Elucidating the Molecular Responses to Waterlogging Stress in Cucumis melo by Comparative Transcriptome Profiling
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Waterlogging Stress Treatment
2.2. Leaf Chlorophyll Content, Malondialdehyde Content, and Relative Electrolyte Leakage (REL) Determination
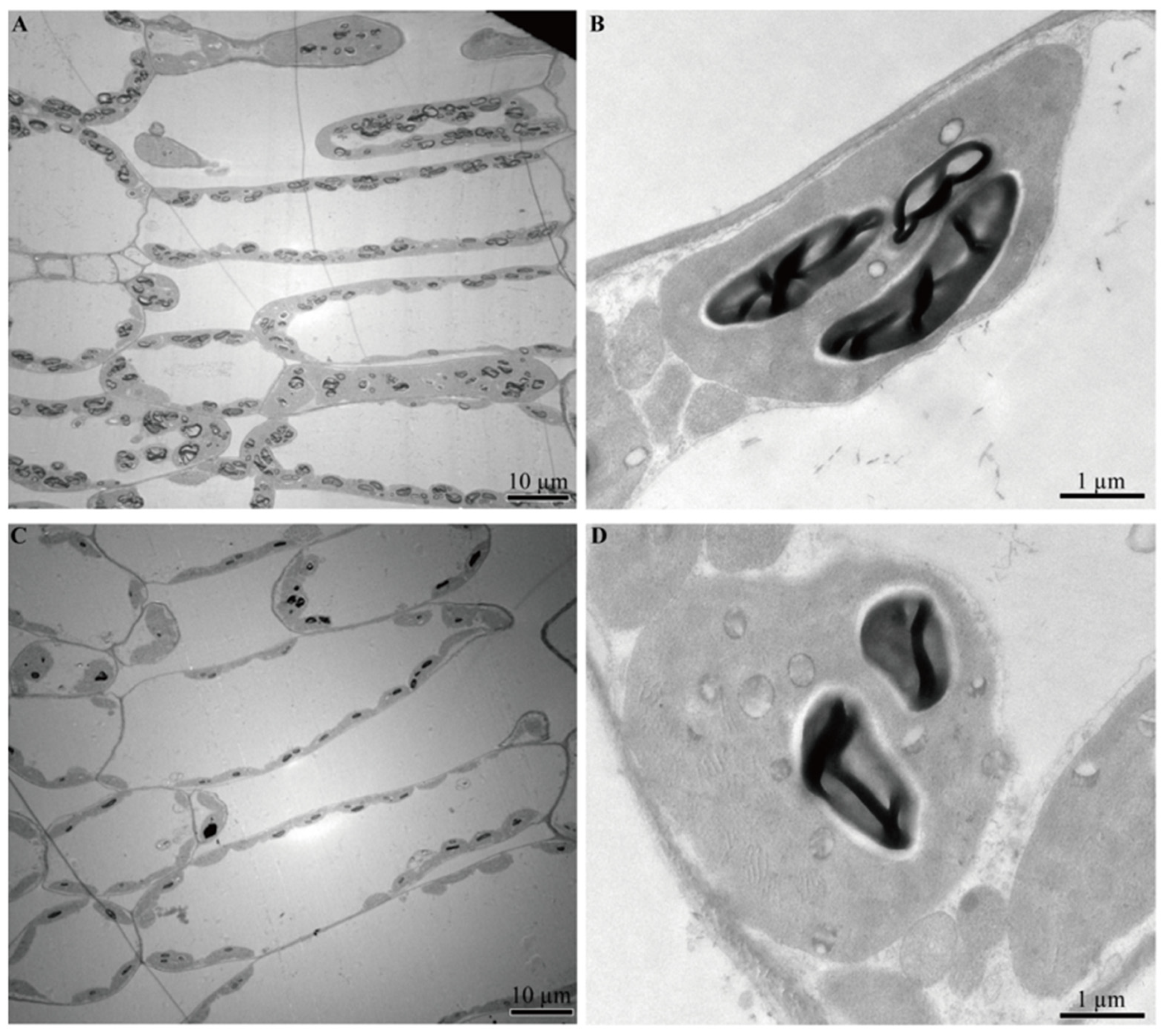
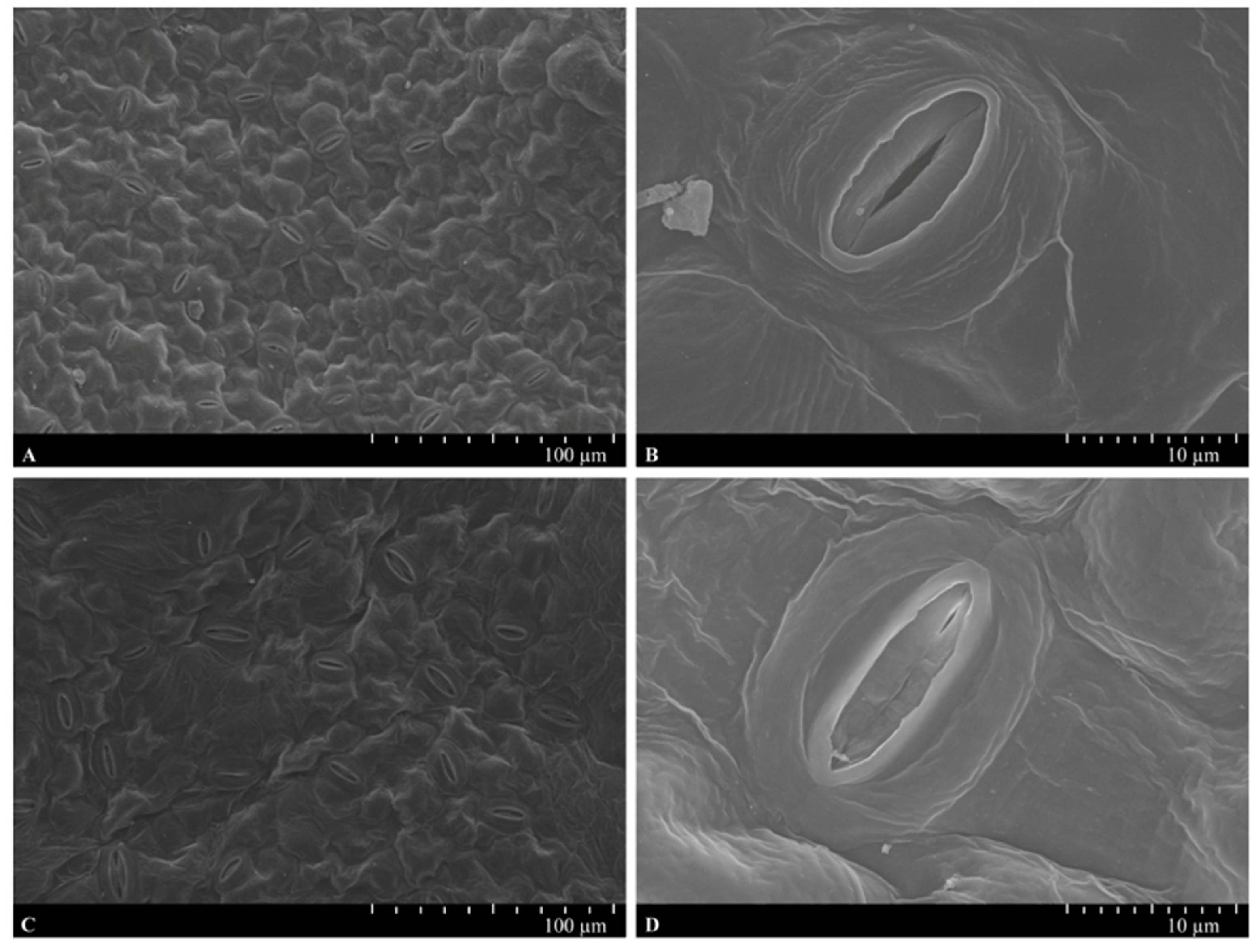
2.3. Ultrastructure Analysis
2.4. RNA Extraction and Transcriptome Sequencing
2.5. Analysis of Transcriptome Data
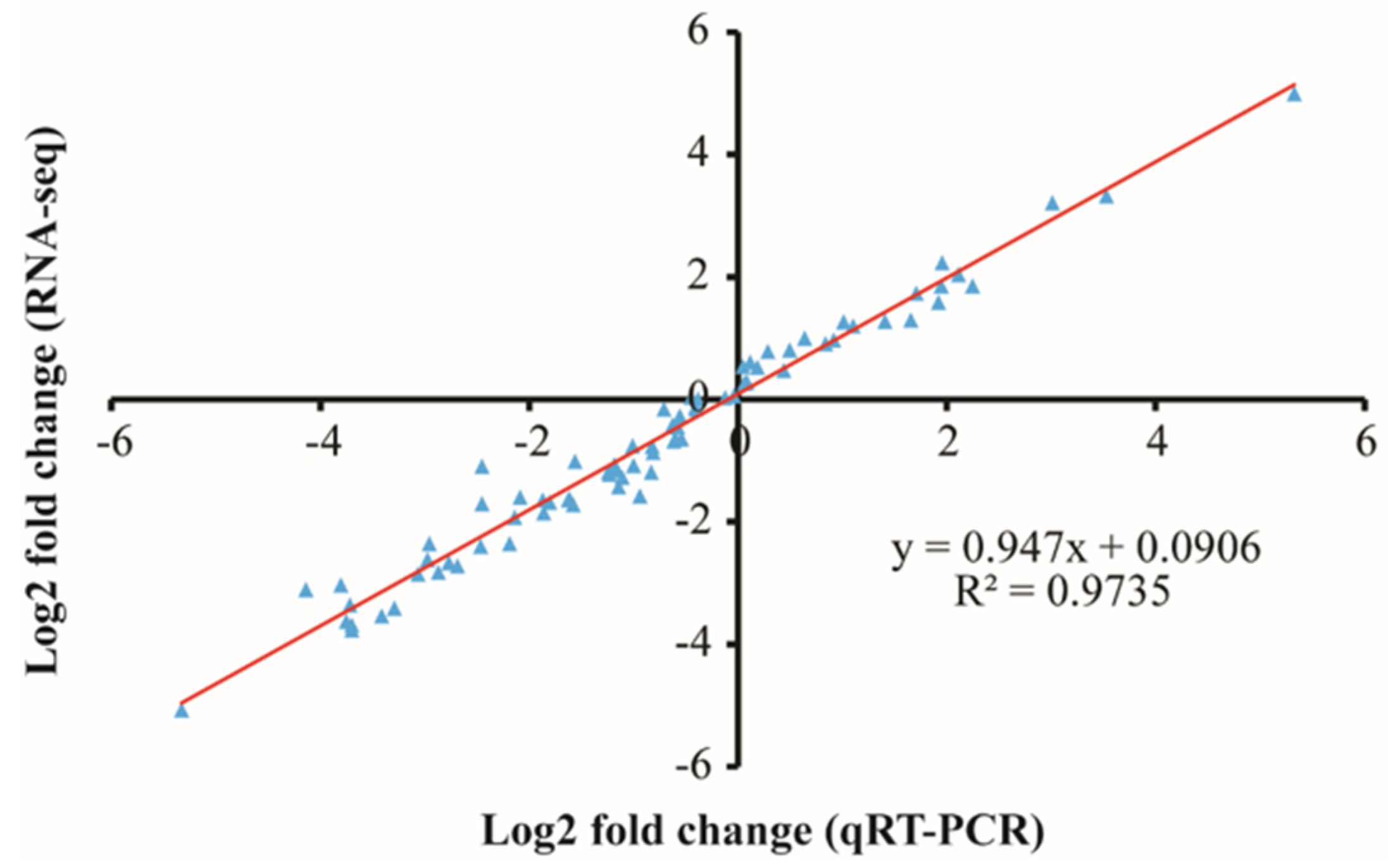
2.6. Quantitative Reverse Transcription-PCR (qRT-PCR) Verification
2.7. Statistical Analysis
3. Results
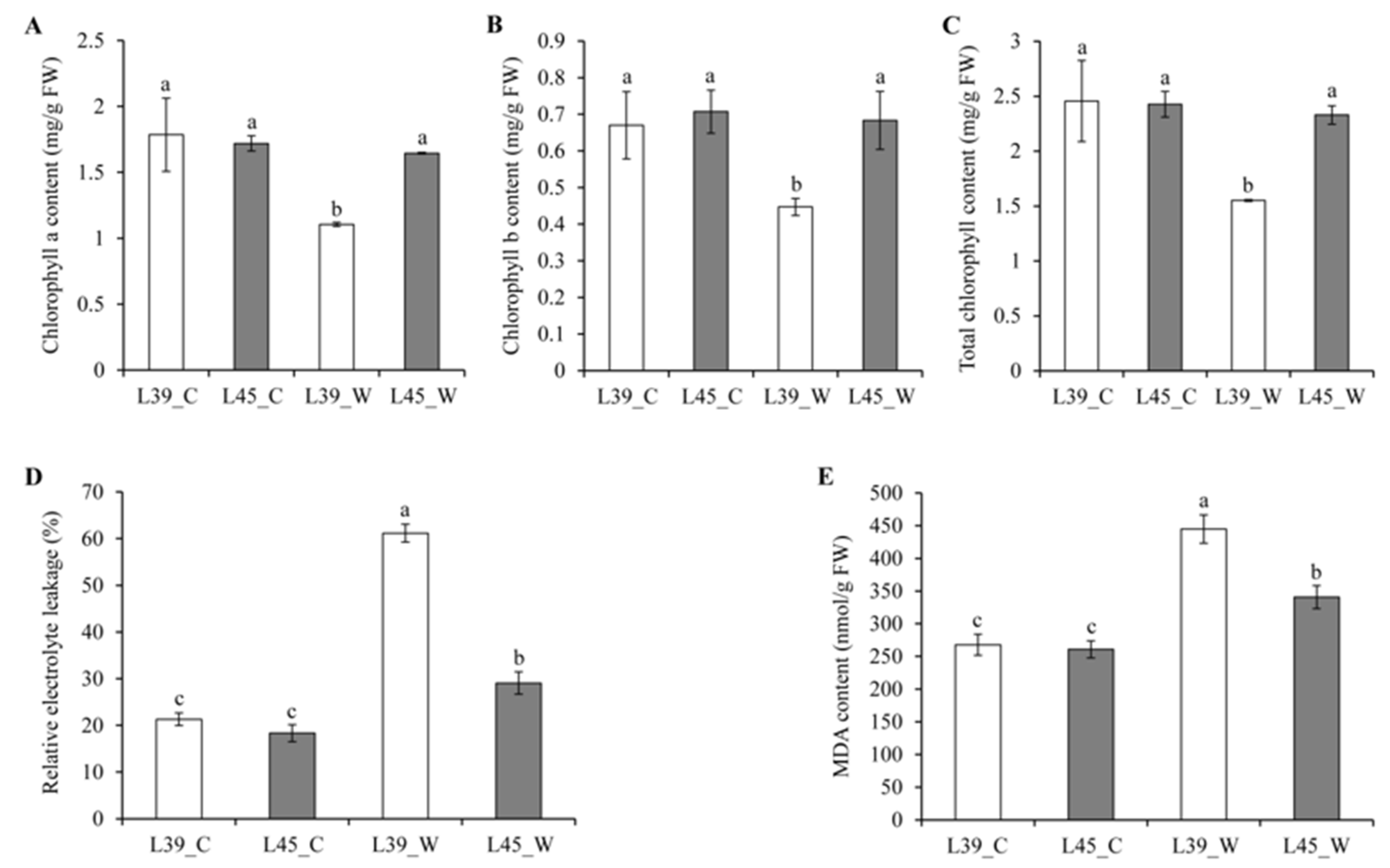
3.1. Physiological Responses to Waterlogging Stress
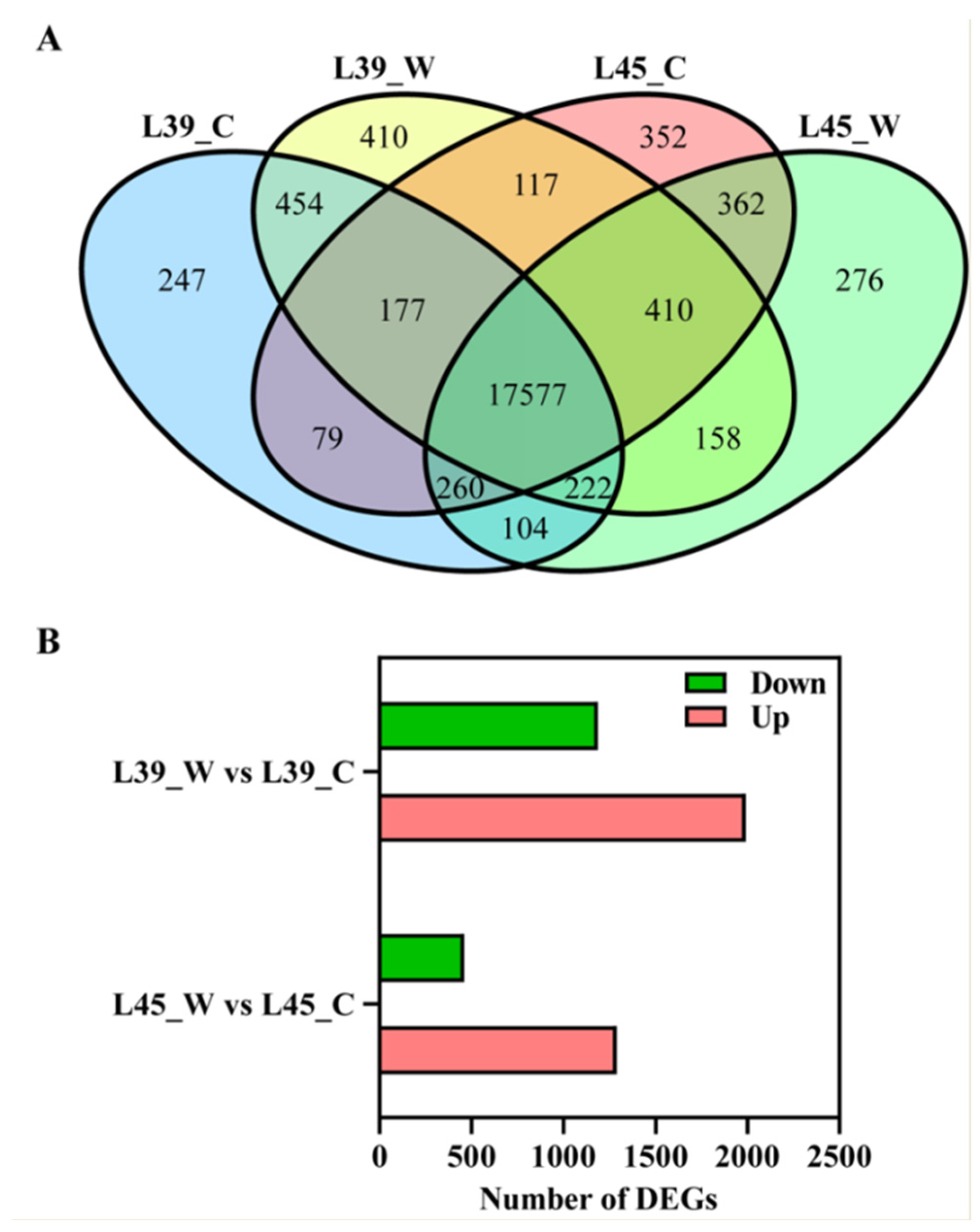
3.2. Transcriptome Data Analysis
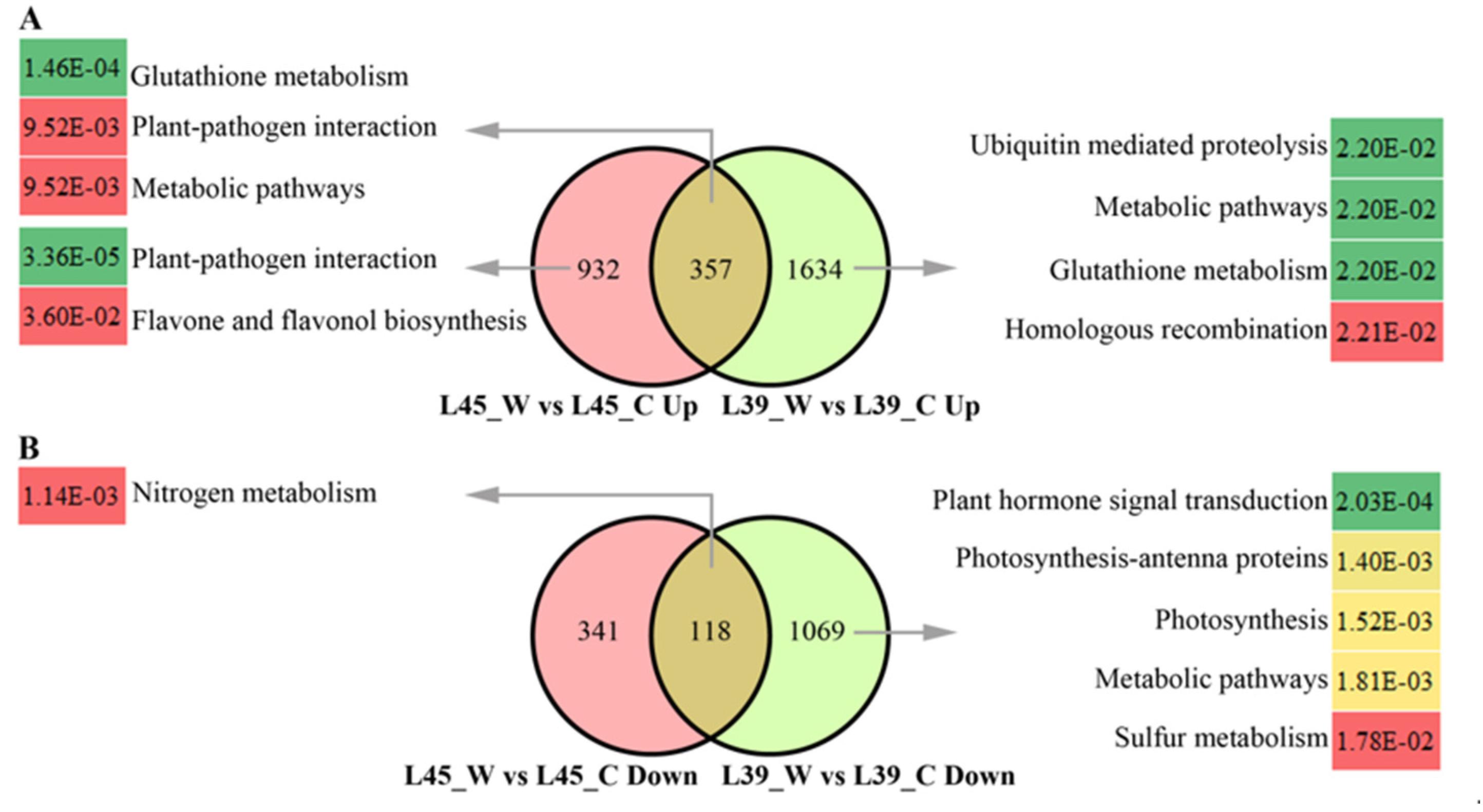
3.3. Identification and Functional Classification of DEGs
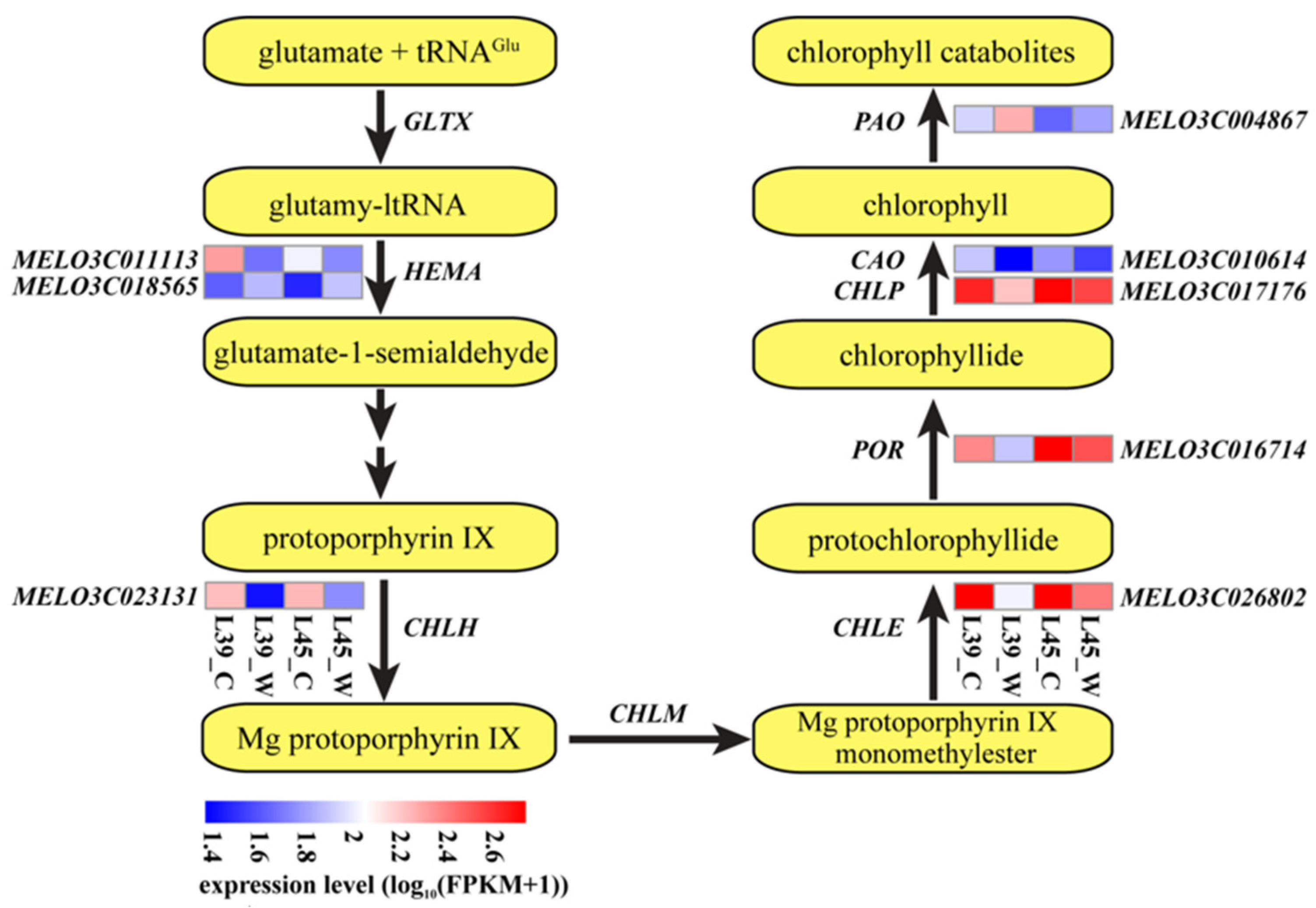
3.4. Analysis of DEGs Related to Chlorophyll Metabolism and Photosynthesis
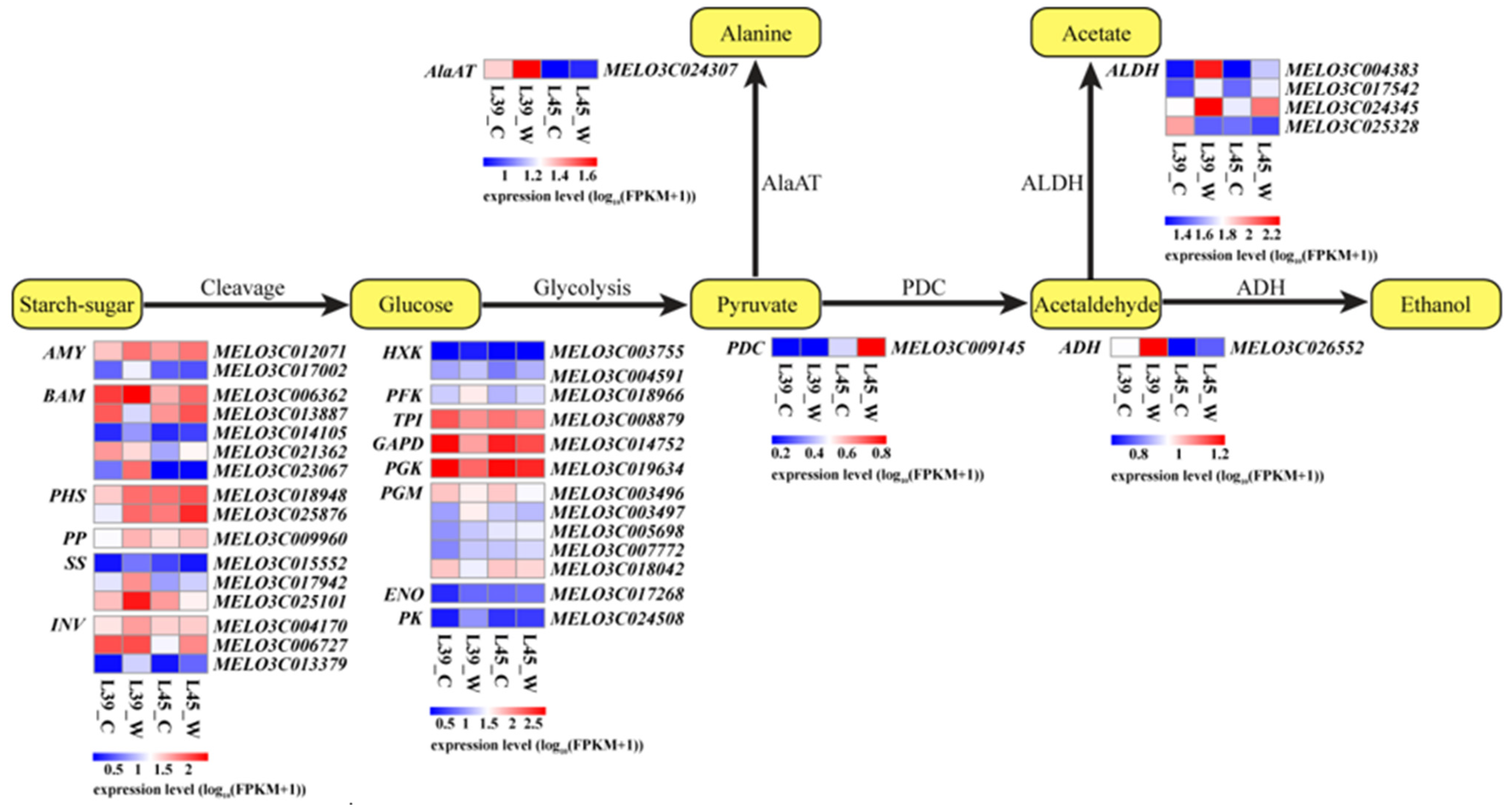
3.5. Analysis of DEGs Related to Energy Generation
3.6. Analysis of DEGs Related to Reactive Oxygen Species Scavenging
3.7. Analysis of DEGs Related to Hormone Metabolism
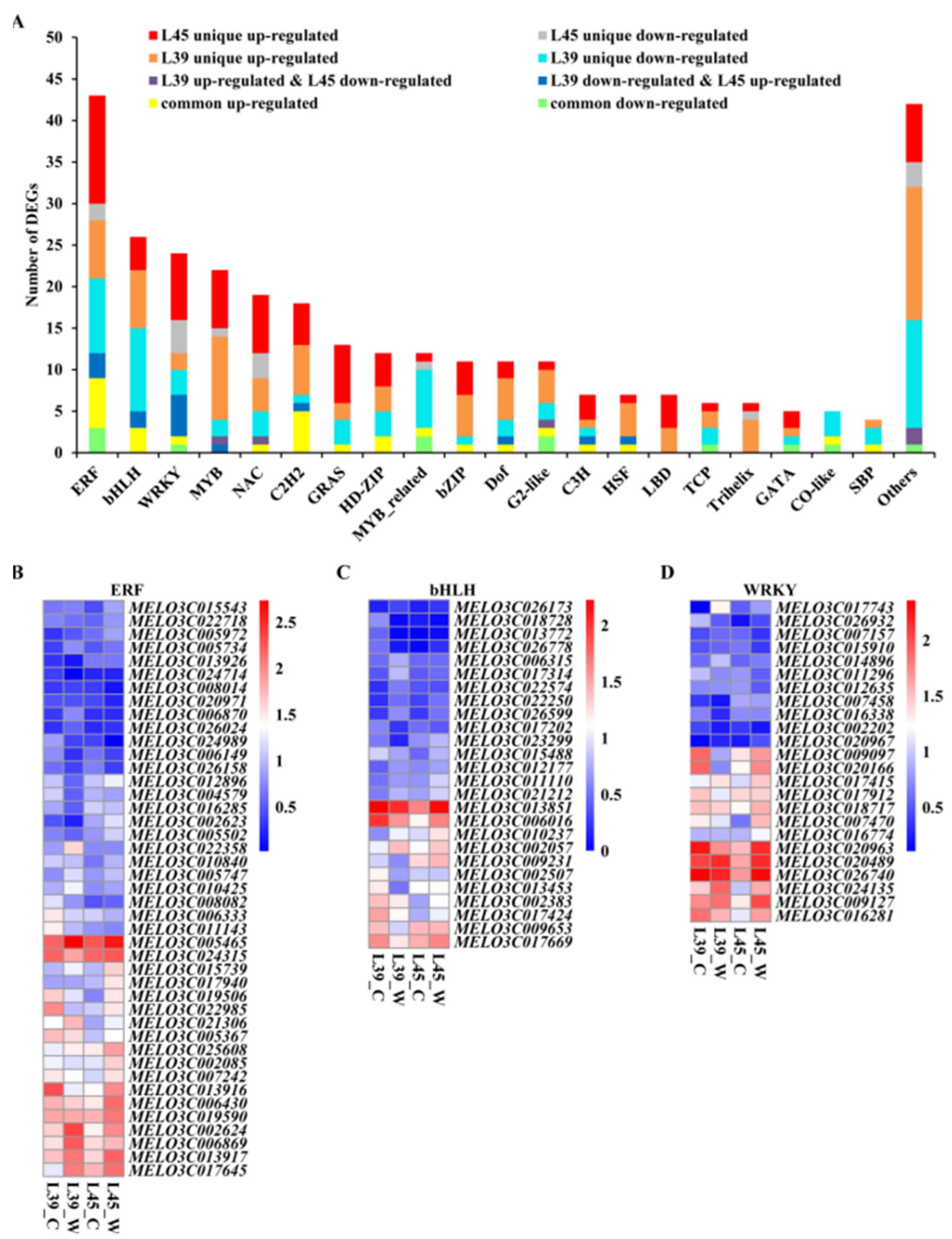
3.8. Analysis of Differentially Expressed Transcription Factors
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Sasidharan, R.; Bailey-Serres, J.; Ashikari, M.; Atwell, B.J.; Colmer, T.; Fagerstedt, K.; Fukao, T.; Geigenberger, P.; Hebelstrup, K.H.; Hill, R.D.; et al. Community recommendations on terminology and procedures used in flooding and low oxygen stress research. New Pythol. 2017, 214, 1403–1407. [Google Scholar] [CrossRef] [PubMed]
- Fukao, T.; Barrera-Figueroa, B.E.; Juntawong, P.; Pena-Castro, J.M. Submergence and waterlogging wtress in plants: A review highlighting research opportunities and understudied aspects. Front. Plant Sci. 2019, 10, 340. [Google Scholar] [CrossRef] [PubMed]
- Butsayawarapat, P.; Juntawong, P.; Khamsuk, O.; Somta, P. Comparative transcriptome analysis of waterlogging-sensitive and tolerant zombi pea (Vigna vexillata) reveals energy conservation and root plasticity controlling waterlogging tolerance. Plants 2019, 8, 264. [Google Scholar] [CrossRef]
- Sreeratree, J.; Butsayawarapat, P.; Chaisan, T.; Somta, P.; Juntawong, P. RNA-seq qeveals waterlogging-triggered root plasticity in mungbean associated with ethylene and jasmonic acid signal integrators for root regeneration. Plants 2022, 11, 930. [Google Scholar] [CrossRef] [PubMed]
- Valliyodan, B.; Ye, H.; Song, L.; Murphy, M.; Shannon, J.G.; Nguyen, H.T. Genetic diversity and genomic strategies for improving drought and waterlogging tolerance in soybeans. J. Exp. Bot. 2017, 68, 1835–1849. [Google Scholar] [CrossRef]
- Visser, E.J.W.; Voesenek, L.; Vartapetian, B.B.; Jackson, M.B. Flooding and plant growth. Ann. Bot. 2003, 91, 107–109. [Google Scholar] [CrossRef]
- Cao, M.; Zheng, L.; Li, J.; Mao, Y.; Zhang, R.; Niu, X.; Geng, M.; Zhang, X.; Huang, W.; Luo, K.; et al. Transcriptomic profiling suggests candidate molecular responses to waterlogging in cassava. PLoS ONE 2022, 17, e0261086. [Google Scholar] [CrossRef]
- Juntawong, P.; Sirikhachornkit, A.; Pimjan, R.; Sonthirod, C.; Sangsrakru, D.; Yoocha, T.; Tangphatsornruang, S.; Srinives, P. Elucidation of the molecular responses to waterlogging in Jatropha roots by transcriptome profiling. Front. Plant Sci. 2014, 5, 658. [Google Scholar] [CrossRef]
- Kim, Y.; Seo, C.W.; Khan, A.L.; Mun, B.G.; Shahzad, R.; Ko, J.W.; Yun, B.W.; Park, S.K.; Lee, I.J. Exo-ethylene application mitigates waterlogging stress in soybean (Glycine max L.). BMC Plant Biol. 2018, 18, 254. [Google Scholar] [CrossRef]
- Dawood, T.; Yang, X.; Visser, E.J.W.; te Beek, T.A.H.; Kensche, P.R.; Cristescu, S.M.; Lee, S.; Floková, K.; Nguyen, D.; Mariani, C.; et al. A co-opted hormonal cascade activates dormant adventitious root primordia upon flooding in Solanum dulcamara. Plant Physiol. 2016, 170, 2351–2364. [Google Scholar] [CrossRef]
- Herzog, M.; Striker, G.G.; Colmer, T.D.; Pedersen, O. Mechanisms of waterlogging tolerance in wheat-a review of root and shoot physiology. Plant Cell Environ. 2016, 39, 1068–1086. [Google Scholar] [CrossRef]
- Ni, X.L.; Gui, M.Y.; Tan, L.L.; Zhu, Q.; Liu, W.Z.; Li, C.X. Programmed cell death and aerenchyma formation in water-logged sunflower stems and its promotion by ethylene and ROS. Front. Plant Sci. 2019, 9, 1928. [Google Scholar] [CrossRef]
- Gibbs, J.; Greenway, H. Mechanisms of anoxia tolerance in plants. I. Growth, survival and anaerobic catabolism. Funct. Plant Biol. 2003, 30, 1–47. [Google Scholar] [CrossRef] [PubMed]
- Ricoult, C.; Echeverria, L.O.; Cliquet, J.B.; Limami, A.M. Characterization of alanine aminotransferase (AlaAT) multigene family and hypoxic response in young seedlings of the model legume Medicago truncatula. J. Exp. Bot. 2006, 57, 3079–3089. [Google Scholar] [CrossRef] [PubMed]
- Narsai, R.; Rocha, M.; Geigenberger, P.; Whelan, J.; van Dongen, J.T. Comparative analysis between plant species of transcriptional and metabolic responses to hypoxia. New Phytol. 2011, 190, 472–487. [Google Scholar] [CrossRef] [PubMed]
- Vidoz, M.L.; Loreti, E.; Mensuali, A.; Alpi, A.; Perata, P. Hormonal interplay during adventitious root formation in flooded tomato plants. Plant J. 2010, 63, 551–562. [Google Scholar] [CrossRef]
- Zhang, H.; Li, G.; Yan, C.; Cao, N.; Yang, H.; Le, M.; Zhu, F. Depicting the molecular responses of adventitious rooting to waterlogging in melon hypocotyls by transcriptome profiling. 3 Biotech 2021, 11, 351. [Google Scholar] [CrossRef]
- Shukla, V.; Lombardi, L.; Pencik, A.; Novak, O.; Weits, D.A.; Loreti, E.; Perata, P.; Giuntoli, B.; Licausi, F. Jasmonate signalling contributes to primary root inhibition upon oxygen deficiency in Arabidopsis thaliana. Plants 2020, 9, 1046. [Google Scholar] [CrossRef]
- Arora, K.; Panda, K.K.; Mittal, S.; Mallikarjuna, M.G.; Rao, A.R.; Dash, P.K.; Thirunavukkarasu, N. RNAseq revealed the important gene pathways controlling adaptive mechanisms under waterlogged stress in maize. Sci. Rep. 2017, 7, 10950. [Google Scholar] [CrossRef]
- Wang, L.; Dossa, K.; You, J.; Zhang, Y.; Li, D.; Zhou, R.; Yu, J.; Wei, X.; Zhu, X.; Jiang, S.; et al. High-resolution temporal transcriptome sequencing unravels ERF and WRKY as the master players in the regulatory networks underlying sesame responses to waterlogging and recovery. Genomics 2021, 113, 276–290. [Google Scholar] [CrossRef]
- Raineri, J.; Ribichich, K.F.; Chan, R.L. The sunflower transcription factor HaWRKY76 confers drought and flood tolerance to Arabidopsis thaliana plants without yield penalty. Plant Cell Rep. 2015, 34, 2065–2080. [Google Scholar] [CrossRef] [PubMed]
- Zeng, N.; Yang, Z.; Zhang, Z.; Hu, L.; Chen, L. Comparative transcriptome combined with proteome analyses revealed key factors involved in alfalfa (Medicago sativa) response to waterlogging stress. Int. J. Mol. Sci. 2019, 20, 1359. [Google Scholar] [CrossRef] [PubMed]
- Burger, Y.; Sa′ar, U.; Paris, H.; Lewinsohn, E.; Katzir, N.; Tadmor, Y.; Schaffer, A. Genetic variability for valuable fruit quality traits in Cucumis melo. Isr. J. Plant Sci. 2006, 54, 233–242. [Google Scholar] [CrossRef]
- Nuñez-Palenius, H.G.; Gomez-Lim, M.; Ochoa-Alejo, N.; Grumet, R.; Lester, G.; Cantliffe, D.J. Melon fruits: Genetic diversity, physiology, and biotechnology features. Crit. Rev. Biotechnol. 2008, 28, 13–55. [Google Scholar] [CrossRef] [PubMed]
- Zhu, F.Y.; Chen, M.X.; Chan, W.L.; Yang, F.; Tian, Y.; Song, T.; Xie, L.J.; Zhou, Y.; Xiao, S.; Zhang, J.; et al. SWATH-MS quantitative proteomic investigation of nitrogen starvation in Arabidopsis reveals new aspects of plant nitrogen stress responses. J. Proteom. 2018, 187, 161–170. [Google Scholar] [CrossRef] [PubMed]
- Shao, P.; Wang, P.; Niu, B.; Kang, J. Environmental stress stability of pectin-stabilized resveratrol liposomes with different degree of esterification. Int. J. Biol. Macromol. 2018, 119, 53–59. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Liu, H.; Sun, C.; Ma, Q.; Bu, H.; Chong, K.; Xu, Y. A C2H2 zinc-finger protein OsZFP213 interacts with OsMAPK3 to enhance salt tolerance in rice. J. Plant Physiol. 2018, 229, 100–110. [Google Scholar] [CrossRef]
- Wang, M.; Jiang, B.; Peng, Q.; Liu, W.; He, X.; Liang, Z.; Lin, Y. Transcriptome analyses in different cucumber cultivars provide novel insights into drought stress responses. Int. J. Mol. Sci. 2018, 19, 2067. [Google Scholar] [CrossRef]
- Zhang, Z.; Cheng, Z.J.; Gan, L.; Zhang, H.; Wu, F.Q.; Lin, Q.B.; Wang, J.L.; Wang, J.; Guo, X.P.; Zhang, X.; et al. OsHSD1, a hydroxysteroid dehydrogenase, is involved in cuticle formation and lipid homeostasis in rice. Plant Sci. 2016, 249, 35–45. [Google Scholar] [CrossRef]
- Cui, L.G.; Shan, J.X.; Shi, M.; Gao, J.P.; Lin, H.X. DCA1 acts as a transcriptional co-activator of DST and contributes to drought and salt tolerance in rice. PLoS Genet. 2015, 11, e1005617. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Alexa, A.; Rahnenfuhrer, J.; Lengauer, T. Improved scoring of functional groups from gene expression data by decorrelating GO graph structure. Bioinformatics 2006, 22, 1600–1607. [Google Scholar] [CrossRef]
- Xie, C.; Mao, X.; Huang, J.; Ding, Y.; Wu, J.; Dong, S.; Kong, L.; Gao, G.; Li, C.-Y.; Wei, L. KOBAS 2.0: A web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 2011, 39, W316–W322. [Google Scholar] [CrossRef]
- Zhang, H.; Cao, N.; Dong, C.; Shang, Q. Genome-wide identification and expression of ARF gene family during adventitious root development in hot pepper (Capsicum annuum). Hortic. Plant J. 2017, 3, 151–164. [Google Scholar] [CrossRef]
- Kong, Q.; Yuan, J.; Niu, P.; Xie, J.; Jiang, W.; Huang, Y.; Bie, Z. Screening suitable reference genes for normalization in reverse transcription quantitative real-time PCR analysis in melon. PLoS ONE 2014, 9, e87197. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Jin, J.; Zhang, H.; Kong, L.; Gao, G.; Luo, J. PlantTFDB 3.0: A portal for the functional and evolutionary study of plant transcription factors. Nucleic Acids Res. 2014, 42, D1182–D1187. [Google Scholar] [CrossRef]
- Gibbs, D.J.; Conde, J.V.; Berckhan, S.; Prasad, G.; Mendiondo, G.M.; Holdsworth, M.J. Group VII ethylene response factors coordinate oxygen and nitric oxide signal transduction and stress responses in plants. Plant Physiol. 2015, 169, 23–31. [Google Scholar] [CrossRef]
- Minami, A.; Yano, K.; Gamuyao, R.; Nagai, K.; Kuroha, T.; Ayano, M.; Nakamori, M.; Koike, M.; Kondo, Y.; Niimi, Y.; et al. Time-course transcriptomics analysis reveals key responses of submerged deepwater rice to flooding. Plant Physiol. 2018, 176, 3081–3102. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Challa, G.S.; Gupta, A.; Gu, L.; Wu, Y.; Li, W. Physiological and transcriptomic characterization of sea-wheatgrass-derived waterlogging tolerance in wheat. Plants 2022, 11, 108. [Google Scholar] [CrossRef]
- Wang, L.; Li, D.; Zhang, Y.; Gao, Y.; Yu, J.; Wei, X.; Zhang, X. Tolerant and susceptible sesame genotypes reveal waterlogging stress response patterns. PLoS ONE 2016, 11, e0149912. [Google Scholar] [CrossRef] [PubMed]
- Zeng, R.; Chen, T.; Wang, X.; Cao, J.; Li, X.; Xu, X.; Chen, L.; Xia, Q.; Dong, Y.; Huang, L.; et al. Physiological and expressional regulation on photosynthesis, starch and sucrose metabolism response to waterlogging stress in peanut. Front. Plant Sci. 2021, 12, 601771. [Google Scholar] [CrossRef]
- Zhao, N.; Li, C.; Yan, Y.; Cao, W.; Song, A.; Wang, H.; Chen, S.; Jiang, J.; Chen, F. Comparative transcriptome analysis of waterlogging-sensitive and waterlogging-tolerant Chrysanthemum morifolium cultivars under waterlogging stress and reoxygenation conditions. Int. J. Mol. Sci. 2018, 19, 1455. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Chen, M.; Ji, J.; Xu, Q.; Qi, X.; Chen, X. Comparative RNA-seq based transcriptome profiling of waterlogging response in cucumber hypocotyls reveals novel insights into the de novo adventitious root primordia initiation. BMC Plant Biol. 2017, 17, 129. [Google Scholar] [CrossRef]
- Gill, M.B.; Zeng, F.; Shabala, L.; Zhang, G.; Fan, Y.; Shabala, S.; Zhou, M. Cell-based phenotyping reveals QTL for membrane potential maintenance associated with hypoxia and salinity stress tolerance in barley. Front. Plant Sci. 2017, 8, 1941. [Google Scholar] [CrossRef]
- Wang, X.; Yan, L.; Wang, B.; Qian, Y.; Wang, Z.; Wu, W. Comparative proteomic analysis of grapevine rootstock in response to waterlogging stress. Front. Plant Sci. 2021, 12, 749184. [Google Scholar] [CrossRef]
- Zhang, Q.; Liu, X.; Zhang, Z.; Liu, N.; Li, D.; Hu, L. Melatonin improved waterlogging tolerance in alfalfa (Medicago sativa) by reprogramming polyamine and ethylene metabolism. Front. Plant Sci. 2019, 10, 44. [Google Scholar] [CrossRef]
- Kęska, K.; Szczesniak, M.W.; Makalowska, I.; Czernicka, M. Long-term waterlogging as factor contributing to hypoxia stress tolerance enhancement in cucumber: Comparative transcriptome analysis of waterlogging sensitive and tolerant accessions. Genes 2021, 12, 189. [Google Scholar] [CrossRef]
- Lyzenga, W.J.; Stone, S.L. Abiotic stress tolerance mediated by protein ubiquitination. J. Exp. Bot. 2012, 63, 599–616. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.; Wu, X.; Sun, M.; Peng, F. Hydrogen sulfide alleviates waterlogging-induced damage in peach seedlings via enhancing antioxidative system and inhibiting ethylene synthesis. Front. Plant Sci. 2020, 11, 696. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Jiménez, M.; Pérez-Tornero, O. Short-term waterlogging in citrus rootstocks. Plants 2021, 10, 2772. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Su, Y.; Yu, M.; Huang, Y.; Wang, F.; Shen, A. Potassium alleviates post-anthesis photosynthetic reductions in winter wheat caused by waterlogging at the stem elongation stage. Front. Plant Sci. 2021, 11, 607475. [Google Scholar] [CrossRef] [PubMed]
- Barickman, T.C.; Simpson, C.R.; Sams, C.E. Waterlogging causes early modification in the physiological performance, carotenoids, chlorophylls, proline, and soluble sugars of cucumber plants. Plants 2019, 8, 160. [Google Scholar] [CrossRef]
- Jurczyk, B.; Pociecha, E.; Janowiak, F.; Kabala, D.; Rapacz, M. Variation in waterlogging-triggered stomatal behavior contributes to changes in the cold acclimation process in prehardened Lolium perenne and Festuca pratensis. Plant Physiol. Biochem. 2016, 109, 280–292. [Google Scholar] [CrossRef]
- Zhu, X.; Li, X.; Jiu, S.; Zhang, K.; Wang, C.; Fang, J. Analysis of the regulation networks in grapevine reveals response to waterlogging stress and candidate gene-marker selection for damage severity. R. Soc. Open Sci. 2018, 5, 172253. [Google Scholar] [CrossRef]
- Kreuzwieser, J.; Hauberg, J.; Howell, K.A.; Carroll, A.; Rennenberg, H.; Millar, A.H.; Whelan, J. Differential response of gray poplar leaves and roots underpins stress adaptation during hypoxia. Plant Physiol. 2009, 149, 461–473. [Google Scholar] [CrossRef]
- Fukao, T.; Xu, K.; Ronald, P.C.; Bailey-Serres, J. A variable cluster of ethylene response factor-like genes regulates metabolic and developmental acclimation responses to submergence in rice. Plant Cell 2006, 18, 2021–2034. [Google Scholar] [CrossRef]
- Sasidharan, R.; Mustroph, A.; Boonman, A.; Akman, M.; Ammerlaan, A.M.; Breit, T.; Schranz, M.E.; Voesenek, L.A.; van Tienderen, P.H. Root transcript profiling of two Rorippa species reveals gene clusters associated with extreme submergence tolerance. Plant Physiol. 2013, 163, 1277–1292. [Google Scholar] [CrossRef]
- Voesenek, L.A.C.J.; Sasidharan, R.; Weber, A. Ethylene- and oxygen signalling—Drive plant survival during flooding. Plant Biol. 2013, 15, 426–435. [Google Scholar] [CrossRef] [PubMed]
- Trebitsh, T.; Goldschmidt, E.E.; Riov, J. Ethylene induces de novo synthesis of chlorophyllase, a chlorophyll degrading enzyme, in Citrus fruit peel. Proc. Natl. Acad. Sci. USA 1993, 90, 9441–9445. [Google Scholar] [CrossRef] [PubMed]
- Jacob-Wilk, D.; Holland, D.; Goldschmidt, E.E.; Riov, J.; Eyal, Y. Chlorophyll breakdown by chlorophyllase: Isolation and functional expression of the Chlase1 gene from ethylene-treated Citrus fruit and its regulation during development. Plant J. 1999, 20, 653–661. [Google Scholar] [CrossRef]
- Valliyodan, B.; Van Toai, T.; Alves, J.; De Fátima, P.; Goulart, P.; Lee, J.; Fritschi, F.; Rahman, M.; Islam, R.; Shannon, J.; et al. Expression of root-related transcription factors associated with flooding tolerance of soybean (Glycine max). Int. J. Mol. Sci. 2014, 15, 17622–17643. [Google Scholar] [CrossRef]
- Ren, C.G.; Kong, C.C.; Yan, K.; Zhang, H.; Luo, Y.M.; Xie, Z.H. Elucidation of the molecular responses to waterlogging in Sesbania cannabina roots by transcriptome profiling. Sci. Rep. 2017, 7, 9256. [Google Scholar] [CrossRef] [PubMed]
- Phukan, U.J.; Jeena, G.S.; Tripathi, V.; Shukla, R.K. MaRAP2-4, a waterlogging-responsive ERF from Mentha, regulates bidirectional sugar transporter AtSWEET10 to modulate stress response in Arabidopsis. Plant Biotechnol. J. 2018, 16, 221–233. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.; Sun, F.; Gao, R.; Dong, H. RAP2.6L overexpression delays waterlogging induced premature senescence by increasing stomatal closure more than antioxidant enzyme activity. Plant Mol. Biol. 2012, 79, 609–622. [Google Scholar] [CrossRef]
- Hinz, M.; Wilson, I.W.; Yang, J.; Buerstenbinder, K.; Llewellyn, D.; Dennis, E.S.; Sauter, M.; Dolferus, R. Arabidopsis RAP2.2: An ethylene response transcription factor that is important for hypoxia survival. Plant Physiol. 2010, 153, 757–772. [Google Scholar] [CrossRef]
- Licausi, F.; van Dongen, J.T.; Giuntoli, B.; Novi, G.; Santaniello, A.; Geigenberger, P.; Perata, P. HRE1 and HRE2, two hypoxia-inducible ethylene response factors, affect anaerobic responses in Arabidopsis thaliana. Plant J. 2010, 62, 302–315. [Google Scholar] [CrossRef]
- Papdi, C.; Pérez-Salamó, I.; Joseph, M.P.; Giuntoli, B.; Bögre, L.; Koncz, C.; Szabados, L. The low oxygen, oxidative and osmotic stress responses synergistically act through the ethylene response factor VII genes RAP2.12, RAP2.2 and RAP2.3. Plant J. 2015, 82, 772–784. [Google Scholar] [CrossRef]
- Borrego-Benjumea, A.; Carter, A.; Tucker, J.R.; Yao, Z.; Xu, W.; Badea, A. Genome-wide analysis of gene expression providesnewinsights into waterlogging responses in barley (Hordeum vulgare L.). Plants 2020, 9, 240. [Google Scholar] [CrossRef] [PubMed]
- Dossa, K.; Mmadi, M.A.; Zhou, R.; Zhang, T.; Su, R.; Zhang, Y.; Wang, L.; You, J.; Zhang, X. Depicting the core transcriptome modulating multiple abiotic stresses responses in sesame (Sesamum indicum L.). Int. J. Mol. Sci. 2019, 20, 3930. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Li, J.; Guo, X.; Ma, Y.; Qiao, Q.; Guo, J. PlWRKY13: A transcription factor involved in abiotic and biotic stress responses in Paeonia lactiflora. Int. J. Mol. Sci. 2019, 20, 5953. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Liu, P.; Yu, J.; Wang, L.; Dossa, K.; Zhang, Y.; Zhou, R.; Wei, X.; Zhang, X. Genome-wide analysis of WRKY gene family in the sesame genome and identification of the WRKY genes involved in responses to abiotic stresses. BMC Plant Biol. 2017, 17, 152. [Google Scholar] [CrossRef]
| Sample Name | Raw Reads Number | Clean Reads Number | Clean Bases (Gb) | Q20 (%) | Q30 (%) | GC (%) | Clean reads Ratio (%) |
|---|---|---|---|---|---|---|---|
| L39_C1 | 47,362,356 | 43,584,000 | 6.54 | 99.98 | 97.97 | 45.00 | 92.02 |
| L39_C2 | 45,108,810 | 41,493,278 | 6.22 | 99.98 | 98.08 | 44.00 | 91.98 |
| L39_W1 | 47,284,196 | 41,121,986 | 6.17 | 99.99 | 97.90 | 45.00 | 86.97 |
| L39_W2 | 45,691,524 | 41,184,558 | 6.18 | 99.98 | 98.07 | 43.50 | 90.14 |
| L45_C1 | 44,275,178 | 38,287,494 | 5.74 | 99.98 | 97.98 | 47.00 | 86.48 |
| L45_C2 | 46,621,728 | 42,792,352 | 6.42 | 99.98 | 98.03 | 45.00 | 91.79 |
| L45_W1 | 45,658,890 | 42,021,172 | 6.30 | 99.98 | 98.03 | 44.00 | 92.03 |
| L45_W2 | 44,756,744 | 41,165,862 | 6.17 | 99.98 | 98.06 | 44.00 | 91.98 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, H.; Li, G.; Yan, C.; Zhang, X.; Cao, N.; Le, M.; Hu, X.; Zhu, F.; Liu, W. Elucidating the Molecular Responses to Waterlogging Stress in Cucumis melo by Comparative Transcriptome Profiling. Horticulturae 2022, 8, 891. https://doi.org/10.3390/horticulturae8100891
Zhang H, Li G, Yan C, Zhang X, Cao N, Le M, Hu X, Zhu F, Liu W. Elucidating the Molecular Responses to Waterlogging Stress in Cucumis melo by Comparative Transcriptome Profiling. Horticulturae. 2022; 8(10):891. https://doi.org/10.3390/horticulturae8100891
Chicago/Turabian StyleZhang, Huanxin, Guoquan Li, Chengpu Yan, Xinlong Zhang, Na Cao, Meiwang Le, Xinlong Hu, Fanghong Zhu, and Wenge Liu. 2022. "Elucidating the Molecular Responses to Waterlogging Stress in Cucumis melo by Comparative Transcriptome Profiling" Horticulturae 8, no. 10: 891. https://doi.org/10.3390/horticulturae8100891
APA StyleZhang, H., Li, G., Yan, C., Zhang, X., Cao, N., Le, M., Hu, X., Zhu, F., & Liu, W. (2022). Elucidating the Molecular Responses to Waterlogging Stress in Cucumis melo by Comparative Transcriptome Profiling. Horticulturae, 8(10), 891. https://doi.org/10.3390/horticulturae8100891






