Abstract
Auxin is an important endogenous plant hormone that is usually present as indole-3-acetic acid (IAA). The flavin monooxygenase YUCCA is the rate-limiting enzyme of IAA biosynthesis and plays an important regulatory role in plant growth and development. To further investigate the function of the YUCCA gene family in weeping peach trees, members of the YUCCA gene family were identified via bioinformatics analysis. The gene structure and conserved domains of the weeping peach YUCCA genes were investigated, and phylogenetic analysis and gene annotation were carried out. Fourteen PpYUCCAs were identified in the weeping peach variety ‘Hongchuizhi’ and were found to be randomly located on five different chromosomes. Moreover, the prediction of subcellular localization showed that most of the YUCCA proteins were localized in the cytoplasm. Based on our transcriptome analysis, only nine PpYUCCAs, including PpYUCCA1, PpYUCCA3/4/5/6, PpYUCCA9, and PpYUCCA12/13/14, were expressed in the weeping peach branches, which could result in the accumulation of auxin. PpYUCCA6/12 may play a critical role in the appearance of the weeping trait, as indicated by the higher expression levels found in the Hongchuizhi variety compared with the Xiahui 6 variety. The results of this study provide a foundation for further research on the biological functions of PpYUCCAs in weeping peach trees.
1. Introduction
The phytohormone auxin plays a vital role in plant growth and development and has been shown to be involved in processes such as vascular bundle differentiation, embryonic polarization, apical dominance, and senescence [1,2,3]. In one study, auxin content was markedly enhanced under shade conditions, and expressions of flavin monooxygenase (YUCCA) genes were upregulated to enable the shade avoidance induced by increases in auxin [4]. Liu et al. [5] found that aluminum stress in Arabidopsis could affect local auxin biosynthesis in the root apex transition zone by regulating YUCCA, resulting in the inhibition of root growth. However, the phytohormone auxin mainly exists in the form of indole-3-acetic acid (IAA). The synthesis of IAA uses tryptophan as a substrate through the indole-3-pyruvate (IPA) synthesis pathway. The IPA pathway contains two steps: (1) Tryptophan is converted into IPA by tryptophan aminotransferase; (2) IPA is then catalyzed to IAA by YUCCA. The second step is the rate-limiting step of IAA synthesis; therefore, YUCCA genes play a crucial role in the biosynthesis of IAA [6].
There is increasing evidence that suggests that auxin is involved in a variety of stress responses in plants, including responses to low temperature, high salt, drought, and wounds [7,8]. Indeed, auxin can induce the transient and rapid expression of auxin response genes, which activates the plant’s defense system. The findings of Wang et al. [8] indicate that the GhYUCCA22 gene plays a key role in modulating ABA homeostasis and regulating the ability of cotton to withstand drought. Cha et al. [9] also confirmed that the improvement of drought tolerance in Arabidopsis lines overexpressing AtYUCCA6 was not caused by elevated IAA levels, but rather by increased activity of peroxidase and upregulated expression of genes involved in redox homeostasis. IAA has been reported to act as a strong accelerator of branch growth in many species, and the high content of IAA in weeping branches may lead to longer branches in weeping progeny [10]. Using a comparative transcriptome analysis, Mao et al. [11] identified 10 candidate genes associated with IAA biosynthesis in Prunus mume that contributed to the weeping trait, including two IAA biosynthesis genes (Pm013243 and Pm030202). The weeping trait appears to be a complex process regulated by a series of metabolic pathways. Genes that control the weeping trait have been found in a few species [11,12,13,14]. In peach and mulberry (Morus alba), the weeping trait is likely controlled by a single recessive gene, according to early cross-breeding results [15].
At present, 11 YUCCA genes have been identified in Arabidopsis, offering some evidence for the function of AtYUCCA1/2/4/5/6/7 in the biosynthesis of IAA [16,17,18,19,20]. In one study, an AtYUCCA6 activation mutant Arabidopsis presented higher IAA levels and the typical high-auxin phenotype, with epinastic cotyledons, elongated petioles, and strong apical dominance [21]. Using transcriptome and qRT-PCR analysis, Yang et al. [22] found that the expression level of TaYUCCA7-A was significantly upregulated in powdery mildew-induced wheat. Studies on YUCCA gene loss-of-function mutants have further demonstrated the important role that the YUCCA gene family plays in auxin synthesis and plant growth and development. It is worth noting that the inactivation of a single YUCCA gene in Arabidopsis does not cause significant developmental defects, whereas the overexpression of any YUCCA gene promotes the biosynthesis of auxin. However, the loss of function of multiple YUCCA genes in Arabidopsis has been shown to cause developmental defects. In particular, AtYUCCA1 and AtYUCCA4 double mutants were shown to have reduced vascular tissue and failed to produce normal inflorescences, whereas the phenotypes of quadruple AtYUCCA1/2/4/6 mutants were more severe [16]. Previous studies on wheat [22], rice [23], tomato [24,25], and strawberry [26] have shown that members of the YUCCA gene family have highly conserved motifs, including the nicotinamide adenine dinucleotide phosphate (NADPH)-binding motif (GxGxxGME), FAD-binding motif (GxGPxGLA), GC motif (ERxxxxASL), etc., which may be key sites for the biological function of YUCCA genes [6]. In addition, the FAD-binding motif and NADPH-binding motif were shown to be identical in FaYUC1 and FaYUC2 and their corresponding Arabidopsis homologues [26].
Peaches (Prunus persica L.) are a popular fruit—cultivated worldwide—and a consumer favorite due to their sweet taste, pleasant aroma, phytonutrients, and attractive color. In addition to the edible fruit, peach blossoms also have high ornamental value. The weeping peach (Prunus persica var. pendula) variant has high ornamental value because of its weeping branches, which are similar to those of willows. Compared with studies on model plants, such as Arabidopsis, and other plants, such as rice and strawberry [16,26,27], there have been few studies focusing on peach YUCCA genes. In this study, with the accomplishment of peach genome sequencing and improvements in the peach genome database [28], a total of 14 peach YUCCA genes were comprehensively identified. In addition, the detailed characteristics, structures, functions, and expression levels of these genes were investigated. These analyses provided useful information for further investigations into the functions of the YUCCA gene family in the weeping peach. This study establishes a foundation for future exploration into the cloning and biological functions of weeping peach; in addition, these genes may be useful for the improvement of weeping peach trees.
2. Materials and Methods
2.1. Identification of PpYUCCA Genes in the Weeping Peach Genome
In order to identify YUCCA family genes in the weeping peach, we downloaded the nucleotide sequences of the YUCCA genes from the model species, Arabidopsis and Solanum lycopersicum, from TAIR (The Arabidopsis Information Resource: http://www.arabidopsis.org/) (accessed on 10 May 2022) and NCBI (https://www.ncbi.nlm.nih.gov/genome/?term=tomato) (accessed on 10 May 2022). As Pyrus bretschneideri, Malus domestica, and Prunus persica belong to the Rosaceae family, and they all have the weeping trait, the nucleotide sequences of the YUCCA genes of Pyrus bretschneideri and Malus domestica were also downloaded from NCBI (https://www.ncbi.nlm.nih.gov/genome/12793) (accessed on 10 May 2022) and the Genome Database for Rosaceae (https://www.rosaceae.org/species/malus/all) (accessed on 10 May 2022). The predicted sequences were acquired using the BLASTp program (a given nucleic acid sequence was aligned with the database), with a threshold of 1 × 10−20, according to peach genome data. Then, the encoded sequences of the PpYUCCA proteins were confirmed using pfamScan (v1.6) and Pfam A (v33.1) software. Subcellular localization was predicted using Softberry (http://linux1.softberry.com/berry.phtml?topic=protcomppl&group=programs&subgroup=proloc) (accessed on 10 May 2022). The chemical and physical properties (i.e., molecular weight, amino acid number, isoelectric point (pI), GRAVY, instability index, and aliphatic index) of the PpYUCCA proteins were calculated with the help of the ExPASy website (http://web.expasy.org/protparam/) (accessed on 10 May 2022). A user-friendly online tool (http://mg2c.iask.in/mg2c_v2.1/ (accessed on 10 May 2022), Chinese Academy of Agricultural Sciences, Qingdao, China) was used to explore the chromosome locations of the PpYUCCA members.
2.2. Classification and Structural Analysis of PpYUCCA Genes
An analysis of YUCCA gene structures was carried out with the aid of the Gene Structure Display Server (http://gsds.gao-lab.org/ (accessed on 10 May 2022), Peking University, Beijing, China). The common motifs were found using the MEME software (v5.0.5, http://meme.nbcr.net/meme (accessed on 10 May 2022), University of Nevada, Reno, NV, USA), where the maximum number of motifs was set to fifteen and the optimum width of the motifs was set from six to twenty. The protein sequences of YUCCA family genes from five species (peach, Arabidopsis, apple, pear, and tomato) were aligned with the help of MAFFT software (v7.427, University of Illinois at Urbana-Champaign, Urbana-Champaign, IL, USA), under the default parameters.
2.3. Phylogenetic Analysis of Weeping Peach YUCCA Genes
The full-length amino acid sequences of YUCCA proteins from five species (peach, Arabidopsis, apple, pear, and tomato) were downloaded from their respective genomic databases. A phylogenetic tree was created with the aid of the MEGA 7.0 software (Mega Limited, Auckland, New Zealand), using the neighbor-joining method and verified with one thousand bootstraps.
2.4. Plant Materials and Treatments
In this study, we selected two varieties of trees: the weeping peach ‘Hongchuizhi’ (herein termed ’HCZ’) and the upright peach ’Xiahui 6’ (herein termed ’XH’, control group). The shoot tips from 6-year-old peach trees were sampled. These trees were grown in the Peach Experimental Garden at the Jiangsu Academy of Agricultural Sciences, Nanjing City, Jiangsu Province, China, and were trained and managed using standard horticultural practices. The shoot tips were harvested during the budding growth period (herein termed ’the early stage of branches growth’, S1), and at the end of the growth period (herein termed ’the late stage of branches growth’, S2). The shoot tips were immediately frozen in liquid nitrogen and stored at −80 °C prior to RNA extraction and gene expression analysis. To provide three replicates, each group consisted of three subgroups of three trees each.
2.5. Gene Expression Analysis
The transcriptomes of the two varieties were determined as described below. RNA quantification and qualification and cDNA library construction and sequencing were performed. Total RNA was extracted from the shoot tips using an RNAprep Pure Plant Kit (DP441, Tiangent, Beijing, China), following the manufacturer’s instructions. The purity and concentration of the obtained RNA were measured using a NanoDrop 2000 (Thermo Fisher Scientific, Waltham, MA, USA). RNA integrity was evaluated using an Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA, USA) with the help of an RNA 6000 Nano Assay Kit (Agilent Technologies, Palo Alto, CA, USA).
A 1 μg RNA sample was used for each examination. Sequencing libraries were generated with the help of Genepioneer Biotechnologies Co. Ltd. (Nanjing, China), and the NEBNextR UltraTM Directional RNA Library Prep Kit for IlluminaR (NEB, Ipswich, MA, USA), according to the manufacturer’s recommendations.
Reads containing the adapter and low-quality sequences were removed prior to analysis of the sequencing data. The resulting high-quality clean data were aligned against the peach reference genome of Prunus persica Genome V2.0.a1 [29] using HISAT2 software tools. Gene expression levels were calculated using the fragments per kilobase of exon model per million mapped fragments (FPKM) method. A differential gene expression analysis of the two varieties was conducted using the DESeq2 R package (1.26.0). The resulting p values were adjusted to control for the false discovery rate using the Benjamini–Hochberg method. Genes with an adjusted p value of <0.05 and an absolute log2(fold change) value of >1, found by DESeq2, were considered to be differentially expressed. KOBAS software was used to test the statistical enrichment of differentially expressed genes (DEGs) in KEGG pathways. The clusterProfiler R package was used to find the KEGG pathways that were significantly enriched compared with the entire genome background. Gene Ontology (GO) enrichment analysis of the DEGs was performed using the clusterProfiler R package.
2.6. Statistical Analysis
A completely randomized design was adopted in the experiments. The data were represented as the mean ± standard error (SE) of three replicates. Next, a one-way ANOVA was performed to establish the differences between the two varieties using Dunnett’s post t-test with a p value of < 0.05 in SPSS 23.0 (SPSS Inc., Chicago, IL, USA).
3. Results
3.1. PpYUCCA Gene Family Analysis in the Weeping Peach Genome
A total of 14 YUCCA genes were identified in Prunus persica var. pendula (Table 1); the genes were named from PpYUCCA1 to PpYUCCA14. The amino acid lengths of these PpYUCCAs were determined to be between 224 and 532 aa. The pIs were between 4.68 and 9.17, and the molecular weights ranged from 24644.57 to 60781.52 Da. Among these values, the pI of PpYUCCA7 was the lowest, whereas the pI of PpYUCCA3 was the highest. In addition, we predicted the subcellular localization of YUCCA proteins in peach cells, and the results showed PpYUCCA5/6 was localized to the plasma membrane, PpYUCCA2/8/11/12 in the extracellular space, and the remaining YUCCA proteins in the cytoplasm. The characteristics of the PpYUCCA gene family are displayed in Table 1, which contains the gene identifier in the genome database, chromosomal location, and some basic physical and chemical properties.

Table 1.
Characteristics of the PpYUCCA gene family in peach trees.
The genetic mapping of PpYUCCA genes to the chromosomes was carried out according to the genome data (Figure 1). These 14 PpYUCCAs were randomly located on five different chromosomes. Chromosome 01 had four PpYUCCAs; chromosomes 05 and 07 had two PpYUCCAs each; chromosome 06 contained only one PpYUCCA; and chromosome 08 contained five PpYUCCAs. It should be noted that PpYUCCA5 and PpYUCCA6 (located on Chr05) and PpYUCCA11 and PpYUCCA12 (located on Chr08) were very close, indicating that the two sets of genes were a pair of tandem duplication genes (Figure 1, Figure 2, Figure 3 and Figure 4).
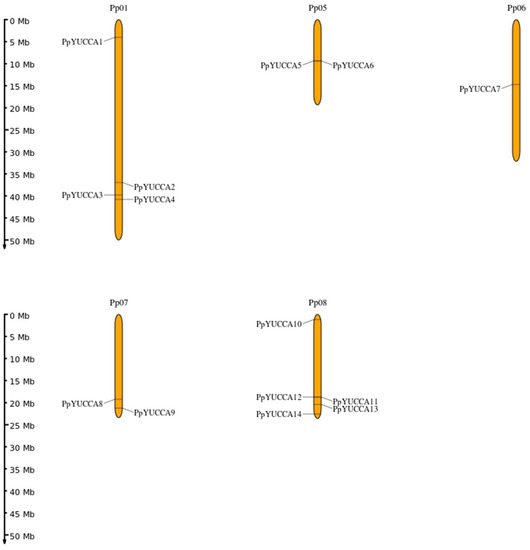
Figure 1.
Chromosomal locations of Prunus persica var. pendula YUCCA genes. The scale represents a 50 Mb chromosomal distance.
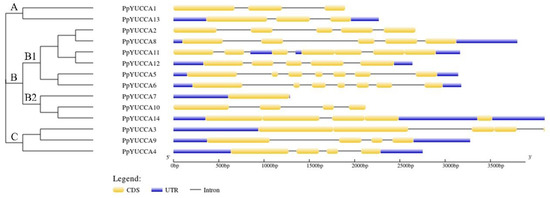
Figure 2.
Structure of Prunus persica var. pendula YUCCA genes. Introns, exons, and untranslated regions (UTRs) are represented by black lines, deep orange boxes, and blue boxes, respectively.
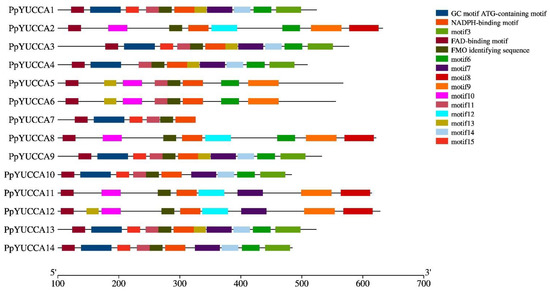
Figure 3.
YUCCA gene family motifs in Prunus persica var. pendula. The various conserved motifs are labeled with colored boxes.
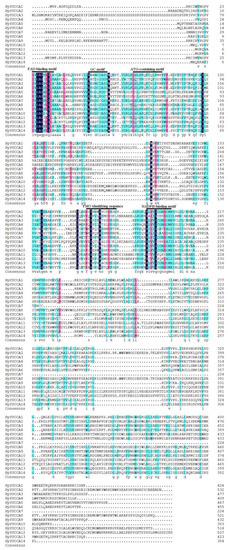
Figure 4.
Multiple sequence alignment of Prunus persica var. pendula YUCCA gene family.
3.2. Classification and Structural Analysis of PpYUCCA Genes
The peach PpYUCCAs were divided into four subfamilies based on their similarities (Figure 2). As shown in Figure 2, PpYUCCA1 and PpYUCCA3 were grouped in subfamily A, and PpYUCCA7/10/14 and PpYUCCA3/9/4 were placed in subfamily B2 and C, respectively. The remaining genes were grouped in subfamily B1 (Figure 2).
In order to study the gene structure of PpYUCCAs, information on the genome and CDS sequence for each YUCCA gene was obtained according to the genome data; the exon–intron gene structure map is shown in Figure 2. The PpYUCCA genes contained between one and seven exons. PpYUCCA7 only contained one exon, whereas PpYUCCA3/5/6/11 contained seven exons. Moreover, PpYUCCA1/3 contained three exons, PpYUCCA4/9/10 contained four exons, and PpYUCCA2/8/12/14 contained five exons (Figure 2).
To further study the conserved domain of YUCCA proteins in the weeping peach, multiple alignment of amino acid sequences of the YUCCA proteins were used to identify the conserved protein motifs (Figure 3 and Figure 4). A total of 15 distinct motifs were discovered in the experiment; among them, five motifs were identified. These identified conserved motifs were the FAD-binding motif, GC motif, ATG-containing motif, FMO-identifying sequence, and NADPH-binding motif. Of these five conserved motifs, the FAD-binding motif, GC motif, and ATG-containing motif were located close to each other at the N-terminus of the amino acid sequences. The FMO-identifying sequence and NADPH-binding motif were located in the middle region of the amino acid sequences.
3.3. Phylogenetic Analysis of Weeping Peach YUCCA Genes
In order to further analyze the evolutionary relationships between YUCCA proteins among different species, the YUCCA proteins of Arabidopsis thaliana, Malus domestica, Pyrus bretschneideri, Solanum lycopersicum, and Prunus persica var. pendula were used to construct a phylogenetic tree using MEGA 7.0 software. As shown in Figure 5, 76 YUCCA proteins from five species were grouped into five categories. Among them, Groups I, III, and V each contained two different weeping peach YUCCA genes: PpYUCCA7 and PpYUCCA10 (Group I); PpYUCCA1 and PpYUCCA13 (Group III); and PpYUCCA3 and PpYUCCA9 (Group V). Group IV only contained one YUCCA protein, namely PpYUCCA4, whereas the remaining YUCCA proteins were all in Group II. Except for the six YUCCA proteins in Group II (PpYUCCA2/5/6/8/11/12), the results of the phylogenetic analysis certified that the YUCCAs of Rosaceae tended to be clustered together, as shown in Figure 5. Therefore, we concluded that the weeping peach YUCCA proteins were highly evolutionarily conserved. These results will provide important support for the further analysis of weeping peach YUCCA genes.
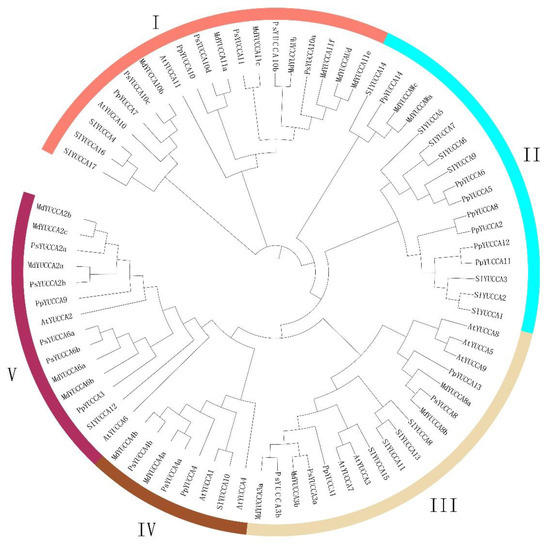
Figure 5.
Phylogenetic tree of YUCCA genes in Prunus persica var. pendula. At—Arabidopsis thaliana; Md—Malus domestica; Ps—Pyrus bretschneideri; Sl—Solanum lycopersicum; Pp—Prunus persica var. pendula.
3.4. Gene Expression Analysis
When comparing the DEGs between HCZ and XH at the same point in time based on KEGG pathways, the most enriched pathway at the early stage of branch growth was ‘carbon metabolism’, followed by ‘metabolism of terpenoids and polyketides’ and ‘biosynthesis of other secondary metabolites’ (Figure 6A); the most enriched pathway at the late stage of branch growth was ‘carbon metabolism’, followed by ‘biosynthesis of other secondary metabolites’ and ‘amino acid metabolism’. Three major categories were identified according to the GO annotations and comparison of DEGs between HCZ and XH at the same point in time, including those associated with cellular components, molecular functions, and biological processes (Figure 7). Genes associated with cell, cell parts, and cellular process were the most abundant.
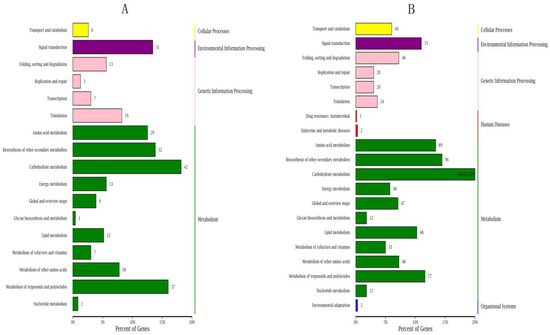
Figure 6.
KEGG pathway classification of DEGs during the growth period of weeping peach branches: (A) comparison of HCZ and XH at the early stage of branch growth (HCZS1 vs. XHS1); (B) comparison of HCZ and XH at the late stage of branch growth (HCZS2 vs. XHS2). HCZ—weeping peach ‘Hongchuizhi’ variety; XH—standard peach ‘Xiahui 6’ variety.
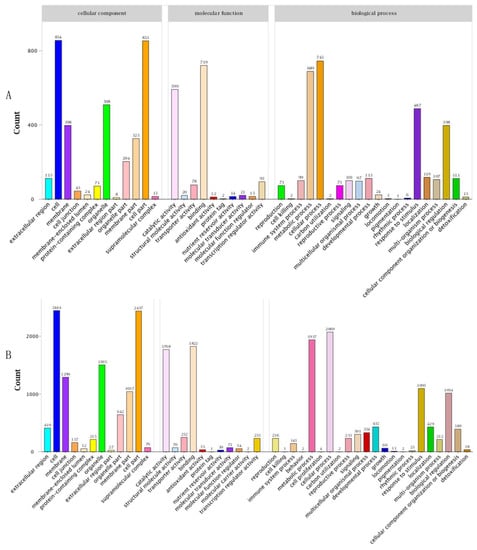
Figure 7.
GO classification of DEGs during the growth period of weeping peach branches: (A) comparison of HCZ and XH at the early stage of branch growth (HCZS1 vs. XHS1); (B) comparison of HCZ and XH at the late stage of branch growth (HCZS2 vs. XHS2). HCZ—weeping peach ‘Hongchuizhi’ variety; XH—standard peach ‘Xiahui 6’ variety.
In this experiment, nine PpYUCCAs were found to be expressed in weeping peach branches (Figure 8), including PpYUCCA1, PpYUCCA3/4/5/6, PpYUCCA9, and PpYUCCA12/13/14. To explore the roles of these genes, the expression patterns of the different varieties of PpYUCCAs during different developmental periods were carried out using a heatmap.
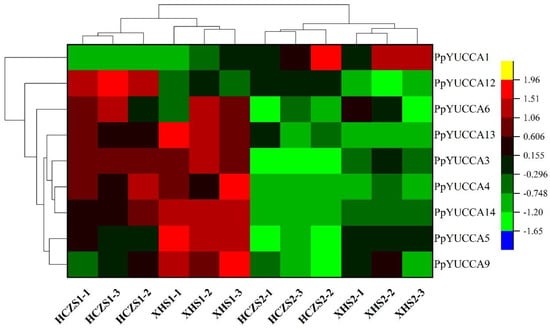
Figure 8.
Heatmap cluster analysis of the FPKM of YUCCA genes during the growth period of weeping peach branches. HCZ—weeping peach ‘Hongchuizhi’ variety; XH—standard peach ‘Xiahui 6’ variety; S1—early stage of branch growth; S2—late stage of branch growth.
As shown in Figure 8, in comparison to the budding growth period, only PpYUCCA1 exhibited higher expression at the end of the growth period in both varieties. The expressions of PpYUCCA3/4/5/6, PpYUCCA9, and PpYUCCA12/13/14 in the two varieties were higher during the budding growth period than at the end of the growth period. In addition, the expressions of PpYUCCA6/12 in HCZ were higher than those in XH during the budding growth period. These results suggest that PpYUCCA6/12 may have greater effects on the weeping branches of peach trees.
4. Discussion
Weeping branches, which act as a special morphological structure of trees, have become a research hotspot, as they are widely used in greenery and garden landscapes worldwide due to their beautiful shape. An increasing number of studies have indicated that auxins, including IAA, play a critical role in the weeping trait of branches [30,31]; however, the molecular information underlying weeping peach trees is still unclear. YUCCA, which converts IPA into IAA, is the rate-limiting enzyme in the IPA pathway [6]. In this paper, to gain insight into peach YUCCA genes, the gene structure and conserved domains of the YUCCA gene family were investigated, and a phylogenetic analysis and gene annotation were carried out. Based on our transcriptome analysis, a total of 14 YUCCA genes were identified in peach trees, only nine of which were expressed in the ‘HCZ’ weeping peach variety.
The structure of proteins determines their biological function, and proteins with similar structures have similar functions. From the subcellular localization analysis, it was found that most of the weeping peach PpYUCCA proteins were localized in the cytoplasm, indicating that the synthesis of auxin by these genes was conducted in the cytoplasm; this is consistent with the results of Yang et al. [22]. The analysis of the conserved motifs of the weeping peach YUCCA proteins showed that the number of motifs in the 14 proteins ranged from 6 to 11, indicating that the functions of the weeping peach YUCCA gene family are diverse. As shown in Figure 2 and Figure 3, the closely related proteins clearly share the same motif profiles, indicating that the weeping peach YUCCA proteins within one branch may have the same conserved function [8]. Furthermore, the conserved motifs were almost identical in number and type. In a way, the specific motifs may result in functional divergences [8,32]. Although the presence or lack of specific motifs may lead to functional variation, this needs to be experimentally demonstrated with precision. Moreover, the specific biological functions of the remaining motifs remain to be further investigated. Similar to the results of studies using wheat and strawberry [22,26], the conserved motifs—including the FAD-binding motif, GC motif, ATG-containing motif, FMO-identifying sequence, and NADPH-binding motif—were also found in the 14 weeping peach YUCCA proteins (Figure 4); thus, these conserved motifs might be critical sites for the biological function of YUCCA genes [6,26].
Phylogenetic analysis showed that the PpYUCCAs could be divided into five subfamilies. Most of the Rosaceae YUCCA proteins were clustered together (Figure 5), indicating that peach YUCCA genes are highly evolutionarily conserved compared with those of Malus domestica and Pyrus bretschneideri [33,34]. In barley and Arabidopsis, high temperature conditions were shown to lead to male sterility via the repression of YUCCA2/6 expression, which decreased the auxin level in the developing anthers [35]. PpYUCCA3/9 were most closely related to AtYUCCA2/6 of Arabidopsis thaliana (Figure 5); it was speculated that they may mediate peach growth and early pollen development by regulating auxin signal transduction [36]. The higher expressions of PpYUCCA3/9 in the two varieties during the budding growth period compared with the end of the growth period also verified the above results (Figure 8). In one study, MdYUCCA11d appeared to be specifically expressed in the apple pistil, and it was found to be responsible for auxin synthesis there [33]. PpYUCCA2/10 appeared to be closely related to MdYUCCA11 in the phylogenetic analysis (Figure 5), indicating specific roles during the development of flower and fruit [33]. This may be the reason why we did not observe the expression of PpYUCCA2/10 in weeping peach branches. The overexpression of MdYUCCA8a in Arabidopsis was shown to increase the level of auxins and produce expected auxin overproduction phenotypes, such as taller plant height and enhanced apical dominance [33]. An increase in the expression of the homologous gene PpYUCCA13 was also successfully detected in weeping peach branches during the budding growth period (Figure 5), indicating that PpYUCCA13 may be a critical gene for IAA biosynthesis in weeping peach trees [37].
Interestingly, the PpYUCCA2/5/6/8/11/12 proteins were not clustered together with the proteins of Rosaceae and Arabidopsis thaliana; instead, they were placed in Group II with the YUCCA proteins of Solanum lycopersicum (Figure 5). These results suggest that the weeping peach YUCCA gene family has been amplified along with whole-genome duplication [38]. The PpYUCCA5/PpYUCCA6 genes and PpYUCCA11/PpYUCCA12 genes were determined to be a pair of tandem duplication genes, indicating that tandem duplication is also a method of gene amplification in the weeping peach YUCCA gene family [39]. As genes can be amplified in a variety of ways, including whole-genome duplication, tandem duplication, retrotransposition, etc., the specific gene amplification mode of the members of the weeping peach YUCCA gene family should be further explored. However, the functions of the Group II genes have not yet been validated in the weeping peach or Solanum lycopersicum. Our transcriptome data suggest that the expression of PpYUCCA6/12 in HCZ was higher than that in XH during the budding growth period (Figure 8); thus, we speculate that PpYUCCA6/12 may contribute to the weeping trait of peach branches.
5. Conclusions
In conclusion, 14 PpYUCCAs were systematically identified in weeping peach branches. In this study, the gene structure and conserved domains of the weeping peach YUCCA gene family were determined, and phylogenetic analysis and gene annotation were carried out. Based on our transcriptome analysis, a total of nine PpYUCCAs, including PpYUCCA1, PpYUCCA3/4/5/6, PpYUCCA9, and PpYUCCA12/13/14, were expressed in the weeping peach branches, which may result in the accumulation of auxin. Among them, PpYUCCA6/12 were more likely to play major roles in the appearance of the weeping shape, as indicated by the higher expression levels in HCZ. However, the functions of these genes require further verification to determine what controls the weeping trait of peach trees more precisely. The useful genetic information presented in this study will be beneficial to the improvement of weeping peach trees.
Author Contributions
Y.Z. performed the majority of this study; Q.M. and J.X. helped to finish the experiments; Y.Z. and M.Y. wrote this manuscript; R.M. and M.Y. designed the experiments. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (31901979) and the National Key Research and Development Program subproject of China (2019YFD1000801-02). The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
The processing and analysis of transcriptome data were completed with the help of Genepioneer Biotechnologies.
Conflicts of Interest
There were no conflicts of interest in the submission of this manuscript.
References
- Feng, H.Q.; Chao, L.I. Research Advances of Auxin Signal Transduction. Biotechnol. Bull. 2018, 34, 24–30. [Google Scholar] [CrossRef]
- Péret, B.; De Rybel, B.; Casimiro, I.; Benková, E.; Swarup, R.; Laplaze, L.; Beeckman, T.; Bennett, M.J. Arabidopsis lateral root development: An emerging story. Trends Plant Sci. 2009, 14, 399–408. [Google Scholar] [CrossRef] [PubMed]
- Andrea, G. The role of auxin in shaping shoot architecture. J. Exp. Bot. 2013, 64, 2593–2608. [Google Scholar] [CrossRef]
- Müller-Moulé, P.; Nozue, K.; Pytlak, M.L.; Palmer, C.M.; Covington, M.F.; Wallace, A.D.; Harmer, S.L.; Maloof, J.N. YUCCA auxin biosynthetic genes are required for Arabidopsis shade avoidance. PeerJ 2016, 4, e2574. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Gao, S.; Tian, H.; Wu, W.; Robert, H.S.; Ding, Z. Local Transcriptional Control of YUCCA Regulates Auxin Promoted Root-Growth Inhibition in Response to Aluminium Stress in Arabidopsis. PLoS Genet. 2016, 12, e1006360. [Google Scholar] [CrossRef]
- Qin, M.; Wang, J.; Zhang, T.; Hu, X.; Liu, R.; Gao, T.e.; Zhao, S.; Yuan, Y.; Zheng, J.; Wang, Z.; et al. Genome-Wide Identification and Analysis on YUCCA Gene Family in Isatis indigotica Fort. and IiYUCCA6-1 Functional Exploration. Int. J. Mol. Sci. 2020, 21, 2188. [Google Scholar] [CrossRef]
- Yuan, H.-Z.; Zhao, M.-Z.; Wu, W.-M.; Yu, H.-M.; Qian, Y.-M.; Wang, Z.-W.; Wang, X.-C. Genome-wide identification and expression analysis of auxin-related gene families in grape. Yi Chuan 2015, 37, 720–730. [Google Scholar] [CrossRef]
- Wang, X.; Chen, B.; Ma, C.; Qiao, K.; Li, Z.; Wang, J.; Peng, R.; Fan, S.; Ma, Q. Systematical characterization of YUCCA gene family in five cotton species, and potential functions of YUCCA22 gene in drought resistance of cotton. Ind. Crops Prod. 2021, 162, 113290. [Google Scholar] [CrossRef]
- Cha, J.-Y.; Kim, W.-Y.; Kang, S.B.; Kim, J.I.; Baek, D.; Jung, I.J.; Kim, M.R.; Li, N.; Kim, H.-J.; Nakajima, M.; et al. A novel thiol-reductase activity of Arabidopsis YUC6 confers drought tolerance independently of auxin biosynthesis. Nat. Commun. 2015, 6, 8041. [Google Scholar] [CrossRef]
- Tanimoto, E. Regulation of Root Growth by Plant Hormones—Roles for Auxin and Gibberellin. Crit. Rev. Plant Sci. 2005, 24, 249–265. [Google Scholar] [CrossRef]
- Mao, T.-Y.; Zhu, H.-H.; Liu, Y.-Y.; Bao, M.-Z.; Zhang, J.-W.; Fu, Q.; Xiong, C.-F.; Zhang, J. Weeping candidate genes screened using comparative transcriptomic analysis of weeping and upright progeny in an F1 population of Prunus mume. Physiol. Plant. 2020, 170, 318–334. [Google Scholar] [CrossRef] [PubMed]
- Sugano, M.; Nakagawa, Y.; Nyunoya, H.; Nakamura, T. Expression of gibberellin 3β-hydroxylase gene in a gravi-response mutant, weeping Japanese flowering cherry. Biol. Sci. Space 2004, 18, 261–266. [Google Scholar] [CrossRef] [PubMed]
- Joshi, C.P.; Thammannagowda, S.; Fujino, T.; Gou, J.-Q.; Avci, U.; Haigler, C.H.; McDonnell, L.M.; Mansfield, S.D.; Mengesha, B.; Carpita, N.C.; et al. Perturbation of Wood Cellulose Synthesis Causes Pleiotropic Effects in Transgenic Aspen. Mol. Plant 2011, 4, 331–345. [Google Scholar] [CrossRef] [PubMed]
- Zhang, N.; Yu, H.; Yu, H.; Cai, Y.; Huang, L.; Xu, C.; Xiong, G.; Meng, X.; Wang, J.; Chen, H.; et al. A Core Regulatory Pathway Controlling Rice Tiller Angle Mediated by the LAZY1-Dependent Asymmetric Distribution of Auxin. Plant Cell 2018, 30, 1461–1475. [Google Scholar] [CrossRef]
- Bassi, D.; Rizzo, M. Peach breeding for growth habit. Acta Hortic. 2000, 538, 411–414. [Google Scholar] [CrossRef]
- Cheng, Y.; Dai, X.; Zhao, Y. Auxin biosynthesis by the YUCCA flavin monooxygenases controls the formation of floral organs and vascular tissues in Arabidopsis. Genes Dev. 2006, 20, 1790–1799. [Google Scholar] [CrossRef]
- Kim, J.I.; Murphy, A.S.; Baek, D.; Lee, S.-W.; Yun, D.-J.; Bressan, R.A.; Narasimhan, M.L. YUCCA6 over-expression demonstrates auxin function in delaying leaf senescence in Arabidopsis thaliana. J. Exp. Bot. 2011, 62, 3981–3992. [Google Scholar] [CrossRef]
- Lee, M.; Jung, J.-H.; Han, D.-Y.; Seo, P.J.; Park, W.J.; Park, C.-M. Activation of a flavin monooxygenase gene YUCCA7 enhances drought resistance in Arabidopsis. Planta 2012, 235, 923–938. [Google Scholar] [CrossRef]
- Li, L.; Ljung, K.; Breton, G.; Schmitz, R.J.; Pruneda-Paz, J.; Cowing-Zitron, C.; Cole, B.J.; Ivans, L.J.; Pedmale, U.V.; Jung, H.-S.; et al. Linking photoreceptor excitation to changes in plant architecture. Genes Dev. 2012, 26, 785–790. [Google Scholar] [CrossRef]
- Woodward, C.; Bemis, S.M.; Hill, E.J.; Sawa, S.; Koshiba, T.; Torii, K.U. Interaction of auxin and ERECTA in elaborating Arabidopsis inflorescence architecture revealed by the activation tagging of a new member of the YUCCA family putative flavin monooxygenases. Plant Physiol. 2005, 139, 192–203. [Google Scholar] [CrossRef]
- Kim, J.I.; Sharkhuu, A.; Jin, J.B.; Li, P.; Jeong, J.C.; Baek, D.; Lee, S.Y.; Blakeslee, J.J.; Murphy, A.S.; Bohnert, H.J.; et al. yucca6, a dominant mutation in Arabidopsis, affects auxin accumulation and auxin-related phenotypes. Plant Physiol. 2007, 145, 722–735. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Xu, T.; Wang, H.; Feng, D. Genome-wide identification and expression analysis of the TaYUCCA gene family in wheat. Mol. Biol. Rep. 2021, 48, 1269–1279. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, Y.; Kamiya, N.; Morinaka, Y.; Matsuoka, M.; Sazuka, T. Auxin Biosynthesis by the YUCCA Genes in Rice. Plant Physiol. 2007, 143, 1362–1371. [Google Scholar] [CrossRef]
- Expósito-Rodríguez, M.; Borges, A.A.; Borges-Pérez, A.; Hernández, M.; Pérez, J.A. Cloning and Biochemical Characterization of ToFZY, a Tomato Gene Encoding a Flavin Monooxygenase Involved in a Tryptophan-dependent Auxin Biosynthesis Pathway. J. Plant Growth Regul. 2007, 26, 329–340. [Google Scholar] [CrossRef]
- Expósito-Rodríguez, M.; Borges, A.A.; Borges-Pérez, A.; Pérez, J.A. Gene structure and spatiotemporal expression profile of tomato genes encoding YUCCA-like flavin monooxygenases: The ToFZY gene family. Plant Physiol. Biochem. 2011, 49, 782–791. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Ying, Y.-Y.; Zhang, L.; Gao, Q.-H.; Li, J.; Zhang, Z.; Fang, J.-G.; Duan, K. Isolation and characterization of two YUCCA flavin monooxygenase genes from cultivated strawberry (Fragaria × ananassa Duch.). Plant Cell Rep. 2012, 31, 1425–1435. [Google Scholar] [CrossRef]
- Abu-Zaitoon, Y.M. Phylogenetic Analysis of Putative Genes Involved in the Tryptophan-Dependent Pathway of Auxin Biosynthesis in Rice. Appl. Biochem. Biotechnol. 2014, 172, 2480–2495. [Google Scholar] [CrossRef]
- Jung, S.; Lee, T.; Cheng, C.H.; Buble, K.; Zheng, P.; Yu, J.; Humann, J.; Ficklin, S.P.; Gasic, K.; Scott, K.; et al. 15 years of GDR: New data and functionality in the Genome Database for Rosaceae. Nucleic Acids Res. 2019, 47, D1137–D1145. [Google Scholar] [CrossRef]
- Verde, I.; Jenkins, J.; Dondini, L.; Micali, S.; Pagliarani, G.; Vendramin, E.; Paris, R.; Aramini, V.; Gazza, L.; Rossini, L.; et al. The Peach v2.0 release: High-resolution linkage mapping and deep resequencing improve chromosome-scale assembly and contiguity. BMC Genom. 2017, 18, 225. [Google Scholar] [CrossRef]
- Li, L.; Zhang, Y.; Zheng, T.; Zhuo, X.; Li, P.; Qiu, L.; Liu, W.; Wang, J.; Cheng, T.; Zhang, Q. Comparative gene expression analysis reveals that multiple mechanisms regulate the weeping trait in Prunus mume. Sci. Rep. 2021, 11, 2675. [Google Scholar] [CrossRef]
- Liu, J.; Zeng, Y.; Yan, P.; He, C.; Zhang, J. Transcriptional and Hormonal Regulation of Weeping Trait in Salix matsudana. Genes 2017, 8, 359. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Liu, H.; Wang, S.; Li, H. Genome-wide identification and expression analysis of the YUCCA gene family in soybean (Glycine max L.). Plant Growth Regul. 2017, 81, 265–275. [Google Scholar] [CrossRef]
- Song, C.; Zhang, D.; Zheng, L.; Shen, Y.; Zuo, X.; Mao, J.; Meng, Y.; Wu, H.; Zhang, Y.; Liu, X.; et al. Genome-wide identification and expression profiling of the YUCCA gene family in Malus domestica. Sci. Rep. 2020, 10, 10866. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Cai, D.; Potter, D.; Postman, J.; Liu, J.; Teng, Y. Phylogeny and evolutionary histories of Pyrus L. revealed by phylogenetic trees and networks based on data from multiple DNA sequences. Mol. Phylogenet. Evol. 2014, 80, 54–65. [Google Scholar] [CrossRef] [PubMed]
- Sakata, T.; Oshino, T.; Miura, S.; Tomabechi, M.; Tsunaga, Y.; Higashitani, N.; Miyazawa, Y.; Takahashi, H.; Watanabe, M.; Higashitani, A. Auxins reverse plant male sterility caused by high temperatures. Proc. Natl. Acad. Sci. USA 2010, 107, 8569–8574. [Google Scholar] [CrossRef]
- Yao, X.; Tian, L.; Yang, J.; Zhao, Y.-N.; Zhu, Y.-X.; Dai, X.; Zhao, Y.; Yang, Z.-N. Auxin production in diploid microsporocytes is necessary and sufficient for early stages of pollen development. PLoS Genet. 2018, 14, e1007397. [Google Scholar] [CrossRef]
- Di, D.-W.; Wu, L.; Zhang, L.; An, C.-W.; Zhang, T.-Z.; Luo, P.; Gao, H.-H.; Kriechbaumer, V.; Guo, G.-Q. Functional roles of Arabidopsis CKRC2/YUCCA8 gene and the involvement of PIF4 in the regulation of auxin biosynthesis by cytokinin. Sci. Rep. 2016, 6, 36866. [Google Scholar] [CrossRef]
- Moghe, G.D.; Hufnagel, D.E.; Tang, H.; Xiao, Y.; Dworkin, I.; Town, C.D.; Conner, J.K.; Shiu, S.-H. Consequences of Whole-Genome Triplication as Revealed by Comparative Genomic Analyses of the Wild Radish Raphanus raphanistrum and Three Other Brassicaceae Species. Plant Cell 2014, 26, 1925–1937. [Google Scholar] [CrossRef]
- Cannon, S.B.; Mitra, A.; Baumgarten, A.; Young, N.D.; May, G. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 2004, 4, 10. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).