The Genes Associated with Jasmonic Acid and Salicylic Acid Are Induced in Tropical Chili Pepper against Ralstonia solanacearum by Applying Arbuscular Mycorrhizal Fungi
Abstract
1. Introduction
2. Materials and Methods
2.1. Chili Pepper Genotypes, Mycorrhizae, and the Ralstonia solanacearum Isolates
2.2. Procedure of Experiments
2.3. Isolation of Leaf RNA
2.4. qRT-PCR for GAPDH-cp and Genes of Interest
2.5. Phenotypic Response of the Genotypes to Mycorrhizae and R. solanacearum
2.6. Data Analysis
3. Results
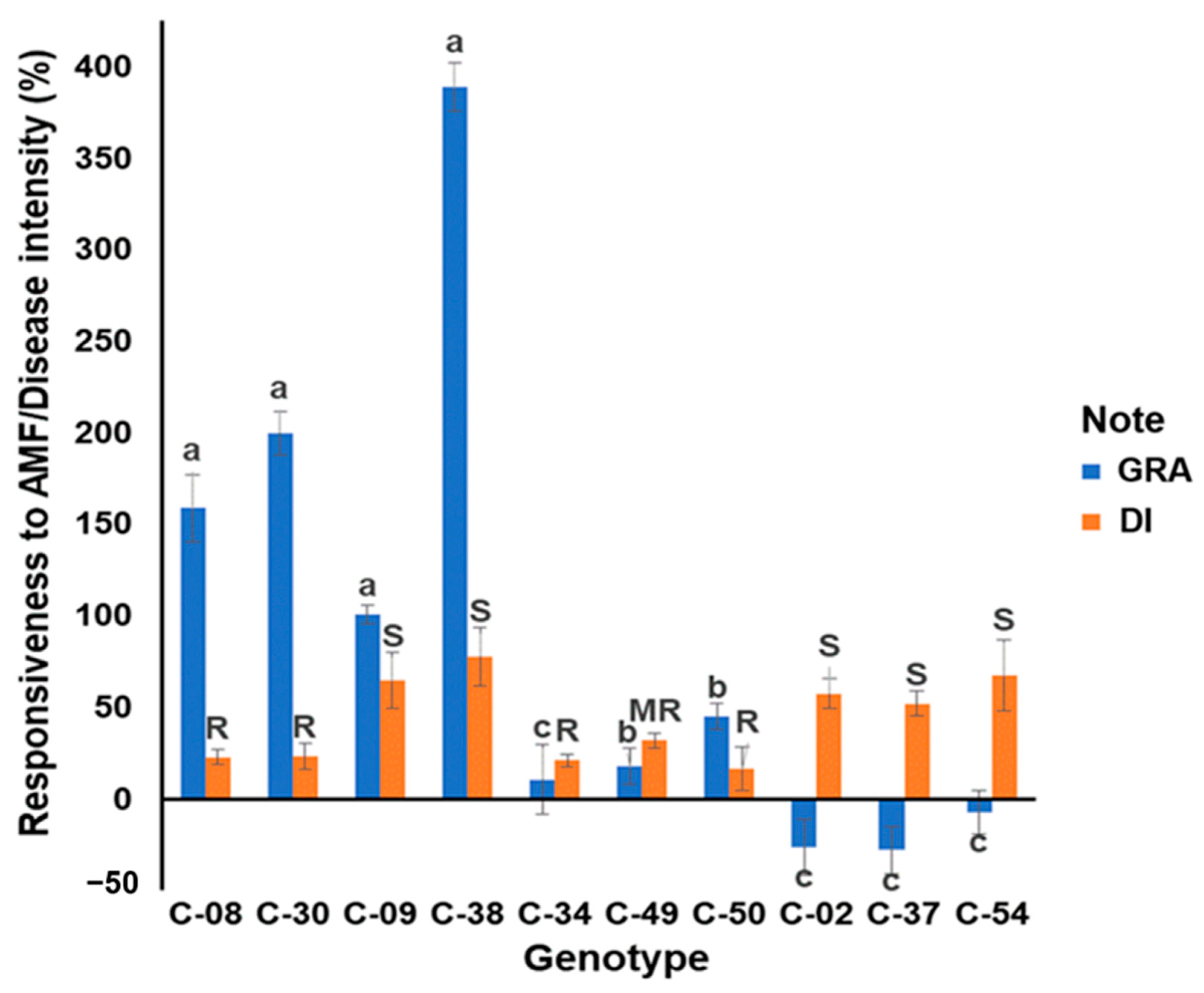
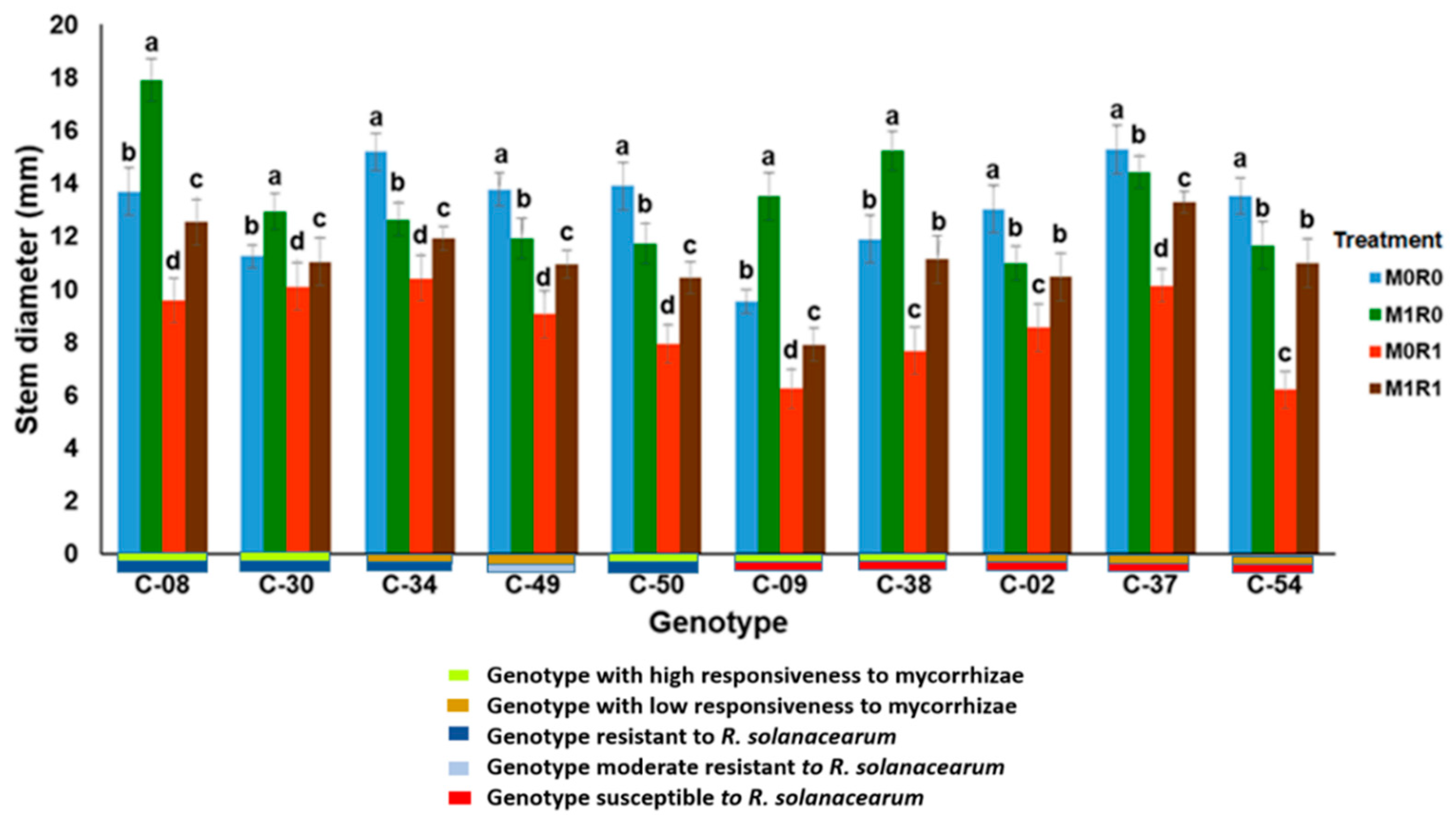
3.1. Evaluation of the Growth Response to Arbuscular Mycorrhizae and Resistance to R. solanacearum
3.2. Effect of Arbuscular Mycorrhizae on the Relative Gene Expression of JA
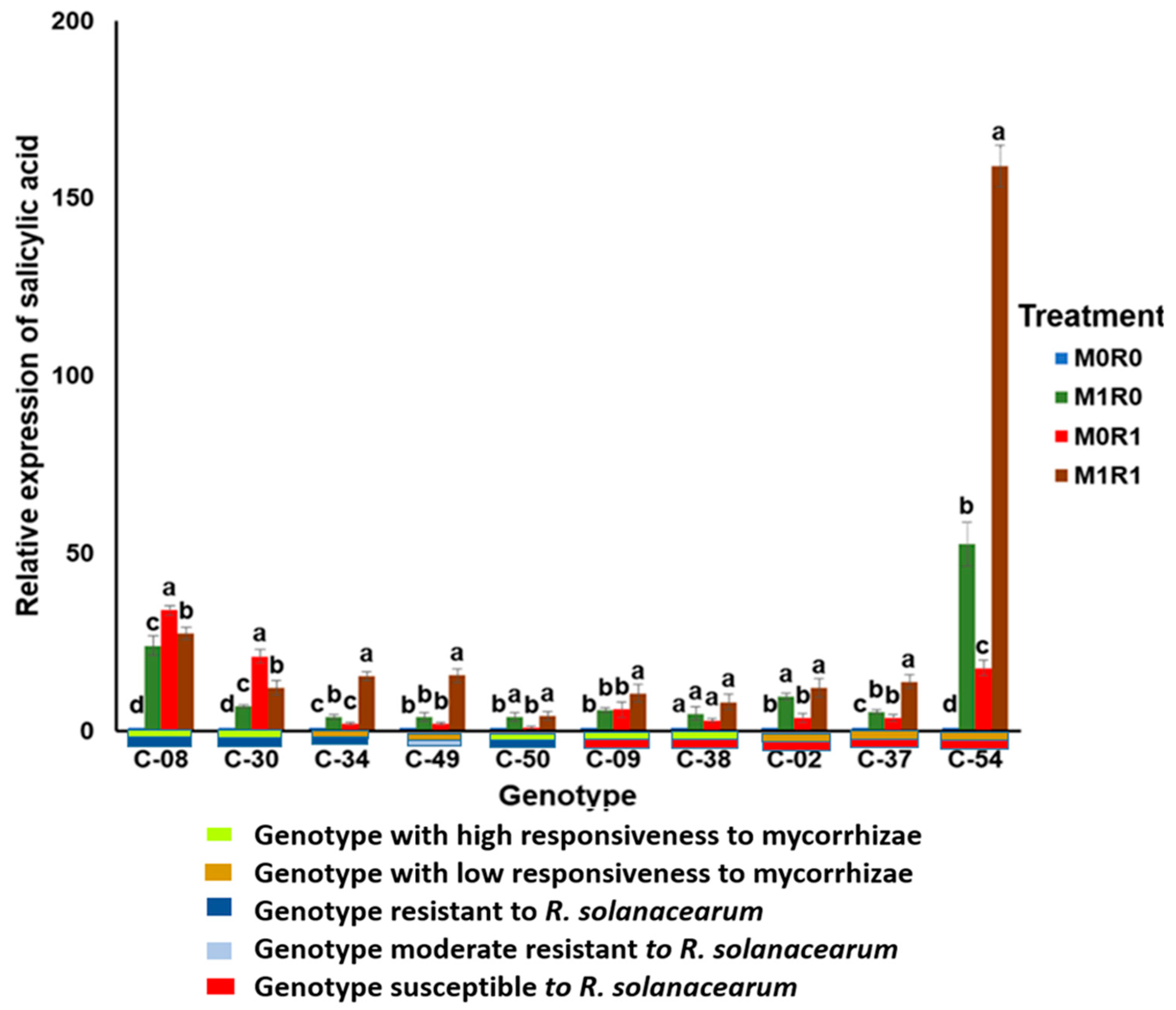
3.3. Effect of Arbuscular Mycorrhizae on the Relative Gene Expression of SA
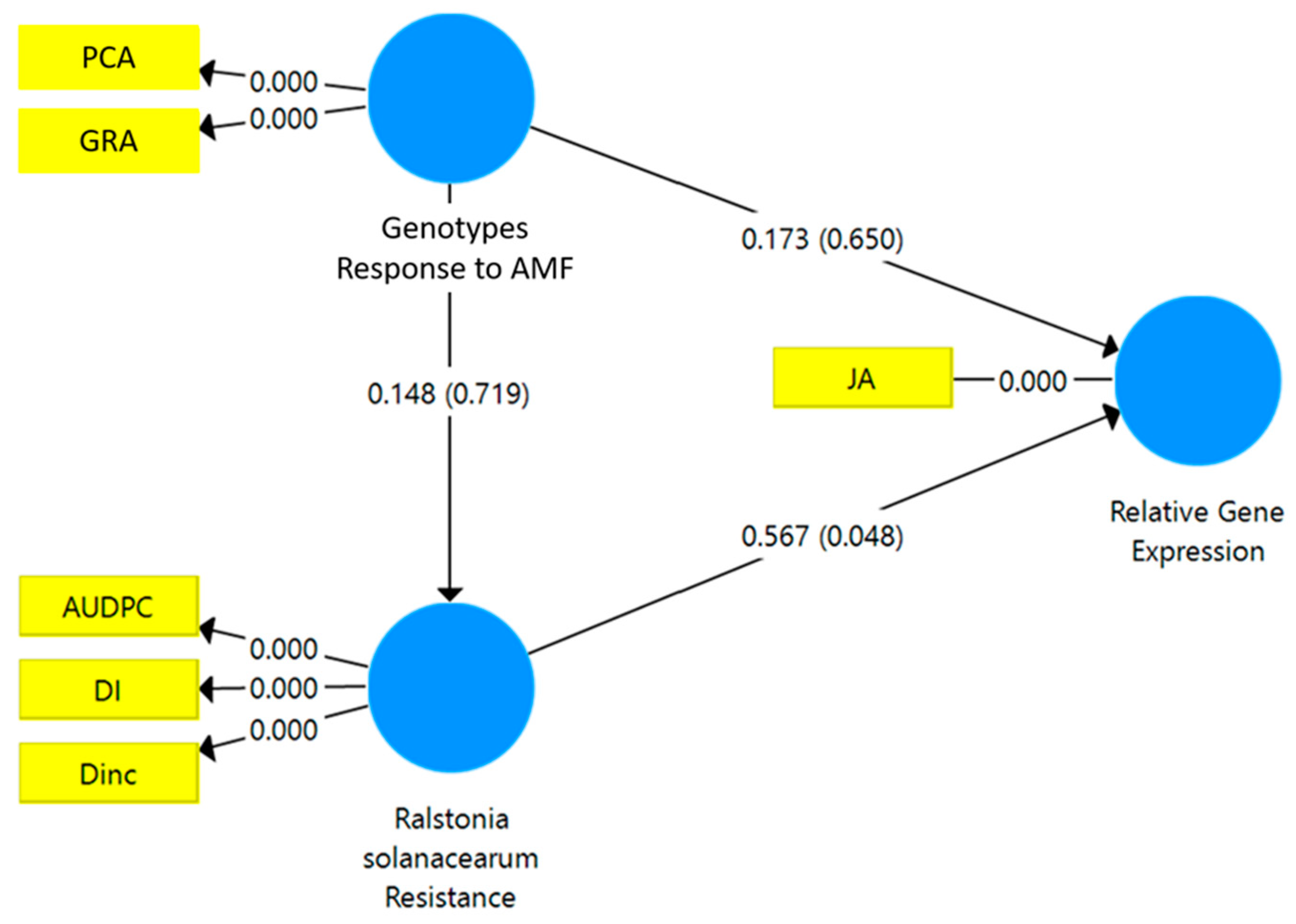
3.4. Structural Equation Modeling-PLS and Standardized Stepwise Regression
3.5. Clustering of the Genotypes Based on the Responsiveness to Mycorrhizae, Resistance to R. solanacearum, and the Relative Gene Expression of JA and SA
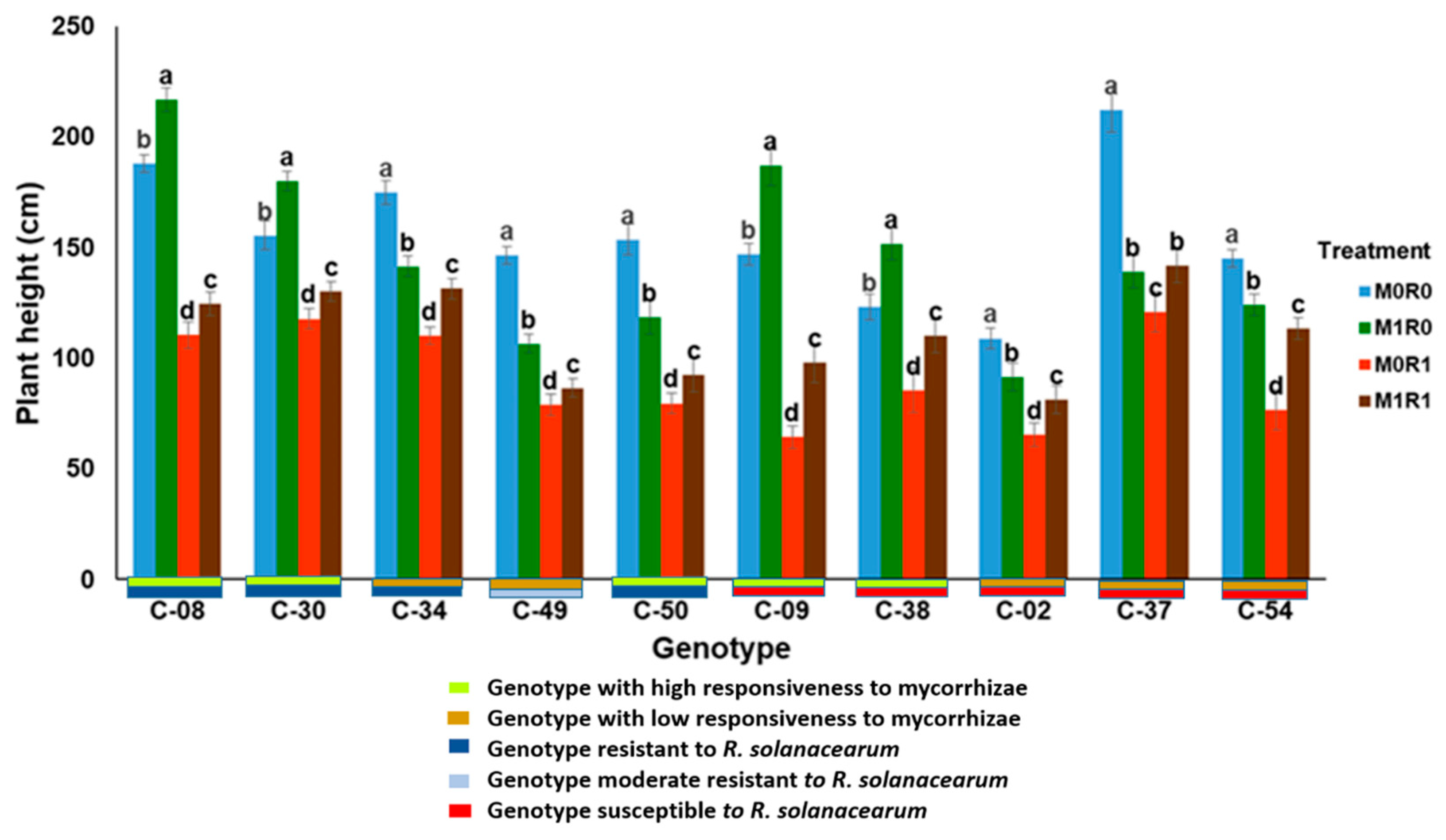
3.6. Phenotypic Response Based on the Responsiveness to Mycorrhizae and Resistance to R. solanacearum
4. Discussion
4.1. Responsiveness to Arbuscular Mycorrhizae and the Resistance to R. solanacearum
4.2. Interaction between the Genotypes of Tropical Chili Pepper, the Arbuscular Mycorrhizae, and R. solanacearum on the Relative Gene Expression of JA and SA, and Phenotypic Response
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bosland, P.W.; Votava, E.J. Peppers: Vegetable and Spice Capsicums, 2nd ed.; CABI Publishing: Willingford, UK; Volume 2, pp. 1–230. [CrossRef]
- López, P.; Gorzalczany, S.; Acevedo, C.; Alonso, R.; Ferraro, G. Chemical Study and Anti-Inflammatory Activity of Capsicum chacoense and C. baccatum. Rev. Bras. Farmacogn. 2012, 22, 455–458. [Google Scholar] [CrossRef]
- Pereira, J.A.P.; Vieira, I.J.C.; Freitas, M.S.M.; Prins, C.L.; Martins, M.A.; Rodrigues, R. Effects of Arbuscular Mycorrhizal Fungi on Capsicum Spp. J. Agric. Sci. 2016, 154, 828–849. [Google Scholar] [CrossRef]
- Sanati, S.; Razavi, B.M.; Hosseinzadeh, H. A Review of the Effects of Capsicum annuum L. and Its Constituent, Capsaicin, in Metabolic Syndrome. Iran. J. Basic Med. Sci. 2018, 21, 439–448. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, W.; Yang, J.; Tan, Y.; Munir, S.; Liu, Q.; Zhang, J.; Ji, G.; Zhao, Z. Ralstonia solanacearum, a Deadly Pathogen: Revisiting the Bacterial Wilt Biocontrol Practices in Tobacco and Other Solanaceae. Rhizosphere 2022, 21, 100479. [Google Scholar] [CrossRef]
- Aguk, J.A.; Karanja, N.; Schulte-Geldermann, E.; Bruns, C.; Kinyua, Z.; Parker, M. Control of Bacterial Wilt (Ralstonia solanacearum) in Potato (Solanum tuberosum) Using Rhizobacteria and Arbuscular Mycorrhiza Fungi. Afr. J. Food Agric. Nutr. Dev. 2018, 18, 13371–13387. [Google Scholar] [CrossRef]
- Du, H.; Chen, B.; Zhang, X.; Zhang, F.; Miller, S.A.; Rajashekara, G.; Xu, X.; Geng, S. Evaluation of Ralstonia solanacearum Infection Dynamics in Resistant and Susceptible Pepper Lines Using Bioluminescence Imaging. Plant Dis. 2017, 101, 272–278. [Google Scholar] [CrossRef]
- Kang, Y.J.; Ahn, Y.K.; Kim, K.T.; Jun, T.H. Resequencing of Capsicum annuum Parental Lines (YCM334 and Taean) for the Genetic Analysis of Bacterial Wilt Resistance. BMC Plant Biol. 2016, 16, 235. [Google Scholar] [CrossRef]
- Mamphogoro, T.P.; Babalola, O.O.; Aiyegoro, O.A. Sustainable Management Strategies for Bacterial Wilt of Sweet Peppers (Capsicum annuum) and Other Solanaceous Crops. J. Appl. Microbiol. 2020, 129, 496–508. [Google Scholar] [CrossRef]
- Thakur, P.P.; Mathew, D.; Nazeem, P.A.; Abida, P.S.; Indira, P.; Girija, D.; Shylaja, M.R.; Valsala, P.A. Identification of Allele-Specific AFLP Markers Linked with Bacterial Wilt [Ralstonia solanacearum (Smith) Yabuuchi et al.] Resistance in Hot Peppers (Capsicum annuum L.). Physiol. Mol. Plant Pathol. 2014, 87, 19–24. [Google Scholar] [CrossRef]
- Mansfield, J.; Genin, S.; Magori, S.; Citovsky, V.; Sriariyanum, M.; Ronald, P.; Dow, M.; Verdier, V.; Beer, S.V.; Machado, M.A.; et al. Top 10 Plant Pathogenic Bacteria in Molecular Plant Pathology. Mol. Plant Pathol. 2012, 13, 614–629. [Google Scholar] [CrossRef]
- Lebeau, A.; Daunay, M.C.; Frary, A.; Palloix, A.; Wang, J.F.; Dintinger, J.; Chiroleu, F.; Wicker, E.; Prior, P. Bacterial Wilt Resistance in Tomato, Pepper, and Eggplant: Genetic Resources Respond to Diverse Strains in the Ralstonia solanacearum Species Complex. Phytopathology 2011, 101, 154–165. [Google Scholar] [CrossRef]
- Álvarez, B.; Biosca, E.G.; López, M.M. On the Life of Ralstonia solanacearum, a Destructive Bacterial Plant Pathogen. In Technology and Education Topics in Applied Microbiology and Microbial Biotechnology; Formatex Research Center: Badajoz, Spain, 2010; pp. 267–279. [Google Scholar]
- Yadeta, K.A.; Thomma, B.P.H.J. The Xylem as a Battleground for Plant Hosts and Vascular Wilt Pathogens. Front. Plant Sci. 2013, 4, 97. [Google Scholar] [CrossRef]
- Jiang, G.; Wei, Z.; Xu, J.; Chen, H.; Zhang, Y.; She, X.; Macho, A.P.; Ding, W.; Liao, B. Bacterial Wilt in China: History, Current Status, and Future Perspectives. Front. Plant Sci. 2017, 8, 1549. [Google Scholar] [CrossRef] [PubMed]
- Agoncillo, E.S. Control Bacterial Wilt Disease Caused by Ralstonia Solanacearum in Pepper Using Arbuscular Mycorrhizal Fungi (Mykovam). J. Nat. Sci. Res. 2018, 8, 62–66. [Google Scholar]
- Namisy, A.; Chen, J.R.; Prohens, J.; Metwally, E.; Elmahrouk, M.; Rakha, M. Screening Cultivated Eggplant and Wild Relatives for Resistance to Bacterial Wilt (Ralstonia Solanacearum). Agriculture 2019, 9, 157. [Google Scholar] [CrossRef]
- Kim, B.-S.; French, E.; Caldwell, D.; Harrington, E.J.; Iyer-Pascuzzi, A.S. Bacterial Wilt Disease: Host Resistance and Pathogen Virulence Mechanisms. Physiol. Mol. Plant Pathol. 2016, 95, 37–43. [Google Scholar] [CrossRef]
- Llorens, E.; Scalschi, L.; Sharon, O.; Vicedo, B.; Sharon, A.; García-Agustín, P. Jasmonic Acid Pathway Is Required in the Resistance Induced by Acremonium sclerotigenum in Tomato against Pseudomonas syringae. Plant Sci. 2022, 318, 111210. [Google Scholar] [CrossRef]
- Yuan, S.; Li, M.; Fang, Z.; Liu, Y.; Shi, W.; Pan, B.; Wu, K.; Shi, J.; Shen, B.; Shen, Q. Biological Control of Tobacco Bacterial Wilt Using Trichoderma harzianum Amended Bioorganic Fertilizer and the Arbuscular Mycorrhizal Fungi Glomus mosseae. Biol. Control 2016, 92, 164–171. [Google Scholar] [CrossRef]
- Latz, M.A.C.; Jensen, B.; Collinge, D.B.; Jørgensen, H.J.L. Endophytic Fungi as Biocontrol Agents: Elucidating Mechanisms in Disease Suppression. Plant Ecol. Divers. 2018, 11, 555–567. [Google Scholar] [CrossRef]
- Enebe, M.C.; Babalola, O.O. The Impact of Microbes in the Orchestration of Plants’ Resistance to Biotic Stress: A Disease Management Approach. Appl. Microbiol. Biotechnol. 2019, 103, 9–25. [Google Scholar] [CrossRef]
- Dowarah, B.; Gill, S.S.; Agarwala, N. Arbuscular Mycorrhizal Fungi in Conferring Tolerance to Biotic Stresses in Plants. J. Plant Growth Regul. 2021, 41, 1429–1444. [Google Scholar] [CrossRef]
- Sanmartín, N.; Pastor, V.; Pastor-Fernández, J.; Flors, V.; Pozo, M.J.; Sánchez-Bel, P. Role and Mechanisms of Callose Priming in Mycorrhiza-Induced Resistance. J. Exp. Bot. 2021, 71, 2769–2781. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Gupta, R.; Pandey, R. Rice Seed Priming with Picomolar Rutin Enhances Rhizospheric Bacillus subtilis CIM Colonization and Plant Growth. PLoS ONE 2016, 11, e01460132. [Google Scholar] [CrossRef] [PubMed]
- Whipps, J.M. Prospects and Limitations for Mycorrhizas in Biocontrol of Root Pathogens. Can. J. Bot. 2004, 82, 1198–1227. [Google Scholar] [CrossRef]
- Jacott, C.N.; Murray, J.D.; Ridout, C.J. Trade-Offs in Arbuscular Mycorrhizal Symbiosis: Disease Resistance, Growth Responses and Perspectives for Crop Breeding. Agronomy 2017, 7, 75. [Google Scholar] [CrossRef]
- Bonfante, P.; Genre, A. Mechanisms Underlying Beneficial Plant—Fungus Interactions in Mycorrhizal Symbiosis. Nat. Commun. 2010, 1, 48. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Garrido, J.M. Regulation of the Plant Defence Response in Arbuscular Mycorrhizal Symbiosis. J. Exp. Bot. 2002, 53, 1377–1386. [Google Scholar] [CrossRef]
- Hause, B.; Mrosk, C.; Isayenkov, S.; Strack, D. Jasmonates in Arbuscular Mycorrhizal Interactions. Phytochemistry 2007, 68, 101–110. [Google Scholar] [CrossRef]
- Hohmann, P.; Messmer, M.M. Breeding for Mycorrhizal Symbiosis: Focus on Disease Resistance. Euphytica 2017, 213, 113. [Google Scholar] [CrossRef]
- Llorens, E.; García-Agustín, P.; Lapeña, L. Advances in Induced Resistance by Natural Compounds: Towards New Options for Woody Crop Protection. Sci. Agric. 2017, 74, 90–100. [Google Scholar] [CrossRef]
- Koo, A.J.K.; Howe, G.A. The Wound Hormone Jasmonate. Phytochemistry 2009, 70, 1571–1580. [Google Scholar] [CrossRef] [PubMed]
- De Geyter, N.; Gholami, A.; Goormachtig, S.; Goossens, A. Transcriptional Machineries in Jasmonate-Elicited Plant Secondary Metabolism. Trends Plant Sci. 2012, 17, 349–359. [Google Scholar] [CrossRef] [PubMed]
- Pieterse, C.M.J.; Van Der Does, D.; Zamioudis, C.; Leon-Reyes, A.; Van Wees, S.C.M. Hormonal Modulation of Plant Immunity. Annu. Rev. Cell Dev. Biol. 2012, 28, 489–521. [Google Scholar] [CrossRef] [PubMed]
- Halim, V.A.; Altmann, S.; Ellinger, D.; Eschen-Lippold, L.; Miersch, O.; Scheel, D.; Rosahl, S. PAMP-Induced Defense Responses in Potato Require Both Salicylic Acid and Jasmonic Acid. Plant J. 2009, 57, 230–242. [Google Scholar] [CrossRef]
- Scalschi, L.; Vicedo, B.; Camañes, G.; Fernandez-Crespo, E.; Lapeña, L.; González-Bosch, C.; García-Agustín, P. Hexanoic Acid Is a Resistance Inducer That Protects Tomato Plants against Pseudomonas Syringae by Priming the Jasmonic Acid and Salicylic Acid Pathways. Mol. Plant Pathol. 2013, 14, 342–355. [Google Scholar] [CrossRef]
- Navarro-Meléndez, A.L.; Heil, M. Symptomless Endophytic Fungi Suppress Endogenous Levels of Salicylic Acid and Interact with the Jasmonate-Dependent Indirect Defense Traits of Their Host, Lima Bean (Phaseolus lunatus). J. Chem. Ecol. 2014, 40, 816–825. [Google Scholar] [CrossRef]
- Hetrick, B.A.D.; Wilson, G.W.T.; Cox, T.S. Mycorrhizal Dependence of Modern Wheat Cultivars and Ancestors: A Synthesis. Can. J. Bot. 1992, 71, 512–518. [Google Scholar] [CrossRef]
- Phillips, J.M.; Hayman, D.S. Improved Procedures for Clearing Roots and Staining Parasitic and Vesicular-Arbuscular Mycorrhizal Fungi for Rapid Assessment of Infection. Trans. Br. Mycol. Soc. 1970, 55, 158–161. [Google Scholar] [CrossRef]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An Open-Source Platform for Biological-Image Analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef]
- Lafortune, D.; Béramis, M.; Daubèze, A.M.; Boissot, N.; Palloix, A. Partial Resistance of Pepper to Bacterial Wilt Is Oligogenic and Stable under Tropical Conditions. Plant Dis. 2005, 89, 501–506. [Google Scholar] [CrossRef]
- Lillemo, M.; Joshi, A.K.; Prasad, R.; Chand, R.; Singh, R.P. QTL for Spot Blotch Resistance in Bread Wheat Line Saar Co-Locate to the Biotrophic Disease Resistance Loci Lr34 and Lr46. Theor. Appl. Genet. 2013, 126, 711–719. [Google Scholar] [CrossRef] [PubMed]
- Geneaid Biotech Ltd. Home Page. Available online: https://www.geneaid.com/ (accessed on 16 March 2022).
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Fukushige, H.; Hildebrand, D.F.; Gan, S. Evidence Supporting a Role of Jasmonic Acid in Arabidopsis Leaf Senescence. Plant Physiol. 2002, 128, 876–884. [Google Scholar] [CrossRef] [PubMed]
- Fu, X.; Feng, Y.Q.; Zhang, X.W.; Zhang, Y.Y.; Bi, H.G.; Ai, X.Z. Salicylic Acid Is Involved in Rootstock–Scion Communication in Improving the Chilling Tolerance of Grafted Cucumber. Front. Plant Sci. 2021, 12, 693344. [Google Scholar] [CrossRef]
- Team R Development Core. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2018; Available online: https://www.R-project.org (accessed on 5 August 2021).
- Ghozali, I. Structural Equation Modeling; Alternative Method with Partial Least Square (PLS), 2nd ed.; Badan Penerbit Universitas Diponegoro: Semarang, Indonesia, 2008. [Google Scholar]
- Fernandes, A. Structural Equation Modeling Approach PLS and SEM; Application of Smart PLS and AMOS Software; Laboratory of Statistics, Faculty of Mathematics and Natural Sciences, Brawijaya University: Malang, Indonesia, 2008. [Google Scholar]
- Krall, J.; Uthoff, V.; Harley, J. A Step-up Procedure for Selecting Variables Associated with Survival. Biometrics 1975, 31, 49–57. [Google Scholar] [CrossRef]
- SAS Institute. SAS System for Windows 9.4; SAS Institute, Inc.: Cary, NC, USA, 2013. [Google Scholar]
- Raivo, K. Package “Pheatmap”: Prety Heatmap. Available online: https://cran.r-project.org/web/packages/pheatmap/index.html (accessed on 6 September 2021).
- Salloum, M.S.; Guzzo, M.C.; Velazquez, M.S.; Sagadin, M.B.; Luna, C.M. Variability in Colonization of Arbuscular Mycorrhizal Fungi and Its Effect on Mycorrhizal Dependency of Improved and Unimproved Soybean Cultivars. Can. J. Microbiol. 2016, 62, 1034–1040. [Google Scholar] [CrossRef]
- Sawers, R.J.H.; Gebreselassie, M.N.; Janos, D.P.; Paszkowski, U. Characterizing Variation in Mycorrhiza Effect among Diverse Plant Varieties. Theor. Appl. Genet. 2010, 120, 1029–1039. [Google Scholar] [CrossRef]
- Mishra, V.; Ellouze, W.; Howard, R.J. Utility of Arbuscular Mycorrhizal Fungi for Improved Production and Disease Mitigation in Organic and Hydroponic Greenhouse Crops. J. Hortic. 2018, 5, 1000237. [Google Scholar] [CrossRef]
- Tawaraya, K. Arbuscular Mycorrhizal Dependency of Different Plant Species and Cultivars. Soil Sci. Plant Nutr. 2003, 49, 655–668. [Google Scholar] [CrossRef]
- Janos, D.P. Plant Responsiveness to Mycorrhizas Differs from Dependence upon Mycorrhizas. Mycorrhiza 2007, 17, 75–91. [Google Scholar] [CrossRef]
- Matsunaga, H.; Saito, T.; Saito, A. Evaluation of Resistance to Bacterial Wilt and Phytophthora Blight in Capsicum Genetic Resources Collected in Myanmar. J. Jpn. Soc. Hortic. Sci. 2011, 80, 426–433. [Google Scholar] [CrossRef][Green Version]
- Mimura, Y.; Kageyama, T.; Minamiyama, Y.; Hirai, M. QTL Analysis for Resistance to Ralstonia Solanacearum in Capsicum Accession “LS2341”. J. Jpn. Soc. Hortic. Sci. 2009, 78, 307–313. [Google Scholar] [CrossRef]
- Aslam, M.N.; Mukhtar, T.; Hussain, M.A.; Raheel, M. Assessment of Resistance to Bacterial Wilt Incited by Ralstonia solanacearum in Tomato Germplasm. J. Plant Dis. Prot. 2017, 124, 585–590. [Google Scholar] [CrossRef]
- Matsunaga, H.; Monma, S. Sources of Resistance to Bacterial Wilt in Capsicum. J. Jpn. Soc. Hort. Sci. 1999, 68, 753–761. [Google Scholar] [CrossRef]
- Pawaskar, J.; Kadam, J.; Navathe, S.; Kadam, J. Response of Chilli Varieties and Genotypes to Bacterial Wilt Caused by Ralstonia solanacearum and Its Management. Indian J. Sci. 2014, 11, 66–72. [Google Scholar]
- Mimura, Y.; Yoshikawa, M.; Hirai, M. Pepper Accession LS2341 Is Highly Resistant to Ralstonia solanacearum Strains from Japan. HortScience 2009, 44, 2038–2040. [Google Scholar] [CrossRef]
- Tripodi, P.; Kumar, S. The Capsicum Crop: An Introduction. In Compendium of Plant Geomes; Springer Nature Switzerland AG: Basel, Switzerland, 2019; pp. 1–8. [Google Scholar] [CrossRef]
- Cameron, D.D.; Neal, A.L.; van Wees, S.C.M.; Ton, J. Mycorrhiza-Induced Resistance: More than the Sum of Its Parts? Trends Plant Sci. 2013, 18, 539–545. [Google Scholar] [CrossRef]
- Jung, S.C.; Martinez-Medina, A.; Lopez-Raez, J.A.; Pozo, M.J. Mycorrhiza-Induced Resistance and Priming of Plant Defenses. J. Chem. Ecol. 2012, 38, 651–664. [Google Scholar] [CrossRef]
- Liu, H.; Wu, M.; Liu, J.; Qu, Y.; Gao, Y.; Ren, A. Tripartite Interactions Between Endophytic Fungi, Arbuscular Mycorrhizal Fungi, and Leymus Chinensis. Microb. Ecol. 2020, 79, 98–109. [Google Scholar] [CrossRef]
- Zhang, L.; Zhou, J.; George, T.S.; Limpens, E.; Feng, G. Arbuscular Mycorrhizal Fungi Conducting the Hyphosphere Bacterial Orchestra. Trends Plant Sci. 2021, 27, 402–411. [Google Scholar] [CrossRef]
- Paszkowski, U. Mutualism and Parasitism: The Yin and Yang of Plant Symbioses. Curr. Opin. Plant Biol. 2006, 9, 364–370. [Google Scholar] [CrossRef]
- Van Wees, S.C.; Van der Ent, S.; Pieterse, C.M. Plant Immune Responses Triggered by Beneficial Microbes. Curr. Opin. Plant Biol. 2008, 11, 443–448. [Google Scholar] [CrossRef] [PubMed]
- Pozo, M.J.; Azcón-Aguilar, C. Unraveling Mycorrhiza-Induced Resistance. Curr. Opin. Plant Biol. 2007, 10, 393–398. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Shi, Y.; Zou, L.; Huang, J.; Shen, L.; Wang, Y.; Guan, D.; He, S. Pepper CaMLO6 Negatively Regulates Ralstonia solanacearum Resistance and Positively Regulates High Temperature and High Humidity Responses. Plant Cell Physiol. 2020, 61, 1223–1238. [Google Scholar] [CrossRef] [PubMed]
- Smith, S.E.; Read, D.J. Mycorrhizal Symbiosis, 3rd ed.; Academic Press: London, UK, 2008; Available online: https://www.scirp.org/(S(351jmbntvnsjt1aadkozje))/reference/referencespapers.aspx?referenceid=1398632 (accessed on 5 August 2021).
- Kaur, S.; Suseela, V. Unraveling Arbuscular Mycorrhiza-Induced Changes in Plant Primary and Secondary Metabolome. Metabolites 2020, 10, 335. [Google Scholar] [CrossRef]



| Target | Primer ID | Sequence (5′–3′) | Ta (°C) | References |
|---|---|---|---|---|
| Jasmonic acid | OPR3 Forward | CTTCTCATGCAGTGTATCAAC | 46.4 | [46] |
| OPR3 Reverse | CGTCCAAGTGATCTATAGCTG | |||
| Salicylic acid | ICL Forward | GCGTCGGAGGAAGTCAAGAA | 50.5 | [47] |
| ICL Reverse | GGATTGGAACTTGGAGCCGA | |||
| Reference gene | GAPDH-Cp Forward | ATGATGATGTGAAAGCAGCG | 46.4/50.5 | - |
| GAPDH-Cp Reverse | TTTCAACTGGTGGCTGCTAC |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ambarwati, E.; Arwiyanto, T.; Widada, J.; Alam, T.; Andika, I.P.; Taryono. The Genes Associated with Jasmonic Acid and Salicylic Acid Are Induced in Tropical Chili Pepper against Ralstonia solanacearum by Applying Arbuscular Mycorrhizal Fungi. Horticulturae 2022, 8, 876. https://doi.org/10.3390/horticulturae8100876
Ambarwati E, Arwiyanto T, Widada J, Alam T, Andika IP, Taryono. The Genes Associated with Jasmonic Acid and Salicylic Acid Are Induced in Tropical Chili Pepper against Ralstonia solanacearum by Applying Arbuscular Mycorrhizal Fungi. Horticulturae. 2022; 8(10):876. https://doi.org/10.3390/horticulturae8100876
Chicago/Turabian StyleAmbarwati, Erlina, Triwidodo Arwiyanto, Jaka Widada, Taufan Alam, Ignatius Putra Andika, and Taryono. 2022. "The Genes Associated with Jasmonic Acid and Salicylic Acid Are Induced in Tropical Chili Pepper against Ralstonia solanacearum by Applying Arbuscular Mycorrhizal Fungi" Horticulturae 8, no. 10: 876. https://doi.org/10.3390/horticulturae8100876
APA StyleAmbarwati, E., Arwiyanto, T., Widada, J., Alam, T., Andika, I. P., & Taryono. (2022). The Genes Associated with Jasmonic Acid and Salicylic Acid Are Induced in Tropical Chili Pepper against Ralstonia solanacearum by Applying Arbuscular Mycorrhizal Fungi. Horticulturae, 8(10), 876. https://doi.org/10.3390/horticulturae8100876









