN4, PbDHN5, and PbDHN6 in the fruit stored at 10 °C were much higher than the fruit stored at 20 °C. The expression levels of the six PbDHNs in the fruit stored at 0 °C rapidly increased at day 3 of storage, among which PbDHN1, PbDHN2, and PbDHN4 reached the peak at day 15 of storage (Figure 6C,G,O), while PbDHN3, PbDHN5, and PbDHN6 increased gradually until 30 days of storage (Figure 6K,S,W). Moreover, the mRNA amounts of PbDHN1, PbDHN3, PbDHN4, and PbDHN6 in the fruit stored at 0 °C were much higher than the fruit stored at 20 °C and 10 °C. These results indicate that the six PbDHNs are low temperature-induced genes.
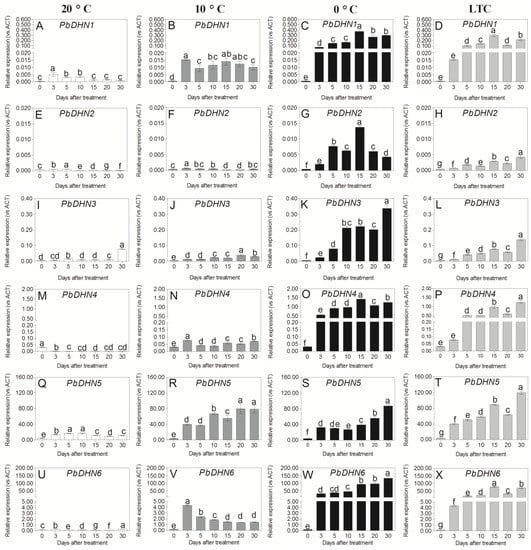
Figure 6.
Effects of 20 °C (A,E,I,M,Q,U), 10°C (B,F,J,N,R,V), 0°C (C,G,K,O,S,W) storage and LTC (D,H,L,P,T,X) treatments on the expression pattern of PbDHNs in the peel of ‘Huangguan’ pear fruit. Data are means of three biological replicates, and error bars indicate SD. Different letters (a–g) above the bar chart indicated significant differences (p < 0.05) by Duncan’s multiple comparison test.
3.6. Expression Patterns of PbDHNs under LTC Treatment
In the LTC-treated fruit, the expression levels of PbDHN1 and PbDHN6 markedly increased till 15 days of storage, and then decreased, while PbDHN2, PbDHN3, PbDHN4 and PbDHN5 increased until day 30 of storage. Compared with the fruit stored at 0 °C, the mRNA amount of PbDHN1, PbDHN2, PbDHN3, PbDHN4 and PbDHN6 were lower (Figure 6D,H,L,P,X), while PbDHN5 was higher (Figure 6T).
4. Discussion
DHNs can protect proteins and membranes from unfavorable structural changes. The K-segment especially is believed to plan an important role in cold stress by interacting with various cell components [5,7,27,28,29]. The structure of two K-fragments connecting with φ fragments is believed to have the function of resisting cold stress [30]. The DHN with cold-responsive function should contain at least three K-fragments and the acidic and neutral type DHN (Kn, SKn, and YnKn) preferentially accumulate in response to cold stress [31]. In the present study, the six DHNs cloned from ‘Huangguan’ pear fruit were classified as YnKn (PbDHN1), SKn (PbDHN5), and YnSKn (PbDHN2, PbDHN3, PbDHN5, and PbDHN6) type, respectively, all of which contained at least three K-segments (Figure 1 and Figure 2A), suggesting that they might be closely related to response to low temperature in ‘Huangguan’ pear fruit. Previous studies have confirmed that the cis-acting elements such as ABRE, DRE, MYB, and MYC were involved in the process of low temperature stress [4,32,33,34,35,36,37], and in this study the promoter regions of the six PbDHNs in ‘Huangguan’ pear fruit contained at least the ABRE and MYB cis-acting elements (Figure 4), so it further suggests that these PbDHNs were associated with the low temperature stress in ‘Huangguan’ pear fruit.
The expression profiles confirmed that the six PbDHNs of ‘Huangguan’ pear fruit were indeed induced by low temperature, among which PbDHN1, PbDHN2, PbDHN3, PbDHN4, and PbDHN6 were up-regulated under 0 °C, while the PbDHN5 was also induced by a moderate temperature treatment (10 °C) (Figure 6). It has been considered that the homologous genes of PbDHNs (Figure 3), such as the EjDHN6, MdDHN6, and PpDHN6 (homolog to PbDHN1), the EjDHN1 (homolog to PbDHN2 and PbDHN3), the VyDHN1, AtRAB18, MdDHN2, and MdDHN4 (homolog to PbDHN4 and PbDHN6), as well as the AtCOR47, AtLTI29, PmLEA19, and EjDHN5 (homolog to PbDHN5), were all involved in the cold stress response of plants [18,19,21,23,24,25,38]. Thus, it is suggested that the six PbDHNs participated in the cold response process of ‘Huangguan’ pear fruit.
The ‘Huangguan’ pear fruit easily suffers from CI when the fruit is directly stored at low temperature, while the LTC treatment significantly inhibits the occurrence of CI [1]. In this study, LTC treatment down-regulated the expression of the cold response genes such as PbDHN1, PbDHN2, PbDHN3, PbDHN4, and PbDHN6 (Figure 6D,H,L,P,X), indicating that the LTC treatment could desensitize or delay the response to low temperature stress in fruit in the early stages of cold storage, and it could make fruit more suitable for enduring long-term cold stress. In addition, LTC-treated fruit displayed greater transcript of PbDHN5 than the fruit at 0 °C storage, which was different from the other five PbDHNs (Figure 6T); therefore, the acting mechanism of PbDHN5 was different [39]. It has been known that over-expression of EjDHN5 and SbDHN2 (homolog to PbDHN5) in tobacco can improve the ability to resist low temperature stress [40]. Furthermore, in Arabidopsis thaliana, knocking out the PbDHN5 homologous gene LTI29 can decrease tolerance to cold stress in mutant seedlings [41]. Thus, up-regulation of PbDHN5 by LTC treatment might be beneficial in reducing CI in ‘Huangguan’ pear fruit during cold storage.
5. Conclusions
Six PbDHNs genes were cloned from ‘Huangguan’ pear fruit, and the six PbDHN proteins belonged to three types: YnKn, SKn and YnSKn. Low temperature (0 °C) easily resulted in CI and markedly up-regulated the expression levels of the six PbDHNs. LTC treatment reduced the occurrence of CI in ‘Huangguan’ pear fruit, decreased the expression levels of PbDHN1, PbDHN2, PbDHN3, PbDHN4, and PbDHN6, while increasing the expression level of PbDHN5. Above all, the six PbDHNs were low temperature-responsive genes; as such, they might be involved in the occurrence of CI under low temperature storage. LTC could deduce the CI by regulating PbDHN1, PbDHN4, PbDHN5, and PbDHN6 expression in ‘Huangguan’ pear fruit.
Author Contributions
Conceptualization, Y.C. and J.G.; methodology, Y.F.; software, J.H.; validation, Y.C., J.G., and J.Z.; formal analysis, J.Z.; investigation, Y.F.; resources, Y.C.; data curation, Y.C. and J.G.; writing—original draft preparation, Y.C.; writing—review and editing, J.G.; project administration, Y.C. and J.G. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Key R & D plan of Hebei Province (22327505D) and the HAAFS Agriculture Science and Technology Innovation (2022KJCXZX−SSS−3).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available within the article.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Li, D.; Cheng, Y.; Dong, Y.; Shang, Z.; Guan, J. Effects of low temperature conditioning on fruit quality and peel browning spot in ‘Huangguan’ pears during cold storage. Postharvest Biol. Technol. 2017, 13, 68–73. [Google Scholar] [CrossRef]
- Ma, Y.; Yang, M.; Wang, J.; Jiang, C.; Wang, Q. Application of exogenous ethylene inhibits postharvest peel browning of ‘Huangguan’ pear. Front. Plant Sci. 2017, 7, 2029. [Google Scholar] [CrossRef] [PubMed]
- Wei, C.; Ma, L.; Cheng, Y.; Guan, Y.; Guan, J. Exogenous ethylene alleviates chilling injury of ‘Huangguan’ pear by enhancing the proline content and antioxidant activity. Sci. Hortic. 2019, 257, 108671. [Google Scholar] [CrossRef]
- Allagulova, C.R.; Gimalov, F.R.; Shakirova, F.M.; Vakhitov, V.A. The plant dehydrins: Structure and putative functions. Biochemistry 2003, 68, 945–951. [Google Scholar]
- Eriksson, S.K.; Kutzer, M.; Procek, J.; Gröbner, G.; Harryson, P. Tunable membrane binding of the intrinsically disordered dehydrin Lti30, a cold-induced plant stress protein. Plant Cell 2011, 23, 2391–2404. [Google Scholar] [CrossRef]
- Hara, M. The multifunctionality of dehydrins: An overview. Plant Signal. Behav. 2010, 5, 503–508. [Google Scholar] [CrossRef]
- Graether, S.P.; Boddington, K.F. Disorder and function: A review of the dehydrin protein family. Front. Plant Sci. 2014, 5, 576. [Google Scholar] [CrossRef]
- Zhu, W.; Zhang, D.; Lu, X.; Zhang, L.; Yu, Z.; Lv, H.; Zhang, H. Characterisation of an SKn-type dehydrin promoter from wheat and its responsiveness to various abiotic and biotic stresses. Plant Mol. Biol. Rep. 2014, 32, 664–678. [Google Scholar] [CrossRef]
- Chen, R.; Jing, H.; Guo, W.; Wang, S.; Ma, F.; Pan, B.; Gong, Z. Silencing of dehydrin CaDHN1 diminishes tolerance to multiple abiotic stresses in Capsicum annuum L. Plant Cell Rep. 2015, 34, 2189–2200. [Google Scholar] [CrossRef]
- Liu, H.; Yu, C.; Li, H.; Ouyang, B.; Wang, T.; Zhang, J.; Wang, X.; Ye, Z. Overexpression of ShDHN, a dehydrin gene from Solanum habrochaites enhances tolerance to multiple abiotic stresses in tomato. Plant Sci. 2015, 231, 198–211. [Google Scholar] [CrossRef]
- Lv, A.; Su, L.; Liu, X.; Xing, Q.; Huang, B.; An, Y.; Zhou, P. Characterization of dehydrin protein, CdDHN4-L and CdDHN4-S, and their differential protective roles against abiotic stress in vitro. BMC Plant Biol. 2018, 18, 299. [Google Scholar] [CrossRef]
- Luo, D.; Hou, X.; Zhang, Y.; Meng, Y.; Zhang, H.; Liu, S.; Wang, X.; Chen, R. CaDHN5, a dehydrin gene from pepper, plays an important role in salt and osmotic stress responses. Int. J. Mol. Sci. 2019, 20, 1989. [Google Scholar] [CrossRef]
- Alsheikh, M.K.; Heyen, B.J.; Randall, S.K. Ion binding properties of the dehydrin ERD14 are dependent upon phosphorylation. J. Biol. Chem. 2003, 278, 40882–40889. [Google Scholar] [CrossRef]
- Close, T.J. Dehydrins: Emergence of a biochemical role of a family of plant dehydration proteins. Physiol. Plant. 1996, 97, 795–803. [Google Scholar] [CrossRef]
- Close, T.J. Dehydrins: A commonality in the response of plants to dehydration and low temperature. Physiol. Plant. 1997, 100, 291–296. [Google Scholar] [CrossRef]
- Fernandez-Caballero, C.; Rosales, R.; Romero, I.; Escribano, M.I.; Merodio, C.; Sanchez-Ballesta, M.T. Unraveling the roles of CBF1, CBF4 and dehydrin 1 genes in the response of table grapes to high CO2 levels and low temperature. J. Plant Physiol. 2012, 169, 744–748. [Google Scholar] [CrossRef]
- Falavigna, V.S.; Miotto, Y.E.; Porto, D.D.; Anzanello, R.; dos Santos, H.P.; Fialho, F.B.; Margis-Pinheiro, M.; Pasquali, G.; Revers, L.F. Functional diversification of the dehydrin gene family in apple and its contribution to cold acclimation during dormancy. Physiol. Plant. 2015, 155, 315–329. [Google Scholar] [CrossRef]
- Bao, F.; Du, D.; An, Y.; Yang, W.; Wang, J.; Cheng, T.; Zhang, Q. Overexpression of prunus mume dehydrin genes in tobacco enhances tolerance to cold and drought. Front. Plant Sci. 2017, 8, 151. [Google Scholar] [CrossRef]
- Xu, H.X.; Li, X.Y.; Xu, C.J.; Chen, J.W. Overexpression of loquat dehydrin gene EjDHN1 promotes cold tolerance in transgenic tobacco. Russ. J. Plant Physiol. 2018, 65, 69–77. [Google Scholar] [CrossRef]
- Guo, X.; Zhang, L.; Wang, X.; Wang, X.; Zhang, M.; Zhu, J. Overexpression of saussurea involucrata dehydrin gene SiDHN promotes cold and drought tolerance in transgenic tomato plants. PLoS ONE 2019, 14, e0225090. [Google Scholar] [CrossRef]
- Puhakainen, T.; Hess, M.W.; Makela, P.; Svensson, J.; Heino, P.; Palva, E.T. Overexpression of multiple dehydrin genes enhances tolerance to freezing stress in Arabidopsis. Plant Mol. Biol. 2004, 54, 743–753. [Google Scholar] [CrossRef] [PubMed]
- Hundertmark, M.; Hincha, D.K. LEA (Late Embryogenesis Abundant) proteins and their encoding genes in Arabidopsis thaliana. BMC Genom. 2008, 9, 118. [Google Scholar] [CrossRef] [PubMed]
- Liang, D.; Xia, H.; Wu, S.; Ma, F.W. Genome-wide identification and expression profiling of dehydrin gene family in Malus domestica. Mol. Biol. Rep. 2012, 39, 10759–10768. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.Z.; He, M.Y.; Zhu, Z.G.; Li, S.X.; Xu, Y.; Zhang, C.H.; Singer, S.D.; Wang, Y.J. Identification of the dehydrin gene family from grapevine species and analysis of their responsiveness to various forms of abiotic and biotic stress. BMC Plant Biol. 2012, 12, 140. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.X.; Yang, Y.; Li, X.; Li, X.Y.; Feng, C.; Chen, J.W.; Xu, C.J. Involvement of multiple types of dehydrins in the freezing response in loquat (Eriobotrya japonica). PLoS ONE 2014, 9, e87575. [Google Scholar] [CrossRef]
- Hara, M.; Terashima, S.; Fukaya, T.; Kuboi, T. Enhancement of cold tolerance and inhibition of lipid peroxidation by citrus dehydrin in transgenic tobacco. Planta 2003, 217, 290–298. [Google Scholar] [CrossRef]
- Gasic, K.; Hernandez, A.; Korban, S.S. RNA extraction from different apple tissues rich in polyphenols and polysaccharides for cDNA library construction. Plant Mol. Biol. Rep. 2004, 22, 437–438. [Google Scholar] [CrossRef]
- Layton, B.E.; Boyd, M.B.; Tripepi, M.S.; Bitonti, B.M.; Dollahon, M.N.; Balsamo, R.A. Dehydration-induced expression of a 31-k Da dehydrin in polypodium polypodioides (polypodiaceae) may enable large, reversible deformation of cell walls. Am. J. Bot. 2010, 97, 535–544. [Google Scholar] [CrossRef]
- Wei, H.; Yang, Y.; Himmel, M.E.; Tucker, M.P.; Ding, S.; Yang, S.; Arora, R. Identification and characterization of five cold stress-related rhododendron dehydrin genes: Spotlight on a FSK-Type dehydrin with multiple F-Segments. Front. Bioeng. Biotech. 2019, 7, 30. [Google Scholar] [CrossRef]
- Hughes, S.; Graether, S.P. Cryoprotective mechanism of a small intrinsically disordered dehydrin protein. Protein Sci. 2011, 20, 42–50. [Google Scholar] [CrossRef]
- Xiao, H.; Nassuth, A. Stress- and development-induced expression of spliced and unspliced transcripts from two highly similar dehydrin 1 genes in V. riparia and V. vinifera. Plant Cell Rep. 2006, 25, 968–977. [Google Scholar] [CrossRef]
- Jiang, C.; Iu, B.; Singh, J. Requirement of a CCGAC cis-acting element for cold induction of the BN115 gene from winter Brassicanapus. Plant Mol. Biol. 1996, 30, 679–684. [Google Scholar] [CrossRef]
- Busk, P.K.; Jensen, A.B.; Pages, M. 1Acting elements in vivo in the promoter of the abscisic acid responsive gene rab17 from maize. Plant J. 1997, 11, 1285–1295. [Google Scholar] [CrossRef]
- Nylander, M.; Svensson, J.; Palva, E.T.; Welin, B.V. Stress-induced accumulation and tissue-specific localization of dehydrins in Arabidopsis thaliana. Plant Mol. Biol. 2001, 45, 263–279. [Google Scholar] [CrossRef]
- Abe, H.; Urao, T.; Ito, T.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell 2003, 15, 63–78. [Google Scholar] [CrossRef]
- Agarwal, M.; Hao, Y.; Kapoor, A.; Dong, C.; Fujii, H.; Zheng, X.; Zhu, J. A R2R3 type MYB transcription factor is involved in the cold regulation of CBF genes and in acquired freezing tolerance. J. Biol. Chem. 2006, 281, 37636–37645. [Google Scholar] [CrossRef]
- Yamaguchi-Shinozaki, K.; Shinozaki, K. Organization of cis-acting elements in osmotic-and cold-stress-responsive promoters. Trends Plant Sci. 2005, 10, 88–94. [Google Scholar] [CrossRef]
- Bassett, C.L.; Fisher, K.M.; Farrell, R.E., Jr. The complete peach dehydrin family: Characterization of three recently recognized genes. Tree Genet. Genomes 2015, 11, 126. [Google Scholar] [CrossRef]
- Agarwal, T.; Upadhyaya, G.; Halder, T.; Mukherjee, A.; Majumder, A.L.; Ray, S. Different dehydrins perform separate functions in Physcomitrella patens. Planta 2017, 245, 101–118. [Google Scholar] [CrossRef]
- Halder, T.; Upadhyaya, G.; Basak, C.; Das, A.; Chakraborty, C.; Ray, S. Dehydrins impart protection against oxidative stress in transgenic tobacco plants. Front. Plant Sci. 2018, 9, 136. [Google Scholar] [CrossRef]
- Kim, S.Y.; Nam, K.H. Physiological roles of ERD10 in abiotic stresses and seed germination of Arabidopsis. Plant Cell Rep. 2010, 29, 203–209. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).