Abstract
The adventitious rooting−related oxygenase (ARRO−1) gene is specifically expressed in the early rooting stage and is considered a molecular marker of rooting. In this study, a PsARRO−1 gene (GenBank accession number KJ620008) was identified from a pre−constructed transcriptome database of root development of Paeonia suffruticosa under sandy loam cultivation. The expression was verified by RT−qPCR, and it was found that the expression trend was consistent with the expression in the transcriptome database. The PsARRO−1 gene was specifically highly expressed during the root primordium germination phase. In addition, the RT−qPCR analysis indicated that the expression of PsARRO−1 in roots was significantly higher than in stems and leaves, its peak expression in vitro was 5 days earlier than in soil, and its expression was higher than that of tree peony in soil. Subcellular localization analysis showed that PsARRO−1 was localized in the plasma membrane. Moreover, the transient silent expression of the PsARRO−1 gene was found in the roots of peony seedlings grown using VIGS technology. The root activity was significantly reduced after transient silencing of the expression of the PsARRO−1 gene. These results indicate that PsARRO−1 has a positive regulatory effect on tree peony root development.
1. Introduction
The tree peony (Paeonia suffruticosa Andr.) is a woody perennial ornamental flower plant popular for its large and graceful flowers [1,2,3,4]. Traditional propagation methods for tree peony include cuttings and engraftment [5]. However, there are some problems associated with these methods, including a limited number of scion sources and long production periods. These propagation problems have limited the industrialization and commercial production of tree peony seedlings [5,6,7]. The problems could be solved by tissue culture technology based on organogenesis, and a large number of Paeonia suffruticosa could be obtained in vitro [1,8]. However, these methods do not currently meet the requirements for mass production because of the low frequency of rooting, low root quality, the connection barrier between the root and stem conducting tissues, along with the low survival rate of transplanted plantlets in vitro, which seriously restrict the scale of production [9,10,11]. Therefore, solving the problem of rooting in vitro would represent a breakthrough in Paeonia suffruticosa tissue culture. Many studies have focused on micropropagation [12,13,14,15], rooting cultivation physiology [16,17], the root microenvironment [18,19,20], stress physiology [21], and metabolic processes [5,22,23]. These studies have shown that the root formation process of Paeonia suffruticosa is complex and cannot be explained by a single factor [10,12,14,16,21,22]. In essence, the root generation of plantlets in vitro should be controlled by the expression and regulation of related genes and protein interactions [24,25]. Therefore, studying the molecular regulation mechanism of Paeonia suffruticosa rooting, i.e., through rooting−related gene cloning and functional identification, should provide a new way to solve the problem of rooting plantlets in vitro.
It has been found that many genes with specific expression are involved in the generation of plant roots, such as the adventitious rooting−related oxygenase 1 (ARRO−1) gene, the lateral root primordium 1 (LRP1) gene, the lateral organ boundaries genes, and transcription factors related to root formation, and that they coordinate with hormone signaling pathways to jointly induce the growth and development of roots [26,27,28,29,30]. ARRO−1 is a type of dioxygenase that acts in the induction stage of root formation and is one of the molecular markers of root formation in woody plants [31,32,33]; thus, it has attracted significant research attention. At present, research on the molecular regulation mechanism of the ARRO−1 gene in woody plant rooting has mainly been conducted on apples, pears, mulberries, and nandina [29,32,34,35,36,37]. These studies indicate that ARRO−1 is involved in the formation of roots, and it may be involved in regulating hormone homeostasis, as it is expressed in response to auxin stimulation to promote root formation [32,37]. However, there have been few studies on the ARRO−1 gene in Paeonia suffruticosa root development. He et al. [33] obtained the full−length PsARRO−1 (KJ620008) sequence and conducted a preliminary analysis, but they have not yet conducted further systematic studies on its protein location and related functional verification, and its relationship with Paeonia suffruticosa root regeneration is still unclear.
Therefore, this study aimed to analyze the spatiotemporal expression differences and subcellular localization of the PsARRO−1 gene, to construct a heterologous transformed overexpression vector of Arabidopsis thaliana, and to analyze its role in root formation. Combined with its effect on the rooting of Paeonia suffruticosa seedlings, we will explore its molecular biological function of improving the rooting ability of Paeonia suffruticosa test tube seedlings, and provide a theoretical basis and technical support for the establishment of an efficient Paeonia suffruticosa plant regeneration system. The Materials and Methods section will be described our approach with sufficient details to allow others to replicate and build on the published results.
2. Materials and Methods
2.1. Plant Materials and Treatments
Axillary buds of Paeonia suffruticosa cultivar ‘Feng Dan Bai’ were sterilized as source materials and then inoculated into an induction medium to induce seedlings for use as test materials. The roots, stems, and leaves of the plantlets in vitro were collected at 0, 5, 10, 15, 20, 25, 30, 35, 40, and 45 days after transfer to rooting culture [Woody plant medium (WPM) + 3−Indolebutyric acid (IBA) 4.0 mg/L + Polyvinyl Pyrrolidone (PVP) 1.0 g/L + Vitamin C (Vc) 50 mg/L + Sucrose 30 g/L + Phytagel (Solarbio, Beijing, China) 2.0 g/L]. The roots, stems, and leaves of the plantlets in soil were collected at 0, 5, 10, 15, 20, 25, 30, 35, 40, and 45 days after 13 March. The roots of Paeonia suffruticosa in different stages of root formation were collected after determining its age through sectioning. All materials were quick−frozen with liquid nitrogen and stored at −80 °C for later use. Axillary buds of Paeonia suffruticosa cultivar ‘Feng Dan Bai’ were obtained from the Shenzhou Tree Peony Garden in LuoYang, China. Agrobacterium GV3101 was preserved in the Garden Plant Biotechnology Laboratory of the College of Forestry, Henan Agricultural University. The plasmid vectors TRV1 and TRV2 were provided by the College of Horticulture and Plant Protection, Yangzhou University.
2.2. Anatomical Observation
The roots of Paeonia suffruticosa seedlings at different stages of root development were taken as experimental materials, the root primordia were observed by the paraffin section method, and the critical period of root occurrence was determined.
2.3. Relative Expression Analysis
Total RNA was isolated using a modified CTAB method [38]. First−strand cDNA was synthesized using a PrimeScript™ RT Reagent Kit with gDNA Eraser (TaKaRa Bio., Otsu, Japan). Real−time quantitative PCR (RT−qPCR) was performed using TB Green™ Premix Ex Taq™ II (Tli RNaseH Plus) (TaKaRa Bio., Kusatsu City, Japan) on a CFX96 Real−Time PCR Detection System (Bio-Rad, Hercules, CA, USA). Specific primers for PsARRO−1 (PsARRO−1−F: AGTCTGGCACATTCGTAGC; PsARRO−1−R: TCTTTGCACATCACTCGGT) were designed and synthesized by Sangon Biotech Co., Ltd. (Shanghai, China). Actin was used as an internal reference gene (Actin−F: GGTCTATTCTTGCTTCCCTCAG; Actin−R: GAACTCACTATCAAACCCTCCAG). The procedure was a standard two−step method with three biological replicates.
2.4. Subcellular Localization
Tobacco seeds were sown and incubated with 12 h of light per day for one month for the experiment. The constructed vector plasmid was transferred into agrobacterium LBA4404 and cultured at 30 °C for 2 days. The cells were then inoculated into 10 mL YEB liquid medium and shaken at 170 rpm for 1 h. The culture was centrifuged at 4000 rpm for 4 min, and the supernatant was removed. The supernatant was resuspended with a 10 mM MgCl2 (including 120 µM AS) suspension and adjusted to OD600 = 0.6. Selected N. benthamiana plants with good growth were infiltrated with the supernatant at the lower epidermis of the leaves with a 1 ml syringe, without the tip, and labeled accordingly. The injected tobacco plants were cultured in low light for 2 days, and then samples were placed on glass slides, observed under a laser confocal microscope, and photographed [39].
2.5. Vector Construction
Forward and reverse primers with XbaI and KpnI restriction sites were designed according to the PsARRO−1 gene sequence (F: TGCTCTAGATTTCTTTGACCTCCCTCTTG; R: CGGGGTACCCCCATTGCTCCATACTGC). PsARRO−1 plasmid DNA was used as the template for PCR amplification. The target gene and TRV2 plasmid were double−digested with XbaI and KpnI, connected with T4 ligase, and transformed into DH5α−competent Escherichia coli cells. After verification by gel electrophoresis and sequencing, the resulting construct was used for subsequent tests.
2.6. Transformation of Paeonia suffruticosa
The vector plasmids TRV1 and TRV2−PsARRO−1 were transferred into agrobacterium GV3101 and inoculated into 10 mL YEB [including 50 mg/L Rifamycins (Rif) + 50 mg/L Kanamycin (Kan)] liquid medium and shaken at 200 rpm for 12–14 h. Then, 5 mL of the culture was added to 200 mL YEB (including 50 mg/L Rif + 50 mg/L Kan) liquid medium, which was shaken at 200 rpm for 6–12 h and adjusted to OD600 = 0.8–1.0. The culture was centrifuged for 4 min at 4000 rpm and the supernatant was removed. The supernatant was resuspended with a 10 mM MgCl2 + 10 mM MES + 150 mM AS suspension and adjusted to OD600 = 1.0. The injured Paeonia suffruticosa roots were immersed in the heavy suspension, and after vacuum infiltration, the roots were rinsed with deionized water to remove the excess bacterial liquid [40]. The expression levels of the target genes and the root activity of Paeonia suffruticosa seedlings were detected after 5 days of hydroponic culture.
2.7. Root Activity Determination
The root activity was determined using the TTC method [41]. Root samples of 0.5 g were immersed in a 10 ml beaker with 0.4% 2,3,5−triphenyltetrazolium chloride (TTC) and 1/15 mol/L phosphate buffer (pH 7.0) solution, then kept at 37 °C for 1 h. A total of 2 ml 1 mol/L H2SO4 was added to end the reaction. The roots were taken out, dried, and ground with 3–4 ml ethyl acetate and a small amount of quartz sand. The red solution was removed from the test tube, washed with a small amount of ethyl acetate two or three times to remove the residue, moved into the test tube, and finally, ethyl acetate was added to increase the total amount to 10 mL. Using a spectrophotometer for 485 nm colorimetry, the reducing strength of TTC was determined as follows:
where TTC is the amount in μg, divided by the root sample weight (g) and time (h).
2.8. Statistical Analysis
All data in this research underwent a two−way analysis of variance (ANOVA), and significant differences between means were tested using Tukey’s test (p < 0.05). All statistical analyses were conducted using the Microsoft Office 2010 and SPSS 19.0 (SPSS, Chicago, IL, USA) programs for Windows.
3. Results
3.1. Identification and Analysis of PsARRO−1 Gene
In a preliminary experiment, we used the root systems of live seedlings of Paeonia suffruticosa variety ‘Fengdanbai’ at the R1 (root primordium ungerminated period), R2 (root primordium induction period), and R3 (lateral root formation period) as materials to establish the transcriptome database of root development of Paeonia suffruticosa under sandy loam cultivation [42]. In this database, we identified a PsARRO−1 gene (GenBank accession number KJ620008), which was differentially expressed at both the R1/R2 and R2/R3 stages, compared to the expression of root−associated genes such as ARFs, IAAs, WRKYs, WUS, and WOXs in this database, as shown in Figure 1. It is specifically highly expressed in the root primordium germination stage, and it may be one of the important regulators of root primordium germination in Paeonia suffruticosa.
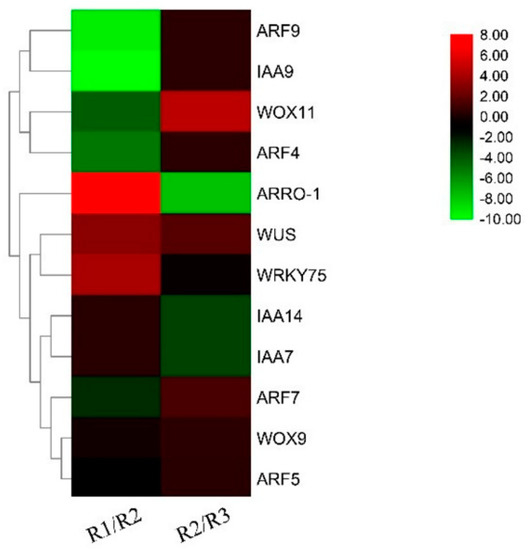
Figure 1.
Root system−related genes in Paeonia suffruticosa root development under sandy soil cultivation (R1: root primordium ungerminated period; R2: root primordium induction period; R3: lateral root formation period).
3.2. Anatomical Observation
Morphological anatomic observation of the root growth process is shown in Figure 2. There was no sign of a root primordium emerging in newly germinated roots with a length of 2–3 cm, but there was an obvious germination phenomenon in new roots, with a length of more than 5 cm and without a convex in the root surface. Thus, the key period of Paeonia suffruticosa root growth and development was divided into four stages: the prophase of root primordium development, the root primordium development period, convex root primordium formation, and root emergence.
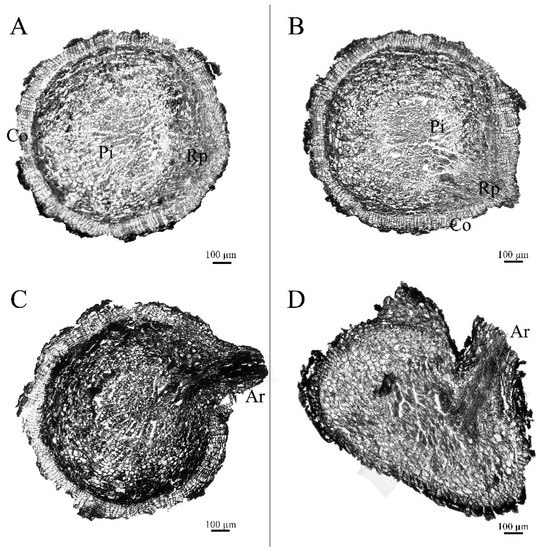
Figure 2.
Morphological anatomic observation of Paeonia suffruticosa adventitious roots: (A). root primordium ungerminated period; (B). root primordium induction period; (C). root primordium convex; (D). root emergence; Co.—cortex; Rp.—adventitious root primordium; Pi.—pith; Ar.—adventitious root). The scale bar is 100 μm.
3.3. Real−Time Fluorescence Quantification of PsARRO−1 of Paeonia suffruticosa in Different Stages of Root Formation
As shown in Figure 3, the relative expression of PsARRO−1 was low at the root primordium ungerminated period, reached a maximum value in the root primordium induction period, and then decreased at the convex root primordium formation and root emergence stages. The expression trend of this figure is the same as the description in Figure 1, showing the specific high expression of the PsARRO−1 gene in the root primordium induction period.
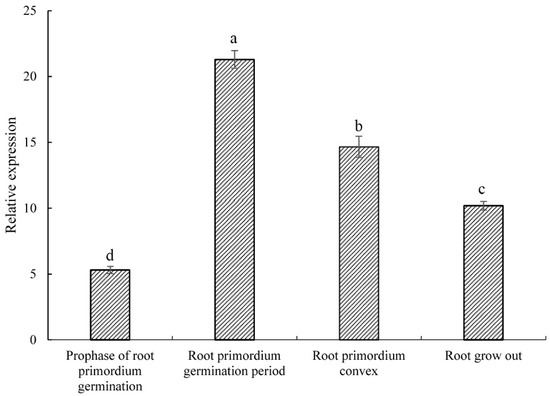
Figure 3.
Relative expression of PsARRO−1 at different stages of rooting. Different letters within the columns indicate significant differences (p < 0.05) according to the Tukey test.
3.4. Real−Time Fluorescence Quantification of PsARRO−1 of Paeonia suffruticosa In Vitro
The relative expression of the PsARRO−1 gene in the root is showed in Figure 4 for the stem and leaf tissues of Paeonia suffruticosa in vitro. The expression was slightly higher in the leaves than in the roots and stems at 0 d, increased linearly from 5 d to 10 d in the roots, reached a maximum value at 10 d, and then fell sharply to the minimum value, which was 20 times lower than the maximum value, at 30 d. A second peak appeared at 35 d, at which point the expression of PsARRO−1 in the roots, stems, and leaves tended to be consistent.
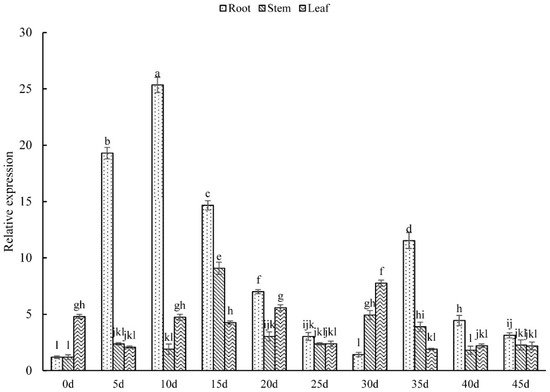
Figure 4.
Relative expression level of PsARRO−1 in different parts of Paeonia suffruticosa in vitro. Different letters within the columns indicate significant differences (p < 0.05) according to the Tukey test.
3.5. Real−Time Fluorescence Quantification of PsARRO−1 of Paeonia suffruticosa in Soil
According to Figure 5, the expression level of PsARRO−1 gene was similar in the root, stem, and leaf tissues of Paeonia suffruticosa seedlings at 0 d, but differences became apparent as the seedlings grew. The expression level in the roots was significantly higher than in stems and leaves, and reached a maximum value at 15 d, which was about 2 times and 9 times that in the stems and leaves, respectively. The expression in stems and leaves was similar, and reached peak values at 15 d and 10 d, respectively.
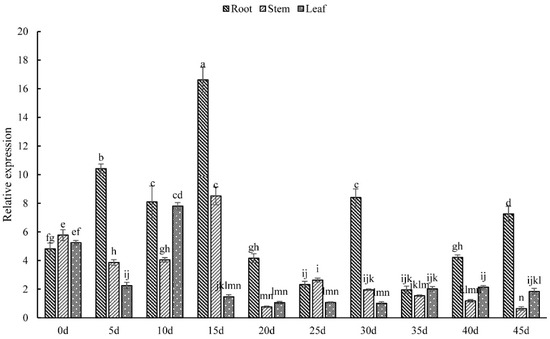
Figure 5.
Relative expression of PsARRO−1 in different parts of tree pony seedlings: different letters within the columns indicate significant differences (p < 0.05) according to the Tukey test.
3.6. Subcellular Localization of PsARRO−1
The fluorescence signal was observed in the plasma membrane, and there was no fluorescence signal in either the chloroplast or cytoplasm (Figure 6). Therefore, it was determined that the PsARRO−1 gene product was localized in the plasma membrane.
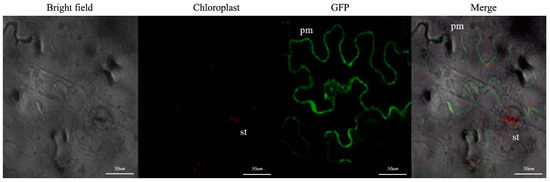
Figure 6.
PsARRO−1 subcellular localization. The scale bar is 50 μm. (pm: plasma membrane; st: stomata).
3.7. Functional Verification of PsARRO−1
The root system of Paeonia suffruticosa was infected with agrobacterium, and the results are shown in Figure 7. The treated Paeonia suffruticosa roots were taken for RT−qPCR, and the results are shown in Figure 8. As can be seen from the figure, compared with wild−type plants and transformed no−load plants, the expression level of PsARRO−1 was significantly reduced, except for in plants 1 and 3. PsARRO−1 expression was significantly inhibited in Paeonia suffruticosa plants 2 and 4–9. The results showed that we successfully obtained the VIGS transient silent expression of PsARRO−1.

Figure 7.
Transgenic Paeonia suffruticosa seedling roots of PsARRO−1. (A): TRV1 + TRV2; (B): TRV1 + TRV2−PsARRO−1. The scale bar is 5 cm.
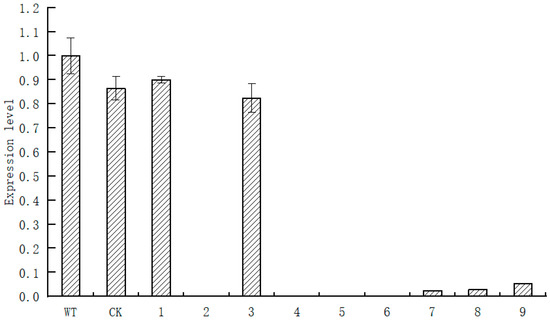
Figure 8.
Relative expression of PsARRO−1 at different treated transgenic Paeonia suffruticosa seedlings.
Through the measurement of the root activity of momentary silent expression plants of Paeonia suffruticosa, it was found that the root activity of PsARRO−1 silent expression lines was significantly lower than that of transgenic plants and transformed empty vector plants (Figure 9). The results showed that PsARRO−1 may promote the root activity of Paeonia suffruticosa, and its silencing expression significantly inhibited the root activity of Paeonia suffruticosa.
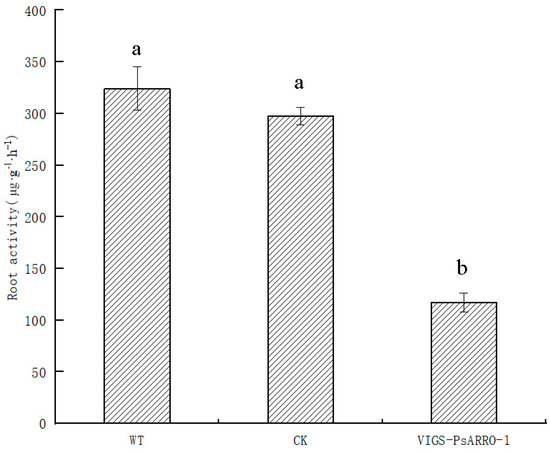
Figure 9.
Root activity of different PsARRO−1 treated transgenic Paeonia suffruticosa seedling. Different letters within the columns indicate significant differences (p < 0.05) according to the Tukey test.
4. Discussion
As the cornerstone of plant growth and development, the root system plays the role of absorbing water and nutrients and supporting the process of plant growth. The root system plays an important role in plant survival. In this study, we identified a PsARRO−1 gene from the transcriptome database of root development of Paeonia suffruticosa, and analysis revealed that its expression was significantly elevated during the root primordium induction period. We identified a PsARRO−1 gene from the transcriptome database of root development of Paeonia suffruticosa. Subsequently, we identified the root primordium ungerminated period, root primordium induction period, root primordium convex period, and root emergence period of live seedling root material by paraffin sectioning for RT−qPCR and found that the expression of PsARRO−1 was similar to the trend of the transcriptome database, consistent with the findings of Smolka et al. that ARRO−1 gene expression is upregulated during root induction [29].
In this research, the expression levels of the PsARRO−1 gene in root, stem, and leaf tissues during different root germination periods of Paeonia suffruticosa ‘Feng Dan Bai’ were investigated in vitro and in seedlings using RT−qPCR. The results showed that the PsARRO−1 expression level in the roots was significantly higher than those in the stems and leaves. The expression level in the roots was 14 and 5 times than in the stems and leaves in vitro, and the level in seedling roots was 2 and 9 times higher than those in the stems and leaves, respectively. In addition, the expression of PsARRO−1 in vitro was higher than that in the soil. It was speculated that this might be because the internal growth and development (totipotency) needed to form a complete plant may be the first organogenesis sequence activated when it is induced in the rooting medium, leading to a higher expression of the PsARRO−1 gene at this stage to meet the needs of root development. He [43] found that the root primordium appeared on the 3rd day of treatment, formed from the 5th to the 15th day, and then broke in the epidermis. In our research, the expression level of PsARRO−1 in vitro rose from 0 d to 10 d, reached a peak value at 10 d, and then decreased. It was speculated that a large number of root primordia were formed during 0–10 d, and the formation of root primordia decreased after 10 d, so the expression level of PsARRO−1 decreased from 10 d on. Root primordia formation began five days earlier and lasted five days longer than in a study by Ji [44]; the reason for this may be due to the use of different environmental factors and materials. The expression of the PsARRO−1 gene showed significant differences in the four root germination periods. It was the lowest at the root primordium ungerminated period, the highest in the root primordium induction period (about four times higher than in the prophase of root primordium germination), and decreased in the root primordium convex and root emergence periods. Therefore, the PsARRO−1 gene was considered to be specifically expressed in the root induction stage, and was mainly active in the root primordium germination stage.
Subcellular localization results showed that the PsARRO−1 gene product was localized in the plasma membrane, which was consistent with the results for apples from Li et al. [37]. PsARRO−1 belongs to the 2−oxoglutarate−dependent dioxygenases (2−ODD) family, which plays important roles in the synthesis of many signal substances, such as gibberellin (GA3), ethylene, flavonoids, and secondary metabolites [45]; thus, this result was consistent with the function in the catalytic hydroxylation, desaturation, and epoxidation reactions of the 2−ODD family. Previous studies have shown that gibberellin treatment can promote root elongation [46,47,48], and GA3 regulates root growth mainly by regulating the proliferation and elongation of root tip meristem cells [49,50]. Thus, it might be speculated that PsARRO−1 is related to GA3, but this requires further research for confirmation.
Root activity is one of the important indicators of plant root growth, and higher root activity shows stronger root germination and growth ability. In this study, the root activity of Paeonia suffruticosa was significantly reduced after transient silencing of the expression of PsARRO−1 in its root system, and PsARRO−1 may play a positive role in the root growth of Paeonia suffruticosa.
5. Conclusions
In summary, the PsARRO−1 gene product was located in the plasma membrane and the gene was specifically expressed in the root induction stage in Paeonia suffruticosa, while mainly acting during the root primordium development period. Its root activity was significantly reduced after the transient silencing of its expression in the Paeonia suffruticosa root system using VIGS technique, and PsARRO−1 played an active role in the development of the Paeonia suffruticosa root system. The mechanism of PsARRO−1 in the development of Paeonia suffruticosa root system requires further study.
Author Contributions
Study conception and design: Z.W. and S.H.; data collection: Y.S. (Yuke Sun), J.Y., Y.S. (Yinglong Song), L.S. and Y.X.; analysis and interpretation of results: Y.S. (Yuke Sun), W.S., J.Y., Y.S. (Yuxiao Shen), J.M. and J.W.; draft manuscript preparation: Y.S. (Yuke Sun) and W.S. All authors have read and agreed to the published version of the manuscript.
Funding
This study was financially supported by the National Key Research and Development Program of China (Grant No. 2020YFD1000503), the Natural Science Foundation of China (Grant No.31870698), the Natural Science Foundation of Henan Province (Grant No. 222300420462), and the Open Research Fund of Shanghai Key Laboratory of Plant Functional Genomics and Resources (Grant No. PFGR202102).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data supporting the reported results are available and will be provided upon request.
Conflicts of Interest
The authors declare that they have no conflicts of interest to report regarding the present study.
References
- Li, Y.L.; Wu, D.Y.; Pan, S.L.; Xu, S.L. The study on micropropagation technology of tree peony plantlets in vitro. Chin. Sci. Bull. 1984, 8, 500–502. [Google Scholar]
- Beruto, M.; Lanteri, L.; Portogallo, C. Micropropagation of tree peony (Paeonia suffruticosa). Plant Cell Tiss Org. 2004, 79, 249–255. [Google Scholar] [CrossRef]
- Zhou, Z.Q. Taxonomy, geographic distribution and ecological habitats of tree peonies. Genet. Resour. Crop. Evol. 2006, 53, 11–22. [Google Scholar] [CrossRef]
- Guo, L.; Guo, S.; Xu, J.; He, L.; Carlson, J.E.; Hou, X. Phylogenetic analysis based on chloroplast genome uncover evolutionary relationship of all the nine species and six cultivars of tree peony. Ind. Crop. Prod. 2020, 153, 112567. [Google Scholar] [CrossRef]
- Meng, X.; Wang, Z.; He, S.; Shi, L.; Song, Y.; Lou, X.; He, D. Endogenous hormone levels and activities of IAA-modifying enzymes during adventitious rooting of tree peony cuttings and grafted scions. Hortic. Environ. Biotechnol. 2019, 60, 187–197. [Google Scholar] [CrossRef]
- Tian, D.K.; Tile, K.M.; Dane, F.; Woods, F.M.; Sibley, J.L. Comparison of shoot induction ability of different explants in herbaceous peony (Paeonia lactiflora Pall.). Sci. Hortic. 2010, 123, 385–389. [Google Scholar] [CrossRef]
- Gao, J.; Xue, J.; Xue, Y.; Liu, R.; Ren, X.; Wang, S.; Zhang, X. Transcriptome sequencing and identification of key callus browning-related genes from petiole callus of tree peony (Paeonia suffruticosa cv. Kao) cultured on media with three browning inhibitors. Plant Physiol. Biochem. 2020, 149, 36–49. [Google Scholar] [CrossRef]
- Wang, H.; Van, S.J. Establishment of in vitro cultures of tree peonies. South Afr. J. Bot. 2001, 67, 358–361. [Google Scholar] [CrossRef][Green Version]
- Bouza, L.; Jacques, M.; Sotta, B.; Miginiac, E. The reactivation of tree peony (Paeonia suffruticosa Andr.) in vitro plants by chilling is correlated with modifcations of abscisic acid, auxin and cytokinin levels. Plant Sci. 1994, 97, 153–160. [Google Scholar] [CrossRef]
- He, D.; Wang, Z.; He, S.L. Adventitious root generating process and hormone and enzyme changes in vitro Paeonia suffruticosa. Acta Hortic. Sin. 2011, 38, 770–776. [Google Scholar]
- Zhu, X.T.; Li, X.Q.; Ding, W.J.; Jin, S.H.; Wang, Y. Callus induction and plant regeneration from leaves of peony. Hortic. Environ. Biotechnol. 2018, 59, 575–582. [Google Scholar] [CrossRef]
- Jaime, A.; Teixeira, D.S.; Shen, M.M.; Yu, X.N. Tissue culture and micropropagation of tree peony (Paeonia suffruticosa Andr.). J. Crop Sci. Biotechnol. 2012, 15, 159–168. [Google Scholar] [CrossRef]
- Du, Y.M.; Cheng, F.Y.; Zhong, Y. Induction of direct somatic embryogenesis and shoot organogenesis and histological study in tree peony (Paeonia sect. Moutan). Plant Cell Tissue Organ 2020, 141, 557–570. [Google Scholar] [CrossRef]
- Ren, X.X.; Liu, Y.; Jeong, B.R. Callus induction and browning suppression in tree peony Paeonia ostii ‘Fengdan’. Hortic. Environ. Biotechnol. 2020, 61, 591–600. [Google Scholar] [CrossRef]
- Wen, S.S.; Chen, L.; Tian, R.N. Micropropagation of tree peony (Paeonia sect. Moutan): A review. Plant Cell Tissue Organ 2020, 141, 1–14. [Google Scholar] [CrossRef]
- Lydia, B.; Monique, J.; Emile, M. Requirements for in vitro rooting of Paeonia suffruticosa Andr. cv. ‘Mme de Vatry’. Sci. Hortic. 1994, 58, 223–233. [Google Scholar] [CrossRef]
- Xiao, Z.; Ji, N.; Zhang, X.; Zhang, Y.; Wang, Y.; Wu, T.; Xu, X.; Han, Z. The lose of juvenility elicits adventitious rooting recalcitrance in apple rootstocks. Plant Cell Tissue Organ 2014, 119, 51–63. [Google Scholar] [CrossRef]
- Han, J.G.; Song, Y.; Liu, Z.G.; Hu, Y.H. Culturable bacterial community analysis in the root domains of two varieties of tree peony (Paeonia ostii). Fems Microbiol. Lett. 2011, 1, 15–24. [Google Scholar] [CrossRef]
- Wang, H.Y.; He, S.L.; Tanaka, M.; Van, P.T.; Jaima, A.T. Effect of IBA Concentration, Carbon Source, Substrate, and Light Source on Root Induction Ability of Tree Peony (Paeonia suffruticosa Andr.) Plantlets in Vitro. Eur. J. Hortic. Sci. 2012, 77, 122–128. [Google Scholar] [CrossRef]
- Xue, D.; Huang, X.D. Changes in soil microbial community structure with planting years and cultivars of tree peony (Paeonia suffruticosa). World J. Microb. Biotechnol. 2014, 30, 389–397. [Google Scholar] [CrossRef]
- Shang, W.; Wang, Z.; He, S.; He, D.; Liu, Y.; Fu, Z. Research on the relationship between phenolic acids and rooting of tree peony (Paeonia suffruticosa) plantlets in vitro. Sci. Hortic. 2017, 224, 53–60. [Google Scholar] [CrossRef]
- Sun, M.; Zhou, W.Y.; Yang, Y.; Lv, M.W. Analysis of chemical components in the roots of eight intersubgeneric hybrids of Paeonia. Chem. Biodivers. 2021, 18, e2000848. [Google Scholar] [CrossRef]
- Zhao, D.Q.; Zhang, X.Y.; Fang, Z.W.; Wu, Y.Q.; Tao, J. Physiological and Transcriptomic Analysis of Tree Peony (Paeonia section Moutan DC.) in Response to Drought Stress. Forests 2019, 10, 135. [Google Scholar] [CrossRef]
- Khan, M.A.; Wang, Y.; Muhammad, B.; Uddin, S.; Saeed, A.; Khan, D.; Ali, M.; Saeed, S.; Kui, J.Z. Morpho-physiological and phytohormonal changes during the induction of adventitious root development stimulated by exogenous IBA application in Magnolia biondii Pamp. Braz. J. Biol. 2022, 84, e255664. [Google Scholar] [CrossRef]
- Xu, X.Z.; Che, Q.Q.; Cheng, C.X.; Yuan, Y.B.; Wang, Y.Z. Genome-wide identification of WOX gene family in apple and a functional analysis of MdWOX4b during adventitious root formation. J. Integr. Agric. 2022, 21, 1332–1345. [Google Scholar] [CrossRef]
- Smith, D.L.; Fedoroff, N.V. LRP1, a gene expressed in lateral and adventitious root primordia of Arabidopsis. Plant Cell 1995, 7, 735–745. [Google Scholar] [CrossRef][Green Version]
- Liu, H.; Wang, S.; Yu, X.; Yu, J.; He, X.; Zhang, S.; Shou, H.; Wu, P. ARL1. a LOB-domain protein required for adventitious root formation in rice. Plant J. 2005, 43, 47–56. [Google Scholar] [CrossRef]
- Lee, H.W.; Kim, N.Y.; Lee, D.J.; Kim, J. LBD18/ASL20 Regulates Lateral Root Formation in Combination with LBD16/ASL18 Downstream of ARF7 and ARF19 in Arabidopsis. Plant Physiol. 2009, 151, 1377–1389. [Google Scholar] [CrossRef]
- Smolka, A.; Welander, M.; Olsson, P.; Holefors, A.; Zhu, L.H. Involvement of the ARRO−1 gene in adventitious root formation in apple. Plant Sci. 2009, 177, 710–715. [Google Scholar] [CrossRef]
- Trupiano, D.; Yordanov, Y.; Regan, S.; Meilan, R.; Tschaplinski, T.; Scippa, G.S.; Busov, V. Identification, characterization of an AP2/ERF transcription factor that promotes adventitious, lateral root formation in Populus. Planta 2013, 238, 271–282. [Google Scholar] [CrossRef]
- Butler, E.D.; Gallagher, T.F. Isolation and characterization of a cDNA encoding a novel 2-oxoacid-dependent dioxygenase which is up-regulated during adventitious root formation in apple (Malus domestica ‘Jork 9’) stem discs. J. Exp. Bot. 1999, 50, 551–552. [Google Scholar]
- Butler, E.D. Characterization of auxin-induced ARRO−1 expression in the primary root of Malus domestica. J. Exp. Bot. 2000, 51, 1765–1766. [Google Scholar] [CrossRef]
- He, D.; Li, R.; Ji, S.Y.; Wu, J.; Wang, Z.; Liu, Y.P.; He, S.L. Cloning and Expression Analysis of Adventitious Rooting Related Gene PsARRO−1 of Tree Peony. Plant Physiol. J. 2014, 8, 1151–1158. [Google Scholar] [CrossRef]
- De, K.G.J.; Krieken, W.V.D.; Jong, J.C.D. Review the formation of adventitious roots: New concepts, new possibilities. Vitr. Cell. Dev. Biol.-Plant 1999, 35, 189–199. [Google Scholar] [CrossRef]
- Moriya, S.; Iwanami, H.; Haji, T.; Okada, K.; Yamada, M.; Yamamoto, T.; Abe, K. Identification and genetic characterization of a quantitative trait locus for adventitious rooting from apple hardwood cuttings. Tree Genet. Genomes 2015, 11, 59. [Google Scholar] [CrossRef]
- Yang, L.; Meng, H.; Ma, H.; Sun, T.; Li, Z.; Zhang, X.; Xu, J. Effect of Constriction on Content of Endogenous Hormones and Expression Rooting Related Genes During Shoot Layering of Apple Dwarfing Rootstock ‘9-3’. Acta Hortic. Sin. 2017, 44, 613–621. [Google Scholar] [CrossRef]
- Li, T.Y.; Wang, Y.; Zhang, X.Z.; Han, Z.H. Isolation and Characterization of ARRO−1 Genes from Apple Rootstocks in Response to Auxin Treatment. Plant Mol. Biol. Rep. 2012, 30, 1408–1414. [Google Scholar] [CrossRef]
- Honaas, L.; Kahn, E. A practical examination of RNA isolation methods for European pear (Pyrus communis). BMC Res. Notes 2017, 10, 237. [Google Scholar] [CrossRef]
- Li, P.; Dong, Q.; Ge, S.; He, X.; Verdier, J.; Li, D.; Zhao, J. Metabolic engineering of proanthocyanidin production by repressing the isoflavone pathways and redirecting anthocyanidin precursor flux in legume. Plant Biotechnol. J. 2016, 14, 1604–1618. [Google Scholar] [CrossRef]
- Tian, J.; Pei, H.; Zhang, S.; Chen, J.; Chen, W.; Yang, R.; Meng, Y.; You, J.; Gao, J.; Ma, N. TRV-GFP: A modified Tobacco rattle virus vector for efficient and visualizable analysis of gene function. J. Exp. Bot. 2014, 65, 311–322. [Google Scholar] [CrossRef]
- Ding, F.; Wang, R.; Chen, B. Effect of exogenous ammonium gluconate on growth, ion flux and antioxidant enzymes of maize (Zea Mays L.) seedlings under nacl stress. Plant Biol. 2019, 21, 643–651. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Shang, W.; Wang, Z.; He, S.; Meng, X.; Shi, L.; Shen, Y.; He, D.; Lou, X.; Sun, Y. Transcriptome analysis and genes function verification of root development of Paeonia suffruticosa under sandy loam cultivation. Phyton-Int. J. Exp. Bot. 2022, 91, 2791–2812. [Google Scholar] [CrossRef]
- He, D. The Control on Rooting Culture of Paeonia Suffruticosa In Vitro. Master’s Thesis, Henan Agricultural University, ZhengZhou, China, 2009. [Google Scholar]
- Ji, S.Y. Temporal and Spatial Expression of Rooting Gene PsARRO−1 in Tree Peony. Master’s Thesis, Henan Agricultural University, ZhengZhou, China, 2013. [Google Scholar]
- Prescott, A.G.; John, P. DIOXYGENASES: Molecular Structure and Role in Plant Metabolism. Annu. Rev. Plant Biol. 1996, 47, 245–271. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.L.; Chen, X.; Li, C.C. Dynamic of physiology and biochemistry during wild Rhododendron scabrifolium cutting propagation. Sci. Silvae Sin. 2013, 49, 45–51. [Google Scholar] [CrossRef]
- Zhu, C.H.; Gan, L.J.; Ng, D.; Xia, K. GA3 enhances root responsiveness to exogenous IAA by modulating auxin transport and signalling in Arabidopsis. Plant Cell Rep. 2015, 34, 483–494. [Google Scholar] [CrossRef]
- Han, Y.L.; Yoon, G.M. Regulation of Ethylene Biosynthesis by Phytohormones in Etiolated Rice e (Oryza sativa L.) Seedlings. Mol. Cell 2018, 41, 4. [Google Scholar] [CrossRef]
- Ubeda-Tomás, S.; Federici, F.; Casimiro, I.; Beemster, G.T.; Bhalerao, R.; Swarup, R.; Doerner, P.; Haseloff, J.; Bennett, M.J. Gibberellin signaling in the endodermis controls Arabidopsis root meristem size. Curr. Biol. 2009, 19, 1194–1199. [Google Scholar] [CrossRef]
- Li, J.; Zhao, Y.; Chu, H.; Wang, L.; Fu, Y.; Liu, P.; Upadhyaya, N.; Chen, C.; Mou, T.; Feng, Y.-Q.; et al. SHOEBOX Modulates Root Meristem Size in Rice through Dose-Dependent Effects of Gibberellins on Cell Elongation and Proliferation. PLoS Genet. 2015, 11, e1005464. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).