The Auxin/Indole-3-Acetic Acid (Aux/IAA) Gene Family Analysis of Four Rosaceae Genomes and Expression Patterns of PmIAAs in Prunus mume
Abstract
1. Introduction
2. Materials and Methods
2.1. Identification of Aux/IAA Genes in Four Rosaceae Species
2.2. Phylogenetic Analysis
2.3. Exon–Intron Structural and Conserved Motif Analysis
2.4. Chromosomal Distribution, Synteny Analysis, and Gene Duplication
2.5. Gene Expression Analysis
2.6. Analysis of Cis-Acting Elements in Promoters
3. Results
3.1. Genome-Wide Identification of Aux/IAA Genes in Rosaceae
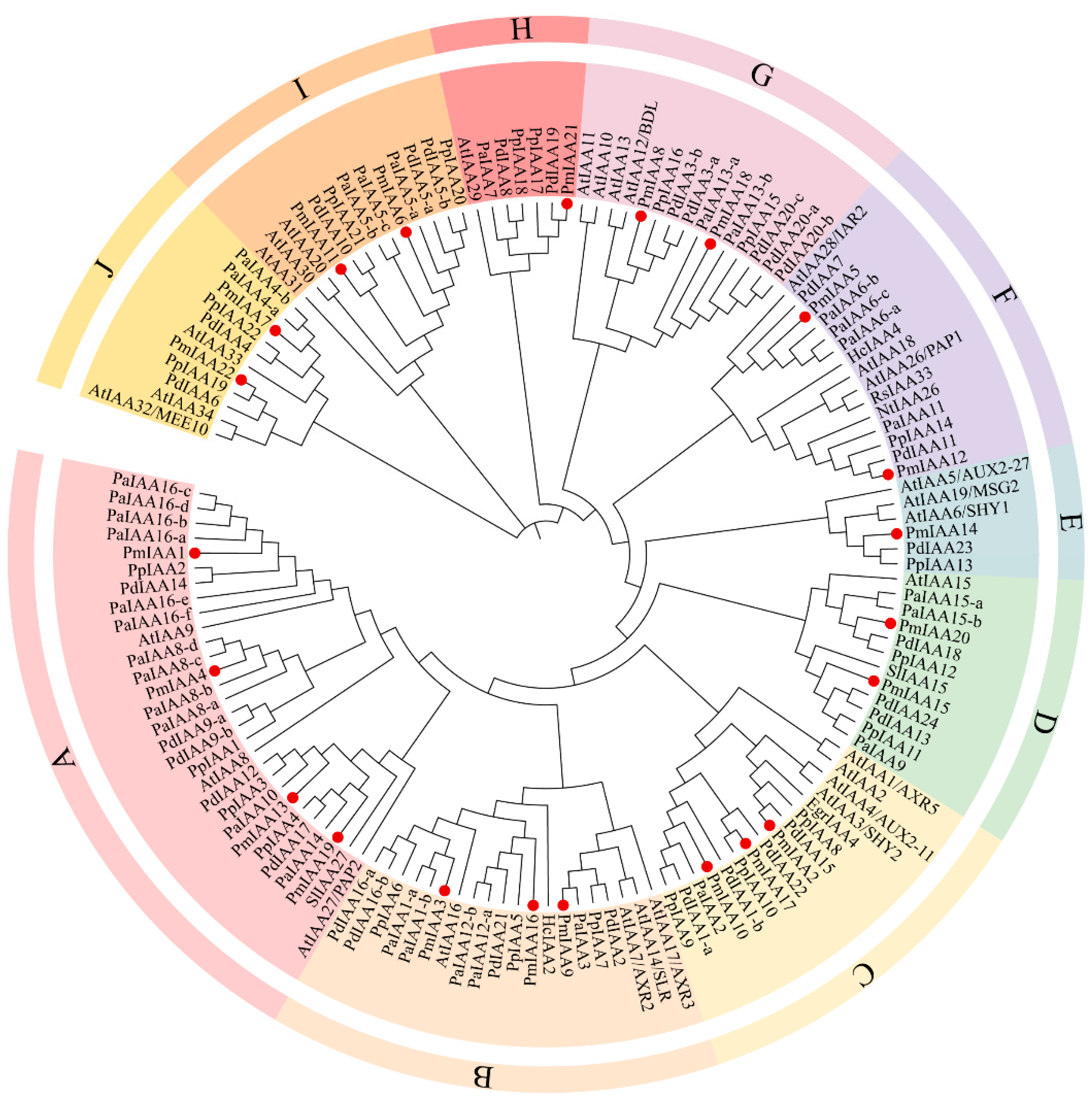
3.2. Phylogenetic Analysis of RoIAAs
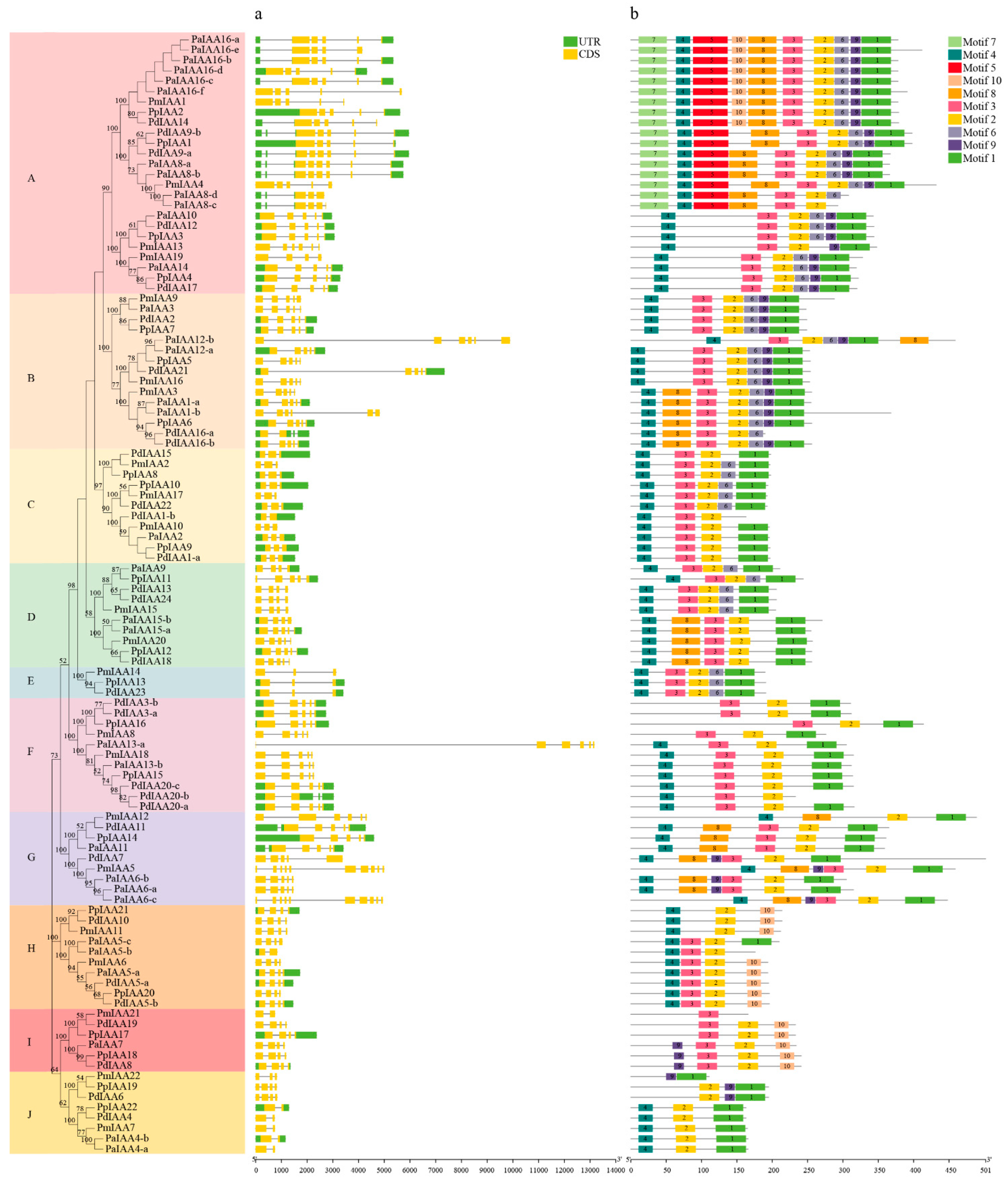
3.3. Gene Structure and Motif Analysis of RoIAAs
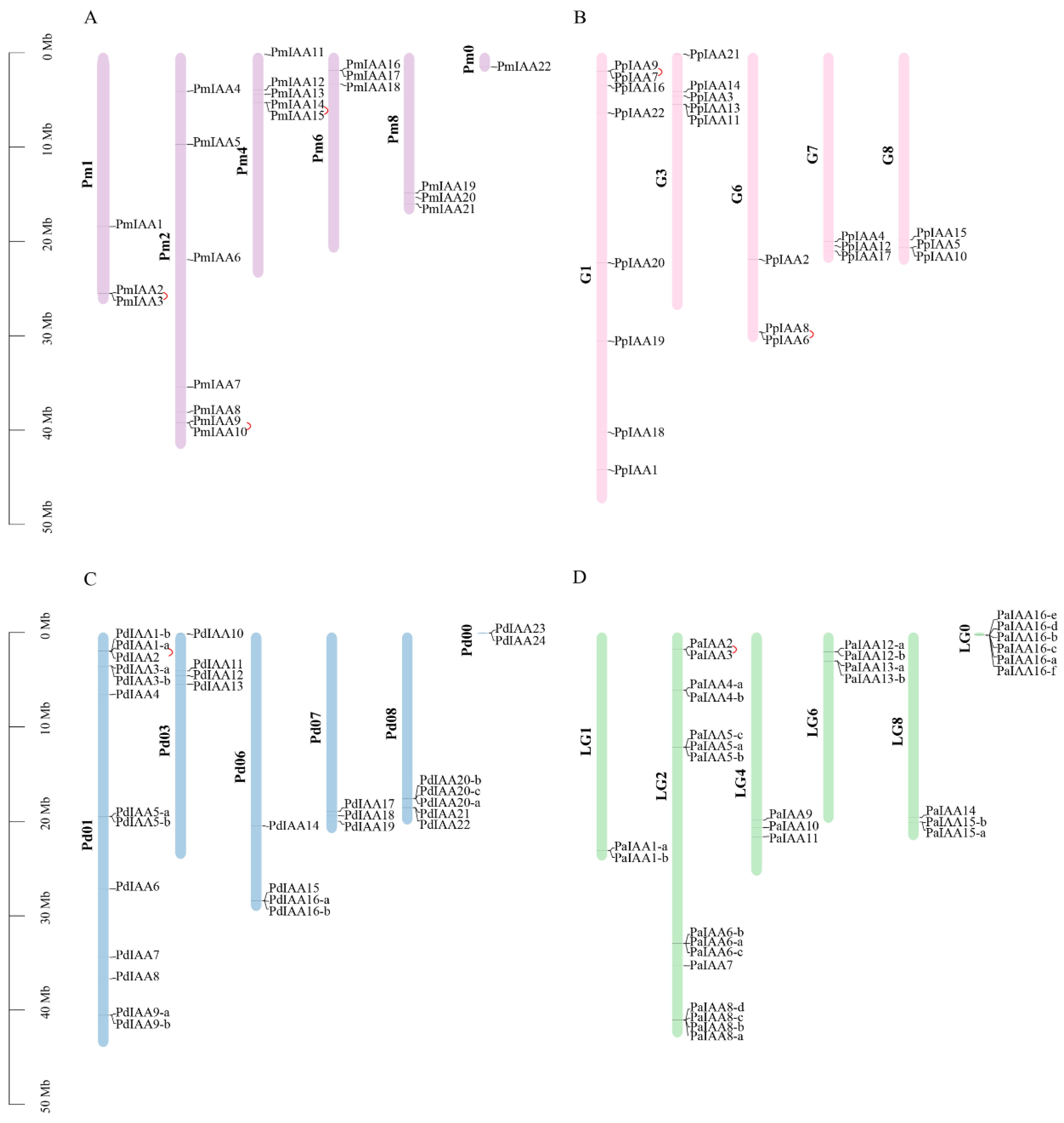
3.4. Chromosomal Location Analyses
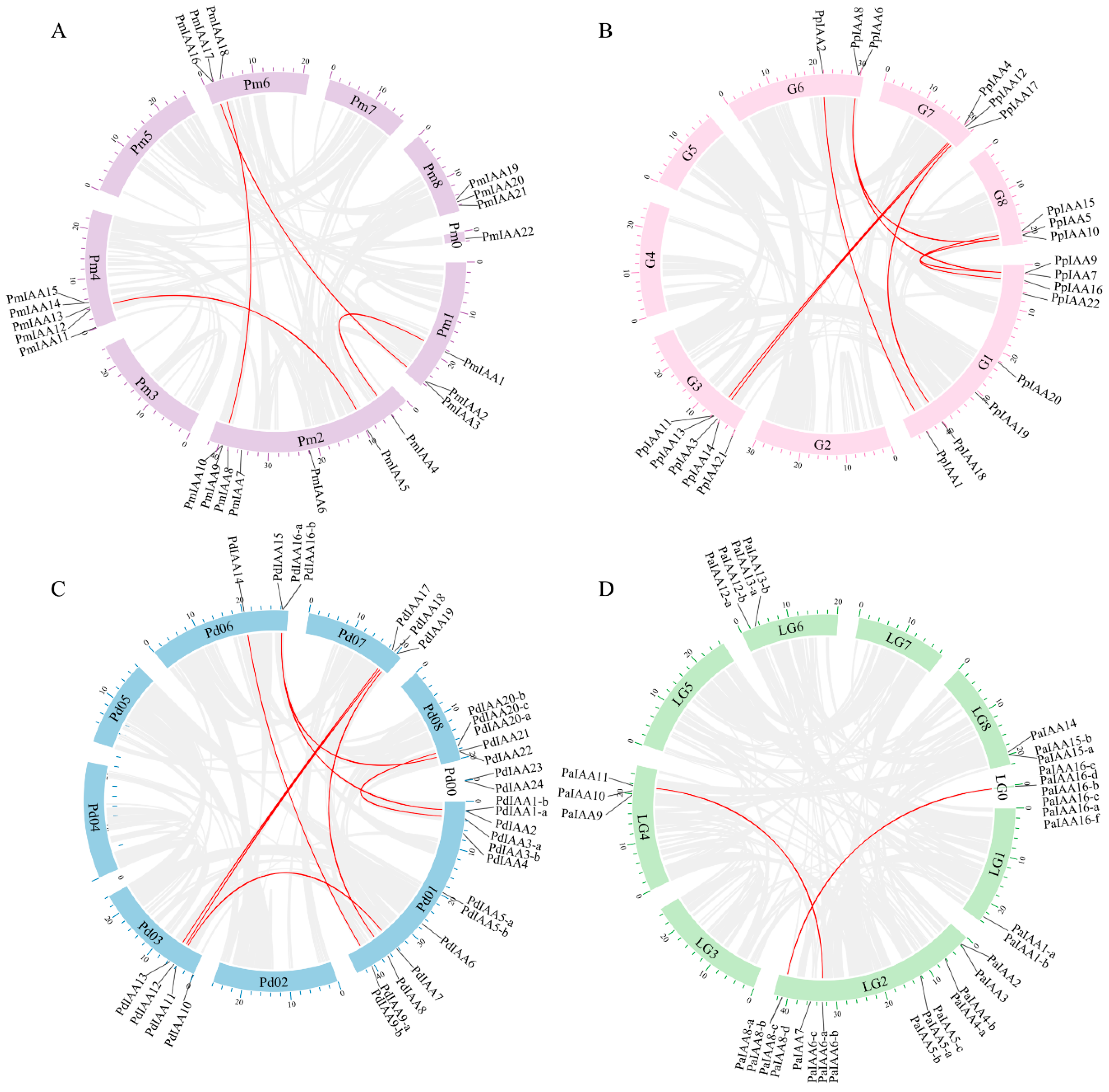
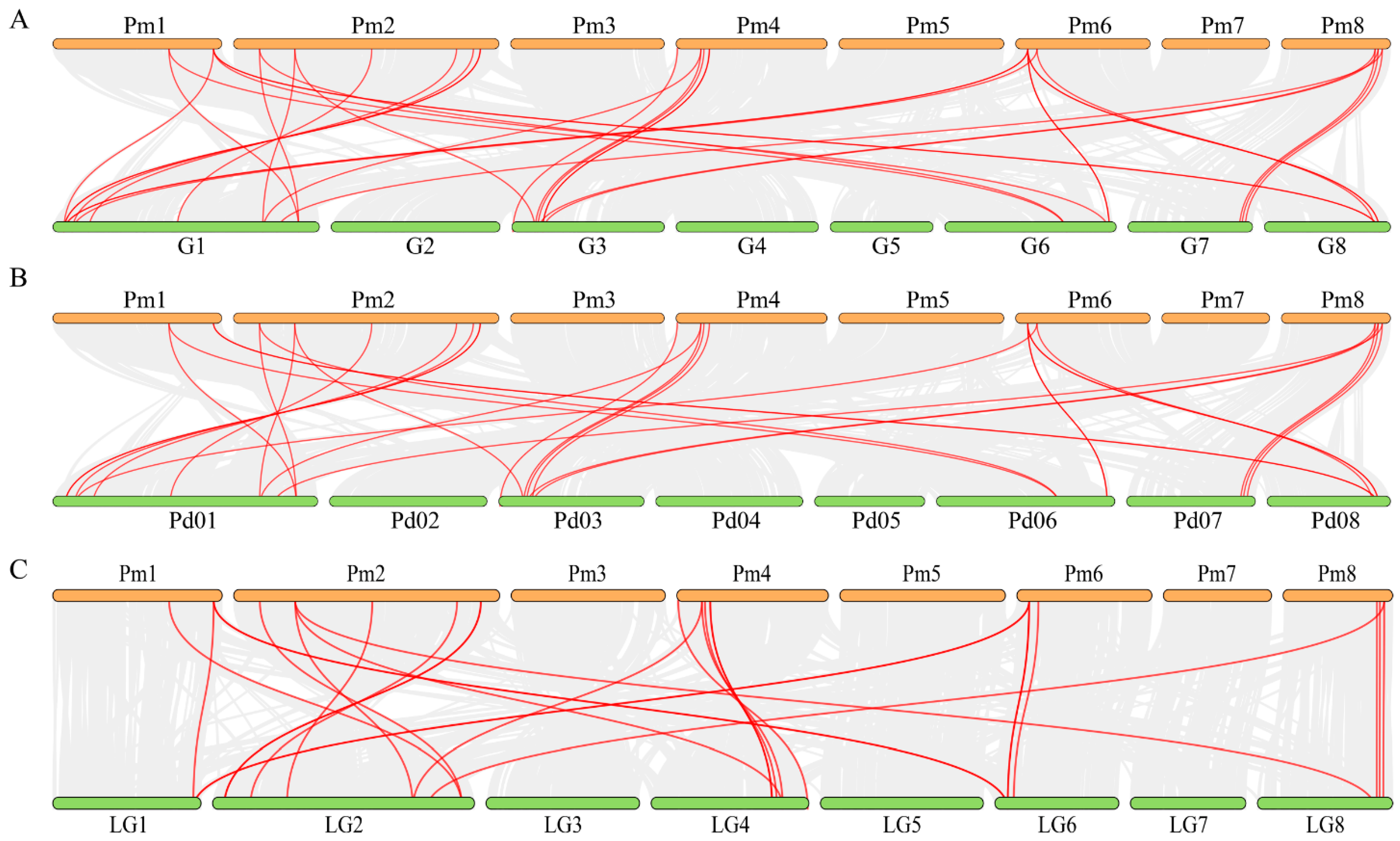
3.5. Synteny Analyses
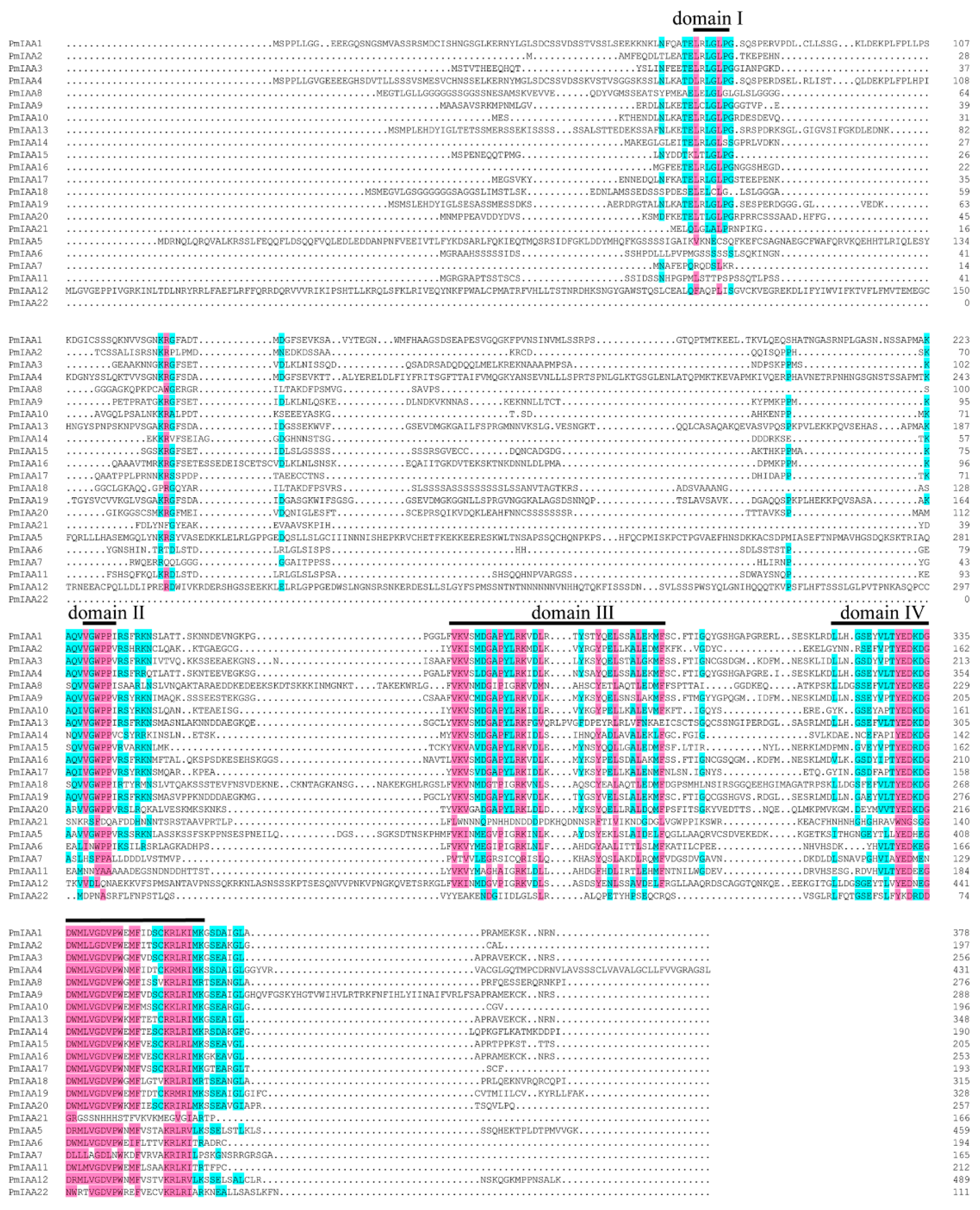
3.6. Sequence Analysis of PmIAAs in P. mume
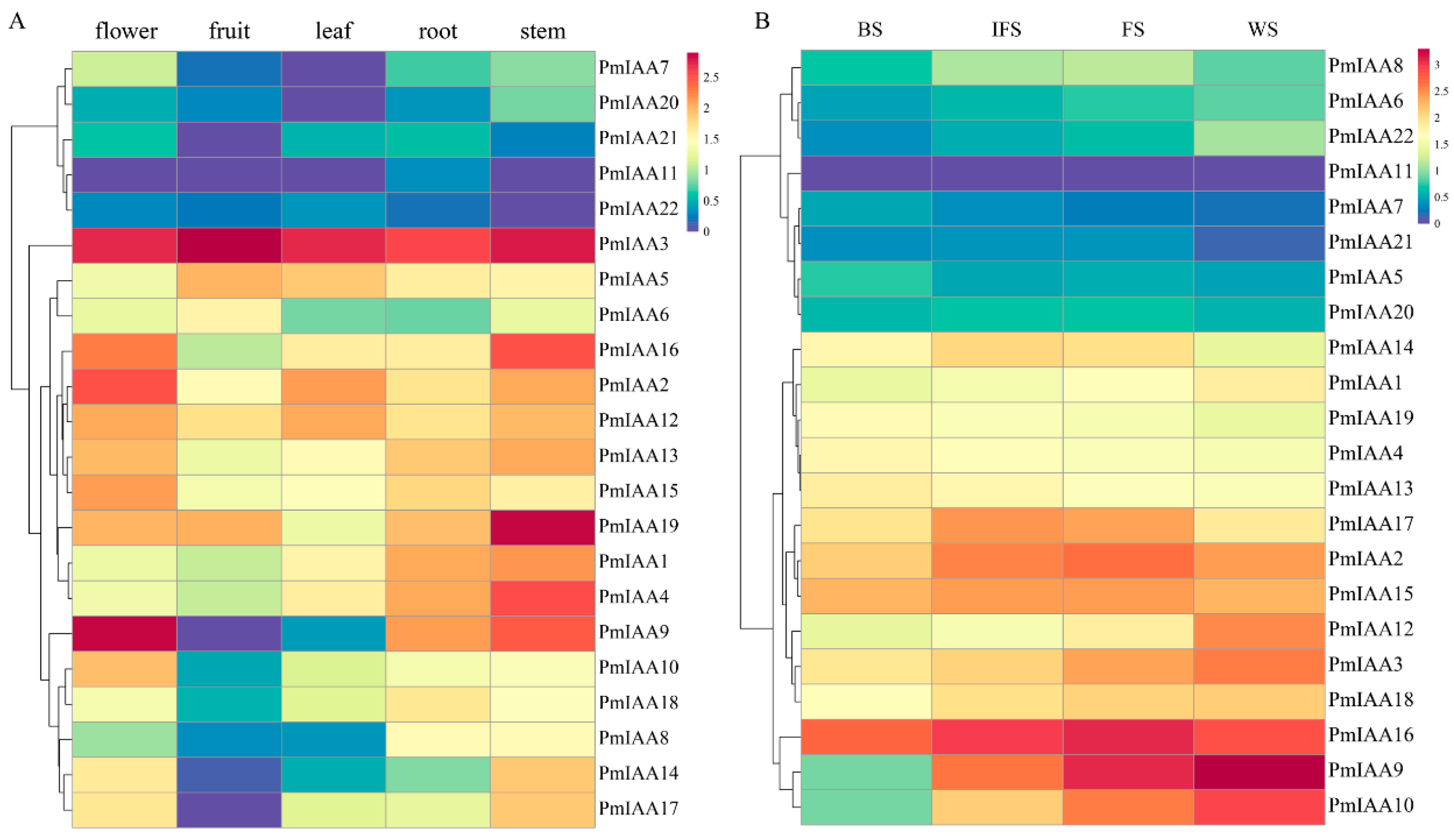
3.7. Expression Profiles of PmIAAs in Different Tissues and at Different Flowering Development Stages
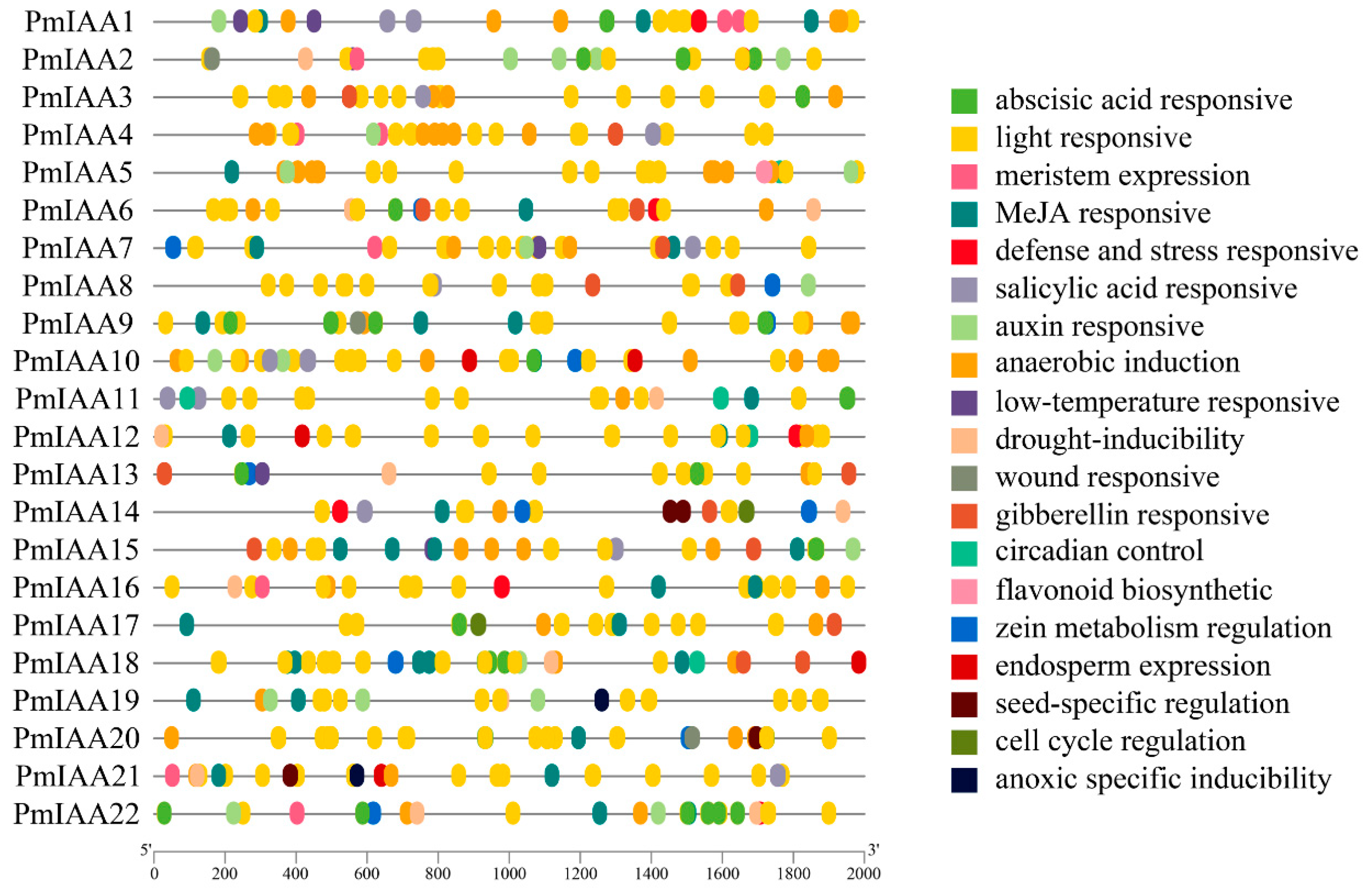
3.8. Promoter Analysis of Cis-Acting Elements of PmIAAs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Goldental-Cohen, S.; Israeli, A.; Ori, N.; Yasuor, H. Auxin Response Dynamics During Wild-Type and entire Flower Development in Tomato. Plant Cell Physiol. 2017, 58, 1661–1672. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.; Tyagi, A.K.; Sharma, A.K. Genome-wide analysis of auxin response factor (ARF) gene family from tomato and analysis of their role in flower and fruit development. Mol. Genet. Genom. 2011, 285, 245–260. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Soler, M.; Clemente, H.S.; Mila, I.; Paiva, J.A.P.; Myburg, A.A.; Bouzayen, M.; Grima-Pettenati, J.; Cassan-Wang, H. Comprehensive Genome-Wide Analysis of the Aux/IAA Gene Family in Eucalyptus: Evidence for the Role of EgrIAA4 in Wood Formation. Plant Cell Physiol. 2015, 56, 700–714. [Google Scholar] [CrossRef]
- Zhang, X.R.; Zhang, K.; Luo, L.; Lv, Y.Y.; Li, Y.Y.; Zhu, S.Q.; Luo, B.; Wan, Y.S.; Zhang, X.S.; Liu, F.Z. Identification of Peanut Aux/IAA Genes and Functional Prediction during Seed Development and Maturation. Plants 2022, 11, 472. [Google Scholar] [CrossRef] [PubMed]
- Aloni, R.; Aloni, E.; Langhans, M.; Ullrich, C.I. Role of auxin in regulating Arabidopsis flower development. Planta 2006, 223, 315–328. [Google Scholar] [CrossRef]
- Luo, J.; Zhou, J.J.; Zhang, J.Z. Aux/IAA Gene Family in Plants: Molecular Structure, Regulation, and Function. Int. J. Mol. Sci. 2018, 19, 259. [Google Scholar] [CrossRef] [PubMed]
- Wei, S.; Chen, Y.; Hou, J.; Yang, Y.; Yin, T. Aux/IAA and ARF Gene Families in Salix suchowensis: Identification, Evolution, and Dynamic Transcriptome Profiling During the Plant Growth Process. Front. Plant Sci. 2021, 12, 666310. [Google Scholar] [CrossRef]
- Abel, S.; Oeller, P.W.; Theologis, A. Early auxin-induced genes encode short-lived nuclear proteins. Proc. Natl. Acad. Sci. USA 1994, 91, 326–330. [Google Scholar] [CrossRef]
- Tiwari, S.B.; Wang, X.J.; Hagen, G.; Guilfoyle, T.J. AUX/IAA proteins are active repressors, and their stability and activity are modulated by auxin. Plant Cell 2001, 13, 2809–2822. [Google Scholar] [CrossRef]
- Reed, J.W. Roles and activities of Aux/IAA proteins in Arabidopsis. Trends Plant Sci. 2001, 6, 420–425. [Google Scholar] [CrossRef]
- Tiwari, S.B.; Hagen, G.; Guilfoyle, T.J. Aux/IAA proteins contain a potent transcriptional repression domain. Plant Cell 2004, 16, 533–543. [Google Scholar] [CrossRef] [PubMed]
- Hagen, G.; Guilfoyle, T. Auxin-responsive gene expression: Genes, promoters and regulatory factors. Plant Mol. Biol. 2002, 49, 373–385. [Google Scholar] [CrossRef] [PubMed]
- Guilfoyle, T.J. The PB1 domain in auxin response factor and Aux/IAA proteins: A versatile protein interaction module in the auxin response. Plant Cell 2015, 27, 33–43. [Google Scholar] [CrossRef] [PubMed]
- Orlova, I.; Marshall-Colon, A.; Schnepp, J.; Wood, B.; Varbanova, M.; Fridman, E.; Blakeslee, J.J.; Peer, W.A.; Murphy, A.S.; Rhodes, D.; et al. Reduction of benzenoid synthesis in petunia flowers reveals multiple pathways to benzoic acid and enhancement in auxin transport. Plant Cell 2006, 18, 3458–3475. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.K.; Han, P.L.; Zhao, Y.W.; Ji, X.L.; Yu, J.Q.; You, C.X.; Hu, D.G.; Hao, Y.J. Auxin regulates anthocyanin biosynthesis through the auxin repressor protein MdIAA26. Biochem. Bioph. Res. Commun. 2020, 533, 717–722. [Google Scholar] [CrossRef] [PubMed]
- Ke, Y.; Abbas, F.; Zhou, Y.; Yu, R.; Yue, Y.; Li, X.; Yu, Y.; Fan, Y. Genome-Wide Analysis and Characterization of the Aux/IAA Family Genes Related to Floral Scent Formation in Hedychium coronarium. Int. J. Mol. Sci. 2019, 20, 3235. [Google Scholar] [CrossRef]
- Deng, W.; Yan, F.; Liu, M.; Wang, X.; Li, Z. Down-regulation of SlIAA15 in tomato altered stem xylem development and production of volatile compounds in leaf exudates. Plant Signal. Behav. 2012, 7, 911–913. [Google Scholar] [CrossRef]
- Zhang, T.; Bao, F.; Yang, Y.; Hu, L.; Ding, A.; Ding, A.; Wang, J.; Cheng, T.; Zhang, Q. A Comparative Analysis of Floral Scent Compounds in Intraspecific Cultivars of Prunus mume with Different Corolla Colours. Molecules 2019, 25, 145. [Google Scholar] [CrossRef]
- Hao, R.; Du, D.; Wang, T.; Yang, W.; Wang, J.; Zhang, Q. A comparative analysis of characteristic floral scent compounds in Prunus mume and related species. Biosci. Biotechnol. Biochem. 2014, 78, 1640–1647. [Google Scholar] [CrossRef]
- Wang, X.; Wu, Y.; Zhu, H.; Zhang, H.; Xu, J.; Fu, Q.; Bao, M.; Zhang, J. Headspace Volatiles and Endogenous Extracts of Prunus mume Cultivars with Different Aroma Types. Molecules 2021, 26, 7256. [Google Scholar] [CrossRef]
- Hao, R.J.; Zhang, Q.; Yang, W.R.; Wang, J.; Cheng, T.R.; Pan, H.T.; Zhang, Q.X. Emitted and endogenous floral scent compounds of Prunus mume and hybrids. Biochem. Syst. Ecol. 2014, 54, 23–30. [Google Scholar] [CrossRef]
- Su, Y.; He, H.; Wang, P.; Ma, Z.; Mao, J.; Chen, B. Genome-wide characterization and expression analyses of the auxin/indole-3-acetic acid (Aux/IAA) gene family in apple (Malus domestica). Gene 2021, 768, 145302. [Google Scholar] [CrossRef] [PubMed]
- Kalluri, U.C.; Difazio, S.P.; Brunner, A.M.; Tuskan, G.A. Genome-wide analysis of Aux/IAA and ARF gene families in Populus trichocarpa. BMC Plant Biol. 2007, 7, 59. [Google Scholar] [CrossRef] [PubMed]
- Hou, Y.M.; Li, H.X.; Zhai, L.L.; Xie, X.; Li, X.Y.; Bian, S.M. Identification and functional characterization of the Aux/IAA gene VcIAA27 in blueberry. Plant Signal. Behav. 2020, 15, 1700327. [Google Scholar] [CrossRef] [PubMed]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Chen, C.J.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.H.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Wang, D.; Zhang, Y.; Zhang, Z.; Zhu, J.; Yu, J. KaKs_Calculator 2.0: A toolkit incorporating gamma-series methods and sliding window strategies. Genom. Proteom. Bioinform. 2010, 8, 77–80. [Google Scholar] [CrossRef]
- Huang, Z.; Duan, W.; Song, X.; Tang, J.; Wu, P.; Zhang, B.; Hou, X. Retention, Molecular Evolution, and Expression Divergence of the Auxin/Indole Acetic Acid and Auxin Response Factor Gene Families in Brassica rapa Shed Light on Their Evolution Patterns in Plants. Genome Biol. Evol. 2015, 8, 302–316. [Google Scholar] [CrossRef]
- Li, F.; Wu, M.; Liu, H.; Gao, Y.; Xiang, Y. Systematic identification and expression pattern analysis of the Aux/IAA and ARF gene families in moso bamboo (Phyllostachys edulis). Plant Physiol. Biochem. PPB 2018, 130, 431–444. [Google Scholar] [CrossRef]
- Zhang, T.; Huo, T.; Ding, A.; Hao, R.; Wang, J.; Cheng, T.; Bao, F.; Zhang, Q. Genome-wide identification, characterization, expression and enzyme activity analysis of coniferyl alcohol acetyltransferase genes involved in eugenol biosynthesis in Prunus mume. PLoS ONE 2019, 14, e0223974. [Google Scholar] [CrossRef]
- Jain, M.; Kaur, N.; Garg, R.; Thakur, J.K.; Tyagi, A.K.; Khurana, J.P. Structure and expression analysis of early auxin-responsive Aux/IAA gene family in rice (Oryza sativa). Funct. Integr. Genom. 2006, 6, 47–59. [Google Scholar] [CrossRef] [PubMed]
- Shi, Q.; Zhang, Y.; To, V.T.; Shi, J.; Zhang, D.; Cai, W. Genome-wide characterization and expression analyses of the auxin/indole-3-acetic acid (Aux/IAA) gene family in barley (Hordeum vulgare L.). Sci. Rep. 2020, 10, 10242. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; Xu, X.; Fang, D.D.; Zhang, T.; Guo, W. Cloning and expression analysis of novel Aux/IAA family genes in Gossypium hirsutum. Gene 2012, 503, 83–91. [Google Scholar] [CrossRef]
- Xie, Y.; Ying, J.; Tang, M.; Wang, Y.; Xu, L.; Liu, M.; Liu, L. Genome-wide identification of AUX/IAA in radish and functional characterization of RsIAA33 gene during taproot thickening. Gene 2021, 795, 145782. [Google Scholar] [CrossRef]
- Singh, V.K.; Jain, M. Genome-wide survey and comprehensive expression profiling of Aux/IAA gene family in chickpea and soybean. Front. Plant Sci. 2015, 6, 918. [Google Scholar] [CrossRef]
- Audran-Delalande, C.; Bassa, C.; Mila, I.; Regad, F.; Zouine, M.; Bouzayen, M. Genome-wide identification, functional analysis and expression profiling of the Aux/IAA gene family in tomato. Plant Cell Physiol. 2012, 53, 659–672. [Google Scholar] [CrossRef]
- Wang, Y.; Deng, D.; Shi, Y.; Miao, N.; Bian, Y.; Yin, Z. Diversification, phylogeny and evolution of auxin response factor (ARF) family: Insights gained from analyzing maize ARF genes. Mol. Biol. Rep. 2012, 39, 2401–2415. [Google Scholar] [CrossRef]
- Bao, F.; Ding, A.Q.; Zhang, T.X.; Luo, L.; Wang, J.; Cheng, T.R.; Zhang, Q.X. Expansion of PmBEAT genes in the Prunus mume genome induces characteristic floral scent production. Hortic. Res. Engl. 2019, 6, 24. [Google Scholar] [CrossRef]
- Zhang, T.X.; Guo, Y.H.; Shi, X.J.; Yang, Y.J.; Chen, J.T.; Zhang, Q.X.; Sun, M. Overexpression of LiTPS2 from a cultivar of lily (Lilium ‘Siberia’) enhances the monoterpenoids content in tobacco flowers. Plant Physiol. Bioch. 2020, 151, 391–399. [Google Scholar] [CrossRef]
- Gao, F.Z.; Liu, B.F.; Li, M.; Gao, X.Y.; Fang, Q.; Liu, C.; Ding, H.; Wang, L.; Gao, X. Identification and characterization of terpene synthase genes accounting for volatile terpene emissions in flowers of Freesia × hybrida. J. Exp. Bot. 2018, 69, 4249–4265. [Google Scholar] [CrossRef]
- Shalit, M.; Guterman, I.; Volpin, H.; Bar, E.; Tamari, T.; Menda, N.; Adam, Z.; Zamir, D.; Vainstein, A.; Weiss, D.; et al. Volatile ester formation in roses. Identification of an acetyl-coenzyme A. Geraniol/citronellol acetyltransferase in developing rose petals. Plant Physiol. 2003, 131, 1868–1876. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Bao, F.; Ding, A.; Yang, Y.; Cheng, T.; Wang, J.; Zhang, Q. Comprehensive Analysis of Endogenous Volatile Compounds, Transcriptome, and Enzyme Activity Reveals PmCAD1 Involved in Cinnamyl Alcohol Synthesis in Prunus mume. Front. Plant. Sci. 2022, 13, 820742. [Google Scholar] [CrossRef] [PubMed]
- Knox, K.; Grierson, C.S.; Leyser, O. AXR3 and SHY2 interact to regulate root hair development. Development 2003, 130, 5769–5777. [Google Scholar] [CrossRef] [PubMed]
| Locus Name | Gene ID | Chr. 1 | Start (bp) | End (bp) | ORF Length (bp) | Protein Length (aa) | MW (kDa) | pI | Num_Exons |
|---|---|---|---|---|---|---|---|---|---|
| PmIAA1 | Pm002286 | Pm1 | 18,410,490 | 18,413,935 | 1137 | 378 | 40.82 | 6.97 | 5 |
| PmIAA2 | Pm003529 | Pm1 | 25,485,479 | 25,486,334 | 594 | 197 | 22.27 | 8.10 | 3 |
| PmIAA3 | Pm003530 | Pm1 | 25,493,715 | 25,495,247 | 771 | 256 | 28.21 | 6.76 | 5 |
| PmIAA4 | Pm004328 | Pm2 | 4,075,856 | 4,078,825 | 1299 | 432 | 46.76 | 6.68 | 5 |
| PmIAA5 | Pm005182 | Pm2 | 9,685,220 | 9,690,217 | 1380 | 459 | 51.86 | 7.59 | 10 |
| PmIAA6 | Pm007165 | Pm2 | 21,921,866 | 21,922,839 | 585 | 194 | 21.31 | 6.79 | 4 |
| PmIAA7 | Pm008719 | Pm2 | 35,394,523 | 35,395,273 | 498 | 165 | 18.24 | 9.20 | 2 |
| PmIAA8 | Pm009031 | Pm2 | 38,082,783 | 38,084,821 | 831 | 276 | 29.46 | 5.52 | 5 |
| PmIAA9 | Pm009169 | Pm2 | 39,166,644 | 39,168,404 | 867 | 288 | 32.5 | 9.27 | 5 |
| PmIAA10 | Pm009170 | Pm2 | 39,182,270 | 39,183,111 | 591 | 196 | 22.03 | 6.76 | 3 |
| PmIAA11 | Pm012868 | Pm4 | 174,190 | 175,421 | 639 | 212 | 23.35 | 5.58 | 4 |
| PmIAA12 | Pm013416 | Pm4 | 3,933,319 | 3,937,638 | 1470 | 489 | 54.98 | 9.34 | 6 |
| PmIAA13 | Pm013483 | Pm4 | 4,405,042 | 4,407,525 | 1047 | 348 | 37.89 | 8.36 | 6 |
| PmIAA14 | Pm013596 | Pm4 | 5,254,018 | 5,257,147 | 573 | 190 | 21.19 | 6.74 | 3 |
| PmIAA15 | Pm013597 | Pm4 | 5,264,672 | 5,265,938 | 618 | 205 | 22.73 | 7.56 | 4 |
| PmIAA16 | Pm020225 | Pm6 | 1,861,906 | 1,863,664 | 762 | 253 | 28.03 | 6.23 | 5 |
| PmIAA17 | Pm020227 | Pm6 | 1,871,062 | 1,871,873 | 582 | 193 | 21.70 | 5.96 | 3 |
| PmIAA18 | Pm020501 | Pm6 | 3,334,930 | 3,337,142 | 948 | 315 | 32.85 | 6.84 | 5 |
| PmIAA19 | Pm027361 | Pm8 | 14,851,155 | 14,853,714 | 987 | 328 | 34.90 | 6.36 | 4 |
| PmIAA20 | Pm027456 | Pm8 | 15,328,176 | 15,329,553 | 774 | 257 | 28.82 | 8.69 | 5 |
| PmIAA21 | Pm027585 | Pm8 | 15,994,145 | 15,994,901 | 501 | 166 | 18.66 | 8.00 | 2 |
| PmIAA22 | Pm029117 | Pm0 2 | 1,482,408 | 1,483,237 | 336 | 111 | 12.87 | 6.57 | 3 |
| PpIAA1 | Prupe.1G540700.1 | G1 | 44,179,629 | 44,185,073 | 1194 | 397 | 43.43 | 6.68 | 5 |
| PpIAA2 | Prupe.6G210500.1 | G6 | 21,881,526 | 21,887,147 | 1137 | 378 | 40.87 | 6.56 | 5 |
| PpIAA3 | Prupe.3G064200.1 | G3 | 4,585,165 | 4,588,237 | 1032 | 343 | 37.24 | 7.56 | 5 |
| PpIAA4 | Prupe.7G225600.1 | G7 | 19,999,625 | 20,002,918 | 966 | 321 | 34.05 | 6.84 | 5 |
| PpIAA5 | Prupe.8G232200.1 | G8 | 20,644,589 | 20,646,341 | 762 | 253 | 27.88 | 6.76 | 5 |
| PpIAA6 | Prupe.6G343800.1 | G6 | 29,582,927 | 29,585,218 | 768 | 255 | 27.91 | 5.86 | 4 |
| PpIAA7 | Prupe.1G027600.1 | G1 | 1,933,812 | 1,936,072 | 747 | 248 | 27.50 | 8.61 | 5 |
| PpIAA8 | Prupe.6G343700.1 | G6 | 29,575,247 | 29,576,745 | 594 | 197 | 22.38 | 7.49 | 3 |
| PpIAA9 | Prupe.1G027500.1 | G1 | 1,919,207 | 1,920,881 | 591 | 196 | 22.03 | 6.22 | 3 |
| PpIAA10 | Prupe.8G232400.1 | G8 | 20,653,624 | 20,655,672 | 582 | 193 | 21.71 | 5.96 | 3 |
| PpIAA11 | Prupe.3G074900.1 | G3 | 5,490,339 | 5,492,764 | 732 | 243 | 27.04 | 8.92 | 5 |
| PpIAA12 | Prupe.7G234800.1 | G7 | 20,438,451 | 20,440,489 | 768 | 255 | 28.41 | 8.22 | 5 |
| PpIAA13 | Prupe.3G074800.1 | G3 | 5,480,786 | 5,484,245 | 573 | 190 | 21.30 | 7.61 | 3 |
| PpIAA14 | Prupe.3G058600.1 | G3 | 4,137,369 | 4,141,973 | 1083 | 360 | 39.74 | 8.96 | 5 |
| PpIAA15 | Prupe.8G215400.1 | G8 | 19,778,009 | 19,780,277 | 942 | 313 | 32.63 | 6.84 | 5 |
| PpIAA16 | Prupe.1G049200.1 | G1 | 3,457,675 | 3,460,523 | 1242 | 413 | 44.22 | 8.95 | 5 |
| PpIAA17 | Prupe.7G247500.1 | G7 | 21,022,925 | 21,025,300 | 699 | 232 | 26.49 | 9 | 4 |
| PpIAA18 | Prupe.1G481700.1 | G1 | 40,205,556 | 40,206,744 | 723 | 240 | 27.03 | 9.3 | 4 |
| PpIAA19 | Prupe.1G317000.1 | G1 | 30,532,265 | 30,533,090 | 585 | 194 | 22.35 | 4.89 | 4 |
| PpIAA20 | Prupe.1G208300.1 | G1 | 22,238,292 | 22,239,265 | 588 | 195 | 21.24 | 6.49 | 4 |
| PpIAA21 | Prupe.3G001800.1 | G3 | 141,629 | 143,336 | 642 | 213 | 23.36 | 5.58 | 4 |
| PpIAA22 | Prupe.1G085900.1 | G1 | 6,379,476 | 6,380,776 | 489 | 162 | 17.98 | 9.24 | 2 |
| PdIAA1-a | Prudul26A001240P1 | Pd01 | 1,947,364 | 1,948,899 | 591 | 196 | 22.06 | 6.22 | 3 |
| PdIAA1-b | Prudul26A001240P2 | Pd01 | 1,947,364 | 1,948,899 | 489 | 162 | 18.30 | 7.83 | 2 |
| PdIAA2 | Prudul26A014193P1 | Pd01 | 1,970,552 | 1,972,937 | 747 | 248 | 27.52 | 8.61 | 5 |
| PdIAA3-a | Prudul26A024018P1 | Pd01 | 3,538,043 | 3,540,784 | 936 | 311 | 32.66 | 6.78 | 5 |
| PdIAA3-b | Prudul26A024018P2 | Pd01 | 3,538,043 | 3,540,784 | 933 | 310 | 32.57 | 6.78 | 5 |
| PdIAA4 | Prudul26A003100P1 | Pd01 | 6,586,891 | 6,587,633 | 489 | 162 | 18.00 | 9.46 | 2 |
| PdIAA5-a | Prudul26A001943P1 | Pd01 | 19,446,848 | 19,448,314 | 585 | 194 | 21.20 | 6.49 | 4 |
| PdIAA5-b | Prudul26A001943P2 | Pd01 | 19,446,848 | 19,448,314 | 588 | 195 | 21.24 | 6.49 | 4 |
| PdIAA6 | Prudul26A002782P1 | Pd01 | 27,098,030 | 27,098,872 | 585 | 194 | 22.30 | 5.14 | 4 |
| PdIAA7 | Prudul26A022660P1 | Pd01 | 34,374,468 | 34,377,847 | 1506 | 501 | 56.07 | 8.37 | 5 |
| PdIAA8 | Prudul26A031526P1 | Pd01 | 36,644,328 | 36,645,685 | 723 | 240 | 27.11 | 9.32 | 4 |
| PdIAA9-a | Prudul26A030270P1 | Pd01 | 40,499,216 | 40,505,180 | 1101 | 366 | 39.74 | 7.05 | 6 |
| PdIAA9-b | Prudul26A030270P2 | Pd01 | 40,499,216 | 40,505,180 | 1194 | 397 | 43.41 | 7.06 | 6 |
| PdIAA10 | Prudul26A009797P1 | Pd03 | 156,374 | 157,587 | 642 | 213 | 23.43 | 5.58 | 4 |
| PdIAA11 | Prudul26A015075P1 | Pd03 | 4,069,233 | 4,073,513 | 1095 | 364 | 40.20 | 8.58 | 5 |
| PdIAA12 | Prudul26A031482P1 | Pd03 | 4,553,562 | 4,556,627 | 1032 | 343 | 37.23 | 8.1 | 5 |
| PdIAA13 | Prudul26A003871P1 | Pd03 | 5,473,504 | 5,474,756 | 618 | 205 | 22.69 | 7.56 | 4 |
| PdIAA14 | Prudul26A019193P1 | Pd06 | 20,456,945 | 20,461,668 | 1137 | 378 | 40.89 | 6.56 | 5 |
| PdIAA15 | Prudul26A024452P1 | Pd06 | 28,374,773 | 28,376,887 | 594 | 197 | 22.20 | 6 | 3 |
| PdIAA16-a | Prudul26A031524P1 | Pd06 | 28,383,480 | 28,385,565 | 570 | 189 | 20.69 | 6.17 | 2 |
| PdIAA16-b | Prudul26A031524P2 | Pd06 | 28,383,480 | 28,385,565 | 768 | 255 | 27.98 | 5.62 | 5 |
| PdIAA17 | Prudul26A009915P1 | Pd07 | 18,894,197 | 18,897,392 | 960 | 319 | 33.89 | 6.36 | 5 |
| PdIAA18 | Prudul26A010551P1 | Pd07 | 19,357,246 | 19,358,569 | 768 | 255 | 28.43 | 7.59 | 5 |
| PdIAA19 | Prudul26A007192P1 | Pd07 | 19,959,797 | 19,961,008 | 699 | 232 | 26.42 | 8.48 | 4 |
| PdIAA20-a | Prudul26A028501P1 | Pd08 | 17,537,269 | 17,540,308 | 948 | 315 | 32.75 | 6.84 | 5 |
| PdIAA20-b | Prudul26A028501P2 | Pd08 | 17,537,269 | 17,540,308 | 699 | 232 | 23.48 | 6.73 | 2 |
| PdIAA20-c | Prudul26A028501P3 | Pd08 | 17,537,269 | 17,540,308 | 945 | 314 | 32.66 | 6.84 | 5 |
| PdIAA21 | Prudul26A032023P1 | Pd08 | 18,504,485 | 18,511,833 | 762 | 253 | 27.89 | 6.85 | 5 |
| PdIAA22 | Prudul26A030184P1 | Pd08 | 18,518,899 | 18,520,734 | 579 | 192 | 21.62 | 6.62 | 3 |
| PdIAA23 | Prudul26A032154P1 | Pd00 3 | 32,753 | 36,165 | 573 | 190 | 21.33 | 7.61 | 3 |
| PdIAA24 | Prudul26A013580P1 | Pd00 3 | 43,975 | 45,227 | 618 | 205 | 22.69 | 7.56 | 5 |
| PaIAA1-a | PARG03096m01 | LG1 | 23,057,220 | 23,059,328 | 768 | 255 | 27.96 | 6.76 | 5 |
| PaIAA1-b | PARG03096m03 | LG1 | 23,057,414 | 23,062,237 | 1107 | 368 | 40.35 | 8.47 | 6 |
| PaIAA2 | PARG03559m01 | LG2 | 1,794,461 | 1,796,003 | 591 | 196 | 21.96 | 6.76 | 3 |
| PaIAA3 | PARG03560m01 | LG2 | 1,808,332 | 1,810,095 | 747 | 248 | 27.53 | 8.83 | 5 |
| PaIAA4-a | PARG04113m01 | LG2 | 6,111,298 | 6,112,049 | 501 | 166 | 18.24 | 9.51 | 2 |
| PaIAA4-b | PARG04113m02 | LG2 | 6,111,103 | 6,112,270 | 501 | 166 | 18.24 | 9.51 | 2 |
| PaIAA5-a | PARG04885m01 | LG2 | 12,147,321 | 12,149,051 | 585 | 194 | 21.27 | 6.54 | 4 |
| PaIAA5-b | PARG04885m02 | LG2 | 12,148,209 | 12,149,051 | 531 | 176 | 19.08 | 9.17 | 2 |
| PaIAA5-c | PARG04885m03 | LG2 | 12,147,871 | 12,148,911 | 633 | 210 | 23.07 | 5.57 | 4 |
| PaIAA6-a | PARG07885m01 | LG2 | 32,902,089 | 32,903,561 | 948 | 315 | 34.85 | 8.31 | 5 |
| PaIAA6-b | PARG07885m02 | LG2 | 32,902,089 | 32,903,561 | 918 | 305 | 33.73 | 8.36 | 5 |
| PaIAA6-c | PARG07885m05 | LG2 | 32,902,089 | 32,907,032 | 1347 | 448 | 50.56 | 6.9 | 10 |
| PaIAA7 | PARG08244m01 | LG2 | 35,263,474 | 35,264,604 | 705 | 234 | 26.49 | 9.33 | 4 |
| PaIAA8-a | PARG09013m01 | LG2 | 41,017,040 | 41,022,781 | 1101 | 366 | 39.72 | 7.05 | 6 |
| PaIAA8-b | PARG09013m02 | LG2 | 41,017,040 | 41,022,781 | 1101 | 366 | 39.72 | 7.05 | 6 |
| PaIAA8-c | PARG09013m03 | LG2 | 41,017,040 | 41,019,779 | 882 | 293 | 31.35 | 6.97 | 4 |
| PaIAA8-d | PARG09013m04 | LG2 | 41,017,040 | 41,019,703 | 927 | 308 | 33.24 | 6.97 | 3 |
| PaIAA9 | PARG15463m01 | LG4 | 19,847,123 | 19,848,823 | 636 | 211 | 23.27 | 7.56 | 4 |
| PaIAA10 | PARG15551m01 | LG4 | 20,614,154 | 20,617,123 | 1032 | 343 | 37.36 | 7.56 | 5 |
| PaIAA11 | PARG15664m01 | LG4 | 21,590,856 | 21,594,273 | 1080 | 359 | 39.46 | 8.58 | 5 |
| PaIAA12-a | PARG20177m01 | LG6 | 2,032,112 | 2,034,813 | 762 | 253 | 27.91 | 6.76 | 5 |
| PaIAA12-b | PARG20177m02 | LG6 | 2,025,721 | 2,035,609 | 1380 | 459 | 50.97 | 9.46 | 6 |
| PaIAA13-a | PARG20341m01 | LG6 | 3,001,500 | 3,014,665 | 918 | 305 | 32.00 | 7.61 | 6 |
| PaIAA13-b | PARG20341m02 | LG6 | 3,012,400 | 3,014,665 | 939 | 312 | 32.50 | 7.61 | 5 |
| PaIAA14 | PARG27893m01 | LG8 | 19,566,836 | 19,570,224 | 960 | 319 | 33.85 | 6.76 | 5 |
| PaIAA15-a | PARG27974m01 | LG8 | 20,043,833 | 20,045,628 | 768 | 255 | 28.46 | 8.55 | 5 |
| PaIAA15-b | PARG27974m02 | LG8 | 20,043,833 | 20,045,224 | 816 | 271 | 30.46 | 9.01 | 4 |
| PaIAA16-a | PARG29743m01 | LG0 4 | 289,408 | 294,763 | 1137 | 378 | 40.79 | 6.62 | 5 |
| PaIAA16-b | PARG29743m02 | LG0 | 289,408 | 294,763 | 1137 | 378 | 40.79 | 6.62 | 5 |
| PaIAA16-c | PARG29743m03 | LG0 | 289,408 | 294,763 | 1137 | 378 | 40.79 | 6.62 | 5 |
| PaIAA16-d | PARG29743m04 | LG0 | 290,432 | 294,763 | 1137 | 378 | 40.79 | 6.62 | 5 |
| PaIAA16-e | PARG29743m05 | LG0 | 289,408 | 293,554 | 1239 | 412 | 44.68 | 6.33 | 4 |
| PaIAA16-f | PARG29743m06 | LG0 | 290,834 | 296,512 | 1176 | 391 | 42.38 | 5.66 | 5 |
| Syntenic Gene Pairs | Type of Duplication | Ka | Ks | Ka/Ks | Divergence Time (Mya) |
|---|---|---|---|---|---|
| PmIAA1&PmIAA4 | Segmental | 0.223 | 1.843 | 0.121 | 61.44 |
| PmIAA2&PmIAA16 | Segmental | 0.427 | 2.963 | 0.144 | 98.76 |
| PmIAA5&PmIAA12 | Segmental | 0.625 | 2.202 | 0.284 | 73.40 |
| PmIAA9&PmIAA18 | Segmental | 0.642 | 2.262 | 0.284 | 75.41 |
| PmIAA2&PmIAA3 | Tandem | 0.393 | 3.138 | 0.125 | 104.60 |
| PmIAA9&PmIAA10 | Tandem | 0.390 | 3.256 | 0.120 | 108.54 |
| PmIAA14&PmIAA15 | Tandem | 0.441 | 2.850 | 0.155 | 95.00 |
| PpIAA1&PpIAA2 | Segmental | 0.208 | 1.599 | 0.130 | 53.30 |
| PpIAA3&PpIAA12 | Segmental | 0.587 | 2.383 | 0.246 | 79.42 |
| PpIAA4&PpIAA13 | Segmental | 0.385 | 3.090 | 0.125 | 102.99 |
| PpIAA5&PpIAA8 | Segmental | 0.400 | 3.069 | 0.130 | 102.30 |
| PpIAA5&PpIAA9 | Segmental | 0.455 | 2.846 | 0.160 | 94.86 |
| PpIAA8&PpIAA9 | Segmental | 0.259 | 1.784 | 0.145 | 59.47 |
| PpIAA11&PpIAA12 | Segmental | NA | NA | NA | |
| PpIAA15&PpIAA16 | Segmental | 0.290 | 1.955 | 0.149 | 65.17 |
| PpIAA17&PpIAA18 | Segmental | 0.493 | 2.316 | 0.213 | 77.21 |
| PpIAA6&PpIAA8 | Tandem | 0.376 | 3.166 | 0.119 | 105.54 |
| PpIAA7&PpIAA9 | Tandem | 0.364 | 3.241 | 0.112 | 108.04 |
| PdIAA1-a&PdIAA15 | Segmental | 0.255 | 1.782 | 0.143 | 59.41 |
| PdIAA3-a&PdIAA20-a | Segmental | 0.309 | 1.729 | 0.179 | 57.62 |
| PdIAA7&PdIAA11 | Segmental | 0.450 | 1.714 | 0.262 | 57.12 |
| PdIAA8&PdIAA19 | Segmental | 0.499 | 2.280 | 0.219 | 75.99 |
| PdIAA9-b&PdIAA14 | Segmental | 0.205 | 1.573 | 0.130 | 52.44 |
| PdIAA12&PdIAA17 | Segmental | 0.200 | 1.334 | 0.150 | 44.48 |
| PdIAA12&PdIAA18 | Segmental | 0.535 | 2.585 | 0.207 | 86.17 |
| PdIAA13&PdIAA17 | Segmental | 0.316 | 3.221 | 0.098 | 107.36 |
| PdIAA13&PdIAA18 | Segmental | 0.370 | 2.013 | 0.184 | 67.09 |
| PdIAA15&PdIAA21 | Segmental | 0.456 | 2.912 | 0.157 | 97.08 |
| PdIAA1-a&PdIAA2 | Tandem | 0.348 | 3.206 | 0.109 | 106.85 |
| PaIAA6-c&PaIAA11 | Segmental | 0.565 | 1.641 | 0.344 | 54.71 |
| PaIAA8-a&PaIAA16-e | Segmental | 0.211 | 2.161 | 0.097 | 72.03 |
| PaIAA2&PaIAA3 | Tandem | 0.396 | 3.192 | 0.124 | 106.41 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, N.; Li, L.; Chen, X.; Zhang, Y.; Zhang, T. The Auxin/Indole-3-Acetic Acid (Aux/IAA) Gene Family Analysis of Four Rosaceae Genomes and Expression Patterns of PmIAAs in Prunus mume. Horticulturae 2022, 8, 899. https://doi.org/10.3390/horticulturae8100899
Liu N, Li L, Chen X, Zhang Y, Zhang T. The Auxin/Indole-3-Acetic Acid (Aux/IAA) Gene Family Analysis of Four Rosaceae Genomes and Expression Patterns of PmIAAs in Prunus mume. Horticulturae. 2022; 8(10):899. https://doi.org/10.3390/horticulturae8100899
Chicago/Turabian StyleLiu, Nuoxuan, Li Li, Xiling Chen, Yanlong Zhang, and Tengxun Zhang. 2022. "The Auxin/Indole-3-Acetic Acid (Aux/IAA) Gene Family Analysis of Four Rosaceae Genomes and Expression Patterns of PmIAAs in Prunus mume" Horticulturae 8, no. 10: 899. https://doi.org/10.3390/horticulturae8100899
APA StyleLiu, N., Li, L., Chen, X., Zhang, Y., & Zhang, T. (2022). The Auxin/Indole-3-Acetic Acid (Aux/IAA) Gene Family Analysis of Four Rosaceae Genomes and Expression Patterns of PmIAAs in Prunus mume. Horticulturae, 8(10), 899. https://doi.org/10.3390/horticulturae8100899






