Abstract
Nucleotide-binding leucine-rich repeat (NLR) genes play a key role in plant immune responses and have co-evolved with pathogens since the origin of green plants. Comparative genomic studies on the evolution of NLR genes have been carried out in several angiosperm lineages. However, most of these lineages come from the dicot clade. In this study, comparative analysis was performed on NLR genes from five Arecaceae species to trace the dynamic evolutionary pattern of the gene family during species speciation in this monocot lineage. The results showed that NLR genes from the genomes of Elaeis guineensis (262), Phoenix dactylifera (85), Daemonorops jenkinsiana (536), Cocos nucifera (135) and Calamus simplicifolius (399) are highly variable. Frequent domain loss and alien domain integration have occurred to shape the NLR protein structures. Phylogenetic analysis revealed that NLR genes from the five genomes were derived from dozens of ancestral genes. D. jenkinsiana and E. guineensis genomes have experienced “consistent expansion” of the ancestral NLR lineages, whereas a pattern of “first expansion and then contraction” of NLR genes was observed for P. dactylifera, C. nucifera and C. simplicifolius. The results suggest that rapid and dynamic gene content and structure variation have shaped the NLR profiles of Arecaceae species.
1. Introduction
The innate immune system can protect plants from the threats of foreign pathogens [1]. One of the core parts of the plant immune system is a set of genes, termed plant disease resistance genes (R genes), which recognize pathogen-derived virulence proteins (called “effectors”) to activate downstream defense responses [1]. Upon the recognition of the invasion of pathogens, R proteins can activate a hypersensitivity reaction and a series of immune responses, and finally cause the cell death of the infected cells, to restrain the proliferation and spread of pathogens [1]. Nucleotide-binding leucine-rich repeat (NLR) genes are the largest type of all the different R genes, accounting for over 60% of the R genes functionally characterized to date [2]. A typical NLR protein contains a variable domain at the N-terminus, a highly conserved NBS domain in the middle, and a diverse leucine-rich repeat (LRR) domain at the C-terminus [3]. As the N-terminal domains in angiosperms are usually annotated as CC, TIR, or RPW8 domain, angiosperm NLR genes were classified into three subclasses: CC-NBS-LRR (CNL), TIR-NBS-LRR (TNL), and RPW8-NBS-LRR (RNL) [4,5]. Functionally, CNL and TNL proteins act as “sensor NLRs” that recognize specific pathogen effectors to trigger downstream immune responses, while RNL proteins serve as downstream signal transduction molecules (“helper NLR”) of CNL and TNL proteins [6,7].
NLR genes constitute a large gene family in plant genomes, usually comprising hundreds of members [8], and they show very fast evolutionary modes in response to the fast-evolving pathogens [4]. With more and more plant genomes being sequenced, genome-wide evolutionary analyses and comparative genomic studies of NLR genes have been performed in many species and taxa, and different taxa exhibited distinct evolutionary patterns. For example, frequent gene losses and limited gene duplications resulted in a small number of NLR genes in the Cucurbitaceae species [9]. A similar pattern of NLR gene contraction, caused by gene losses or frequent gene deletions, was also reported for Poaceae species [10,11]. In contrast, NLR genes in Fabaceae and Rosaceae species exhibited a “consistent expansion” evolutionary pattern [5,12], while the five Brassicaceae species exhibited a “first expansion and then contraction” of NLR genes [13]. Moreover, species belonging to the same family may also show distinct patterns of NLR gene evolution [14,15]. For example, in four orchid species, Phalaenopsis equestris and Dendrobium catenatum exhibited an “early contraction to recent expansion” evolutionary pattern, while Gastrodia elata and Apostasia shenzhenica showed a “contraction” evolutionary pattern [14].
The distinct evolutionary patterns of these angiosperm lineages provide valuable resources to the understanding of the fast evolutionary modes of R genes due to threats from different pathogens. However, most of these investigated angiosperm lineages are from the dicot clade, while only two monocot lineages have been surveyed [10,11,14]. Because the monocot and dicot clades are different in NLR subclass composition [8], investigating more monocot lineages would provide new insights into the NLR gene dynamics among monocot evolution.
The Arecaceae consists of 183 genera and 2450 species, which are distributed throughout the tropical and subtropical areas in Africa, the Americas, Asia, Madagascar, and the Pacific, and widely grown as ornamentals in botanical gardens (Flora of China, www.iplant.cn/foc/, accessed on 2 August 2021). Recently, the genome of five horticultural plants from the Arecaceae family of the Arecales, including Elaeis guineensis (2n = 32), Phoenix dactylifera (2n = 36), Daemonorops jenkinsiana (2n = 24), Cocos nucifera (2n = 32) and Calamus simplicifolius (2n = 26), were sequenced and made available [16,17,18,19]. Among them, oil palm (E. guineensis) is a source of vegetable oil and has very important economic value [20], date palm (P. dactylifera) is the most popular fruit in the Middle East and North Africa, and C. nucifera is widely distributed on Earth and has considerable food and medicinal value [21]. These horticultural plants are faced with infection by various pathogens during their lifespan. However, the composition and evolutionary pattern of NLR genes in the Arecaceae family have rarely been investigated [22,23]. Deciphering the evolutionary pattern of NLR genes among the five Arecaceae species would provide an additional example of dynamic NLR gene evolution across species speciation in the monocot lineage. Additionally, the obtained NLR information may serve as a primary resource for the disease resistance breeding of the Arecaceae species.
2. Materials and Methods
2.1. Identification and Classification of the NLR Genes
The five whole genomes of the E. guineensis, P. dactylifera, D. jenkinsiana, C. nucifera and C. simplicifolius were used in this study. Genomic sequences and annotation files were obtained from the GigaScience database. NLR genes of the five genomes were retrieved from the ANNA database (https://biobigdata.nju.edu.cn/ANNA/, accessed on 10 August 2021). All the identified NLR genes were subjected to NCBI’s conserved domain database (https://www.ncbi.nlm.nih.gov/ Structure/cdd/wrpsb.cgi, accessed on 30 August 2021) using the default settings to determine whether they encoded CC, RPW8, LRR and other integrated domains (E value: 10−4). The domains that commonly encoded by the NLR genes, such as NBS, LRR, TIR, RPW8, CC, AAA+ and DUF1863 were removed from the integrated domain list.
2.2. Cluster Arrangement of the Identified NLR Genes
Gene clustering was determined according to the criterion used for Medicago truncatula [24]: if two neighboring NLR genes were located within 250 kb on a chromosome, these two genes were regarded as members of the same gene cluster. Based on this criterion, the NLR genes in the five Arecaceae genomes were assigned to clustered loci and singleton loci.
2.3. Sequence Alignment and Phylogenetic Analysis of NLR Genes
The amino acid sequences of the NBS domain were extracted from the identified NLR genes and used for multiple alignments using ClustalW integrated in MEGA 7.0 with default settings [25]. Sequences that were too short (<190 amino acids, less than two-thirds of a regular NBS domain) or too divergent were removed to prevent interference with the alignments and subsequent phylogenetic analysis. The resulting alignments were manually corrected and improved using MEGA 7.0. The phylogenetic tree was constructed using IQ-TREE (version 1.6.12) with the maximum likelihood method, following the selection of best-fit model by ModelFinder [26,27]. Branch support values were assessed using SH-aLRT and UFBoot2 tests with 1000 replications [28].
2.4. Gene Loss/Duplication Analysis of the NLR Genes
In order to identify the gene duplication/loss events during the speciation of the five Arecaceae species, the NLR gene phylogenetic tree was reconciled with the species tree using Notung-2.9 software [29]. The types of NLR gene duplication within a genome were determined using the MCScanX package [30] based on a pair-wise all-against-all blast of protein sequences.
3. Results
3.1. Comparative Analysis of NLR Gene Composition in the Genomes of Five Arecaceae Species
A total of 399, 536, 85, 262, and 135 NLR genes from the genomes of C. simplicifolius, D. jenkinsiana, P. dactylifera, E. guineensis and C. nucifera, respectively, were retrieved from the ANNA database. Among them, D. jenkinsiana possessed the largest number of NLR genes and was as 1.34-, 6.31-, 2.05- and 3.97-times bigger than that of C. simplicifolius, P. dactylifera, E. guineensis and C. nucifera, respectively. All NLR genes were divided into the CNL and RNL subclasses based on the classification provided in ANNA, with no TNL genes found. Among the two NLR subclasses, CNL genes overwhelmingly outnumbered RNL genes, with 99.50%, 99.25%, 100%, 99.62% and 98.52% of NLRs in C. simplicifolius, D. jenkinsiana, P. dactylifera, E. guineensis and C. nucifera, respectively, being CNLs. There were only two, four, one, and two RNL genes in C. simplicifolius, D. jenkinsiana, E. guineensis and C. nucifera, and no RNL genes were identified in P. dactylifera. Domain composition analysis revealed that less than half of NLR genes in C. simplicifolius, D. jenkinsiana and C. nucifera encode intact NLR proteins possessing all three domains (CC/RPW8-NBS-LRR), with the rest of NLR, either lacking the CC/RPW8 domain at the N-terminus, the LRR domain at the C-terminus, or domains at both termini (Table 1). The proportion of intact NLR genes in the E. guineensis genome is much higher, with 144 (143 CNL and 1 RNL) of the 261 genes having all three domains. Some NLR genes were classified as “other” in CNL due to their atypical structural domain compositions (Table 1 and Table S1). For example, the D. jenkinsiana genome encodes one “other” gene in CNL(NCCLNCCL), and the C. nucifera genome encodes two “other” genes, including NCCLNCC and CNCNL.

Table 1.
The number of identified NLR genes in the five Arecaceae genomes.
Integration of alien domains in addition to the three typical domains was detected for NLR genes from the five genomes, including eight, 15, five, three and five distinct integrated domains (IDs) found in 13, 14, six, seven and seven NLR genes of the C. simplicifolius, D. jenkinsiana, P. dactylifera, E. guineensis and C. nucifera genomes, respectively (Table S2). All the NLR-ID genes belong to the CNL subclass. The numbers of NLR genes with fused IDs (NLR-ID gene) in the five genomes show a significant positive correlation with total NLR gene numbers (Figure 1a). An average of 4.15% of NLR genes in each genome possess the NLR-ID structure. The comparison of the ID diversity in the five species shows that a total of 32 non-redundant IDs were present in the five genomes (Table S2). Some of these IDs have been detected in proteins with immune function, including the v-SNARE domain and the PKinase domain. Plant SNARE domain-containing proteins are targets of filamentous fungi effectors and are monitored by NLRs for programmed cell death [31]. PKinases are known to function in the immune pathways in both plants and mammals and are also often found in the receptor-like PKinases that transduce PAMP-triggered immunity [32]. Among the 32 different types of IDs, one of them was found in NLR genes from three species and two were found in NLR genes from two species (Table S2). In contrast, the majority of IDs were found only in one genome, suggesting frequent occurrence of species-specific domain fusions (Figure 1b).
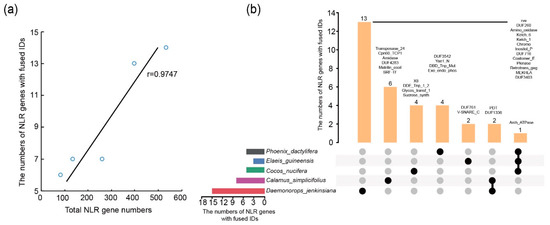
Figure 1.
Exogenous fusion domains of NLR genes in D. jenkinsiana, C. nucifera, C. simplicifolius, E. guineensis and P. dactylifera. (a) Spearman correlation between the total number of species’ NLR genes and the number of NLR genes fused IDs. r represents the correlation coefficient, p < 0.05. (b) Extraneous domains of NLR gene-specific fusions or convergent fusions among different species in the Arecaceae species. Black circles indicate exogenous fusion domain presence, and gray circles indicate exogenous fusion domains non- presence.
3.2. Organization of NLR Genes in Arecaceae Genomes
Clustering organization of NLR genes has been proposed as an important mechanism of generating NLR diversity and functional members. Our results show that the majority of NLR genes were organized into clusters rather than singletons in C. nucifera, D. jenkinsiana, and E. guineensis genomes, with 54.8%, 70.7% and 77.7% NLR genes detected in clusters, respectively (Table 2). However, there were more singleton genes in the other two Arecaceae genomes, with only 15 (17.6%) and 176 (44.1%) NLR genes organized into clusters in the P. dactylifera and C. simplicifolius genomes, respectively. Among the five Arecaceae genomes, the clustered loci in E. guineensis genome contained the most genes (3.37 genes/locus) on average. The largest gene clusters of C. nucifera, D. jenkinsiana, E. guineensis, C. simplicifolius, and P. dactylifera were in locus 47 (eight genes), locus 34 (10 genes) and 126 (10 genes), locus 80 (11 genes), locus 237 (five genes), and locus 47 (three genes), respectively (Table 2).

Table 2.
The organization of NLR genes in five Arecaceae species.
NLR genes may undergo duplication via different mechanisms [33]. We surveyed the duplication patterns of NLR genes from the five Arecaceae species by using MCScanX software. The results showed that amplification of NLR genes in the five genomes was dominated by different types. The majority of NLR genes in C. nucifera (57.0%) and E. guineensis (70.1%) were generated by tandem/proximal duplications, whereas most NLR genes in the remaining three genomes were characterized as dispersed duplication. Only a small proportion of NLR genes in C. nucifera, D. jenkinsiana and E. guineensis were generated by whole genome duplications (WGD)/segmental duplication, whereas no WGD/segmental duplicated NLR genes were found in C. simplicifolius or P. dactylifera (Figure 2). However, the proportion of segmental duplicated genes might have been underestimated because the syntenic relationship of NLR genes would be disrupted during long-term evolution.
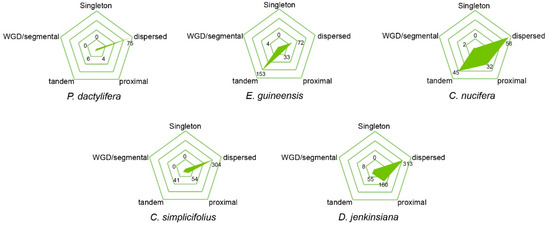
Figure 2.
Type of gene duplication in C. simplicifolius, D. jenkinsiana, P. dactylifera, E. guineensis and C. nucifera, respectively.
3.3. Phylogenetic Analysis of the NLR Genes
To trace the evolutionary history of the NLR genes in Arecaceae, a phylogenetic tree was constructed based on the amino acid sequences of the NBS domain, using three Amborella TNL proteins as the outgroups (Figure 3 and Data S1). The results show that NLR genes from the five species form two monophyletic clades with high support values (>90%) (Figure S1). The two deeply diverged clades correspond to the RNL and CNL subclasses, respectively (Figure 3a), supporting the ancient separation of the two NLR clades (Figure S1; Data S1). Compared to the CNL clade, the branch lengths of the RNL clade were relatively short (Figure 3a), suggesting that RNL genes had a slow evolutionary rate. Within the CNL clade, clustering of NLR genes from a single species was frequently observed (Figure 3a), which is caused by species-specific gene duplications.
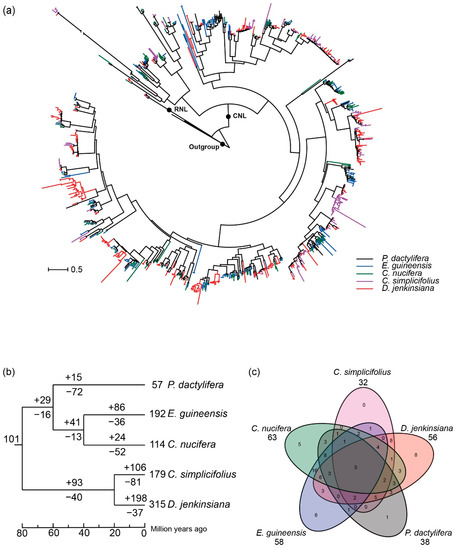
Figure 3.
Phylogenetic tree of CNL and RNL genes based on conserved NBS domain sequences. (a) A total of 1408 CNL and 9 RNL sequences were used: 536 sequences from D. jenkinsiana (shown in red), 135 from C. nucifera (green), 399 from C. simplicifolius (purple), 262 from E. guineensis (blue), and 85 from P. dactylifera (black). Four TNL sequences from Arabidopsis thaliana were used as an outgroup. (b) Number variations of NLR genes at different stages of Arecaceae species evolution. Differential gene losses and gains are indicated by numbers with − or + on each branch. (c) Arecaceae ancestral lineage genes were inherited by C. simplicifolius, D. jenkinsiana, P. dactylifera, E. guineensis and C. nucifera, respectively.
To gain insight into the evolution of the NLR genes during the speciation of the five Arecaceae species, Notung software was used to reconcile gene duplication/loss events of the NLR genes at each node of the phylogenetic tree [34]. The result reveals that at least 101 ancestral NLR lineages (2 RNL and 99 CNL) may have existed in the common ancestor of the five Arecaceae species (Figure S1 and Data S1). We term these ancestral NLR lineages as “Arecaceae NLR lineage”. The common ancestor of P. dactylifera, E. guineensis and C. nucifera (Pd-Eg-Cn) inherited 85 Arecaceae NLR lineages but lost 16. These Arecaceae NLR lineages further expanded to 114 NLR lineages before the divergence of the P. dactylifera, E. guineensis and C. nucifera (Pd-Eg-Cn). The common ancestor of C. simplicifolius and D. jenkinsiana (Cs-Dj) inherited 61 Arecaceae NLR lineages and duplicated to 154 Cs-Dj NLR lineages (Figure 3b). Subsequent gene loss and gain during the divergence of Pd-Eg-Cn and Cs-Dj lineages further shaped the NLR content in the five Arecaceae species, resulting in the current distinct NLR profile in these species. By assigning the NLR genes in each species to the 114 ancestral Arecaceae NLR lineages, the results showed that 32, 56, 38, 58 and 63 Arecaceae NLR lineages were inherited by C. simplicifolius, D. jenkinsiana, P. dactylifera, E. guineensis and C. nucifera, respectively (Figure 3c). Among these ancestral NLR lineages, only five of them were reserved in all five genomes, and 20 lineages were species-specific (D. jenkinsiana, eight; P. dactylifera, one; C. simplicifolius, zero; E. guineensis, six; C. nucifera, five). The remaining 89 lineages were differentially preserved in two to four species (Figure 3c).
3.4. Tracing the Trajectory of NLR Gene Evolution in Different Arecaceae Species
Based on the traced loss and gain of NLR genes at each species divergence node, the evolutionary trajectory of NLR genes for each of the five species can be traced. After separation from the common ancestor of the five Arecaceae species (Cs-Dj-Pd-Eg-Cn node), 40 gene-loss and 93 gain events were detected at the Cs-Dj node (Figure 3b), suggesting a pattern of NLR expansion during this period. The Cs-Dj node then diverged to generate C. simplicifolius and D. jenkinsiana. In C. simplicifolius, 81 NLR genes inherited from the Cs-Dj were lost, while the remaining NLR genes underwent intensive gene duplication, leading to the generation of 106 new members (Figure 3b). Again, a pattern of NLR expansion occurred during the period from Cs-Dj to C. simplicifolius. Taken together, a “consistent expansion” of NLR genes occurred during the evolution of C. simplicifolius from the common ancestor of the five Arecaceae species (Figure 4d). Using this strategy, a similar pattern of “consistent expansion” was also observed for NLR genes in D. jenkinsiana and E. guineensis (Figure 4b,e). However, a different pattern of “first expansion and then contraction” was observed for the other species, P. dactylifera and C. nucifera (Figure 4a,c).
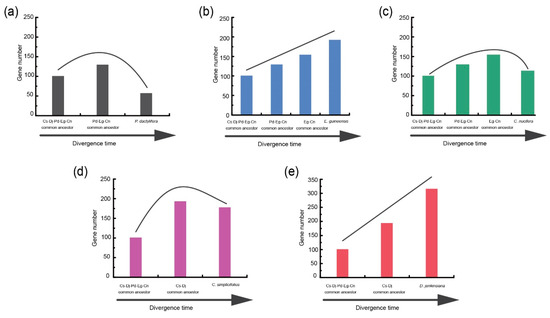
Figure 4.
Dynamic evolutionary patterns of NLR genes among different evolutionary states of five Arecaceae species. (a) P. dactylifera. (b) E. guineensis. (c) C. nucifera. (d) C. simplicifolius. (e) D. jenkinsiana.
4. Discussion
The NLR genes constitute one of the largest gene families in angiosperms, with an average of about 300 genes per genome [8]. Genome-wide identification and comparative analysis of NLR genes has greatly accelerated the mining of functional NLR genes from various crops and ecological or economic important plants in recent years. The lack of NLR information has greatly hampered identification of functional disease resistance genes by using a genome-guided method for the Arecaceae species. Taking advantage of the recently released genomes from the five Arecaceae species, the NLR profile across the five species was compared in several aspects of this study, which may serve as a primary resource for molecular breeding of the Arecaceae species.
Different proportions of NLR genes in the Arecaceae species form clusters of varied size. The members within each of these NLR gene clusters provides large candidates for positive selection to act on. Additionally, high polymorphism can be maintained between NLR genes within the gene cluster through recombination, facilitating the generation of new NLR genes [35,36]. Comparative analysis of NLR genes in the five Arecaceae species showed that two of the Arecaceae genomes have more than 300 NLR genes, whereas the other three have fewer than 300 NLRs, suggesting that species-specific NLR gain and loss have occurred. NLR content dynamics is an important mechanism for plants coping with the varied environments and ecological adaption [8,37]. The species-specific NLR contraction or expansion for the five Arecaceae species suggest they may have faced different selection pressure from environmental microbes after having separated from the common ancestor, although the exact trajectory of environmental microbe diversity dynamic could hardly be traced. Reconstruction of the ancestral states of NLR genes at several divergence nodes of Arecaceae revealed 101 ancestral NLR lineages in the common ancestor, including two RNL lineages and 99 CNL lineages. The number of recovered ancestral NLR lineage in the Arecaceae family is much larger than that in the Orchidaceae family (29), but lower than that of the Poaceae (456) in the monocot clade [4,14]. The high difference of ancestral NLR lineage numbers in the three monocot families suggests that dramatic NLR contraction and amplification consistently occurred in the monocot lineage. Compared with several investigated dicot families, the ancestral NLR lineage in the Arecaceae family is fewer than the 119 ancestral NLR lineages in Fabaceae, and 166 ancestral NLR lineages in Solanaceae, and far fewer than the 228 ancestral NLR lineages found in Brassicaceae [13]. The large ancestral NLR lineage numbers in these dicot families may have benefitted from having an additional NLR subclass, TNL, in their genomes.
The current NLR profile in a genome is contributed by differential inheritance and amplification of ancestral NLR lineages [4]. Previous studies in Cucurbitaceae and Poaceae revealed that species in the two families experienced a similar pattern of NLR “contraction” [9,10,11], whereas Fabaceae and Rosaceae species exhibited “consistent expansion” of NLR genes after the families’ radiation [5,12]. A distinct “first expansion and then contraction” pattern of NLR genes was observed in the five Brassicaceae [13]. However, species in the Arecaceae family exhibited two different modes of NLR gene evolution after diverging from the common ancestor. D. jenkinsiana and E. guineensis have experienced “consistent expansion” of the ancestral NLR lineages, whereas NLR genes in P. dactylifera, C. nucifera. and C. simplicifolius show “first expansion and then contraction”. Such a pattern of species belonging to the same family having distinct NLR gene evolution patterns has also been observed in another monocot lineage, Orchidaceae, and two dicot families [15,16,17]. The results provide additional evidence to support that rapid NLR gene content variation could occur to facilitate plant adaption to changed environments.
The high diversity of NLR could be detected not only for gene content, but also for gene structure. For example, the three NLR subclasses have distinct N-terminal domains to support distinct functional mechanisms, either by making holes in the cell membrane, or by action as an enzyme [38]. Loss of characteristic domains is detected for many NLR genes in the Arecaceae species. This pattern has also been observed in several angiosperms by previous studies. For example, only a small proportion of NLR genes with intact structures were reported in C. annuum (23.2%), S. lycopersicum (42.7%), S. tuberosum (28.2%), P. trichocarpa (46.2%), M. truncatula (39.1%), Lotus japonicus (31.0%), and Oryza sativa (30.6%) genomes [5,39,40,41]. The remaining large proportion of NLR genes further expanded the diversity of NLR genes through the loss of the N-terminal, C-terminal, or both domains (Figure S2). Notably, several studies have reported that NLR genes with atypical structures also function in plant immunity [42,43,44,45], suggesting the loss of the N-terminal or C-terminal domains may also be a mechanism to generated NLR functional diversity. It is worthy of note that a deeply diverged CNL lineage showed the feature of widespread loss of the N-terminal CC domain (Figure S2). The long-term maintenance and expansion of this CNL lineage suggest that the loss of the CC domain did not completely abolish the function of genes on this lineage. Considering the CC domain had been shown to be indispensable for multimerization and forming pores on the plant cell membrane of CNL proteins, the CC-lacking structure of this CNL lineage may suggest a different functional mechanism.
Different from the domain loss found in many NLR genes, we also detected fusion of alien domains for a small proportion of Arecaceae NLR genes to form the NLR-ID structure. This provides another way to expand the structure and functional diversity. In recent years, studies have increasingly found that alien domains can be fused to plant NLR proteins to act as target proteins for pathogens’ effector factors. The research on RGA4/RGA5 and Pik-1/Pik-2 of the NLR with rice blast resistance provides the first experimental evidence for this model. Both RGA5 and Pik-1 genes are fused with an HMA domain, which serves as a decoy to interact directly with pathogenic effectors to stimulate the disease resistance of RGA4 and Pik-2 [46,47]. Arabidopsis NLR genes RPS4/RRS1 provides another example to support the important functions of alien domains. The RRS1 protein fused with WRKY domain can directly interact with the pathogenic effectors. AvrRPS4 interacts to stimulate the disease resistance activity of RPS4 [48]. In this study, different NLR alien domains were found in five species. These domains may be the “baits” proteins of pathogenic effectors in plant cells. Among them, v-SNARE and the PKinase domains have been detected in many proteins that play a role in plant disease resistance [32,33], but SRF-TF, DUF761and DUF4283, etc. have no direct evidence of being related to plant immunity. The discovery of these alien domains is helpful to explore more potential plant immune-related proteins.
5. Conclusions
This study compared the NLR gene profiles in five Arecaceae species and revealed a high NLR content and structure variation, which serve as a resource for the evolution of functional NLR genes. Phylogenetic analysis revealed that NLR genes from the five genomes were derived from 101 ancestral genes, and distinct “consistent expansion” and “first expansion and then contraction” patterns were observed for NLR genes from the five species. The results show that dynamic gene content and structure variation have shaped the NLR profiles of different Arecaceae species. The obtained NLR profiles serve as a valuable resource for molecular breeding of these species and for further exploration of the NLR evolutionary pattern.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/horticulturae7120539/s1, Figure S1. 101 NLR gene families divided based on NLR phylogeny from C. simplicifolius, D. jenkinsiana, P. dactylifera, E. guineensis and C. nucifera. Figure S2. Phylogenetic distribution of NLRgenes with different domain compositions in five Arecaceae species. Table S1. The gene list and classification of NLR genes identified from C. simplicifolius, D. jenkinsiana, P. dactylifera, E. guineensis and C. nucifera. Table S2. Exogenous fusion domains of NLR identified from five Arecaceae species. Data S1. An NEXUS format phylogenetic tree of NLRs in five Arecaceae species with branch support values.
Author Contributions
Z.-Q.S. and Y.L. conceived and designed the study. X.-T.L., G.-C.Z., X.-Y.F., Z.Z. and Y.L. obtained and analyzed the data. X.-T.L. and G.-C.Z. wrote the manuscript. Z.-Q.S. and Y.L. revised the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Conflicts of Interest
The authors have no conflict of interest to declare.
References
- Wang, W.; Feng, B.; Zhou, J.M.; Tang, D. Plant immune signaling: Advancing on two frontiers. J. Integr. Plant Biol. 2020, 62, 2–24. [Google Scholar] [CrossRef] [Green Version]
- Kourelis, J.; van der Hoorn, R.A.L. Defended to the Nines: 25 Years of Resistance Gene Cloning Identifies Nine Mechanisms for R Protein Function. Plant Cell 2018, 30, 285–299. [Google Scholar] [CrossRef] [Green Version]
- Meyers, B.C.; Kozik, A.; Griego, A.; Kuang, H.; Michelmore, R.W. Genome-wide analysis of NBS-LRR-encoding genes in Arabidopsis. Plant Cell 2003, 15, 809–834. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shao, Z.Q.; Xue, J.Y.; Wu, P.; Zhang, Y.M.; Wu, Y.; Hang, Y.Y.; Wang, B.; Chen, J.Q. Large-Scale Analyses of Angiosperm Nucleotide-Binding Site-Leucine-Rich Repeat Genes Reveal Three Anciently Diverged Classes with Distinct Evolutionary Patterns. Plant Physiol. 2016, 170, 2095–2109. [Google Scholar] [CrossRef] [Green Version]
- Shao, Z.Q.; Zhang, Y.M.; Hang, Y.Y.; Xue, J.Y.; Zhou, G.C.; Wu, P.; Wu, X.Y.; Wu, X.Z.; Wang, Q.; Wang, B.; et al. Long-term evolution of nucleotide-binding site-leucine-rich repeat genes: Understanding gained from and beyond the legume family. Plant Physiol. 2014, 166, 217–234. [Google Scholar] [CrossRef] [Green Version]
- Saile, S.C.; Jacob, P.; Castel, B.; Jubic, L.M.; Salas-Gonzales, I.; Backer, M.; Jones, J.D.G.; Dangl, J.L.; El Kasmi, F. Two unequally redundant “helper” immune receptor families mediate Arabidopsis thaliana intracellular “sensor” immune receptor functions. PLoS Biol. 2020, 18, e3000783. [Google Scholar] [CrossRef] [PubMed]
- Castel, B.; Ngou, P.M.; Cevik, V.; Redkar, A.; Kim, D.S.; Yang, Y.; Ding, P.; Jones, J.D.G. Diverse NLR immune receptors activate defence via the RPW8-NLR NRG1. New Phytol. 2019, 222, 966–980. [Google Scholar] [CrossRef]
- Liu, Y.; Zeng, Z.; Zhang, Y.M.; Li, Q.; Jiang, X.M.; Jiang, Z.; Tang, J.H.; Chen, D.; Wang, Q.; Chen, J.Q.; et al. An angiosperm NLR Atlas reveals that NLR gene reduction is associated with ecological specialization and signal transduction component deletion. Mol. Plant 2021, 14, 17. [Google Scholar] [CrossRef]
- Wan, H.; Yuan, W.; Bo, K.; Shen, J.; Pang, X.; Chen, J. Genome-wide analysis of NBS-encoding disease resistance genes in Cucumis sativus and phylogenetic study of NBS-encoding genes in Cucurbitaceae crops. BMC Genom. 2013, 14, 109. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luo, S.; Zhang, Y.; Hu, Q.; Chen, J.; Li, K.; Lu, C.; Liu, H.; Wang, W.; Kuang, H. Dynamic nucleotide-binding site and leucine-rich repeat-encoding genes in the grass family. Plant Physiol. 2012, 159, 197–210. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, J.; Ding, J.; Zhang, W.; Zhang, Y.; Tang, P.; Chen, J.Q.; Tian, D.; Yang, S. Unique evolutionary pattern of numbers of gramineous NBS-LRR genes. Mol. Genet. Genom. MGG 2010, 283, 427–438. [Google Scholar] [CrossRef]
- Jia, Y.; Yuan, Y.; Zhang, Y.; Yang, S.; Zhang, X. Extreme expansion of NBS-encoding genes in Rosaceae. BMC Genet. 2015, 16, 48. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.M.; Shao, Z.Q.; Wang, Q.; Hang, Y.Y.; Xue, J.Y.; Wang, B.; Chen, J.Q. Uncovering the dynamic evolution of nucleotide-binding site-leucine-rich repeat (NBS-LRR) genes in Brassicaceae. J. Integr. Plant Biol. 2016, 58, 165–177. [Google Scholar] [CrossRef] [PubMed]
- Xue, J.Y.; Zhao, T.; Liu, Y.; Liu, Y.; Zhang, Y.X.; Zhang, G.Q.; Chen, H.; Zhou, G.C.; Zhang, S.Z.; Shao, Z.Q. Genome- Wide Analysis of the Nucleotide Binding Site Leucine-Rich Repeat Genes of Four Orchids Revealed Extremely Low Numbers of Disease Resistance Genes. Front. Genet 2019, 10, 1286. [Google Scholar] [CrossRef] [Green Version]
- Zhou, G.C.; Li, W.; Zhang, Y.M.; Liu, Y.; Zhang, M.; Meng, G.Q.; Li, M.; Wang, Y.L. Distinct Evolutionary Patterns of NBS-Encoding Genes in Three Soapberry Family (Sapindaceae) Species. Front. Genet 2020, 11, 737. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.; Ong-Abdullah, M.; Low, E.T.; Manaf, M.A.; Rosli, R.; Nookiah, R.; Ooi, L.C.; Ooi, S.E.; Chan, K.L.; Halim, M.A.; et al. Oil palm genome sequence reveals divergence of interfertile species in Old and New worlds. Nature 2013, 500, 335–339. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hazzouri, K.M.; Gros-Balthazard, M.; Flowers, J.M.; Copetti, D.; Lemansour, A.; Lebrun, M.; Masmoudi, K.; Ferrand, S.; Dhar, M.I.; Fresquez, Z.A.; et al. Genome-wide association mapping of date palm fruit traits. Nat. Commun. 2019, 10, 4680. [Google Scholar] [CrossRef]
- Lantican, D.V.; Strickler, S.R.; Canama, A.O.; Gardoce, R.R.; Mueller, L.A.; Galvez, H.F. De Novo Genome Sequence Assembly of Dwarf Coconut (Cocos nucifera L. ‘Catigan Green Dwarf’) Provides Insights into Genomic Variation Between Coconut Types and Related Palm Species. G3-Genes Genom. Genet. 2019, 9, 2377–2393. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, H.S.; Wang, S.B.; Wang, J.L.; Chen, C.H.; Hao, S.J.; Chen, L.F.; Fei, B.H.; Han, K.; Li, R.S.; Shi, C.C.; et al. The chromosome-level genome assemblies of two rattans (Calamus simplicifolius and Daemonorops jenkinsiana). Gigascience 2018, 7, giy097. [Google Scholar] [CrossRef]
- Duarte Ferreira Ribeiro, C.; Barbosa Schappo, F.; da Silva Sales, I.; Santos Assuncao, L.; Murowaniecki Otero, D.; Teixeira Magalhaes-Guedes, K.; Aparecida Souza Machado, B.; Mara Block, J.; Izabel Druzian, J.; Larroza Nunes, I. Novel bioactive nanoparticles from crude palm oil and its fractions as foodstuff ingredients. Food Chem. 2021, 373, 131252. [Google Scholar] [CrossRef] [PubMed]
- Lima, E.B.; Sousa, C.N.; Meneses, L.N.; Ximenes, N.C.; Santos Junior, M.A.; Vasconcelos, G.S.; Lima, N.B.; Patrocinio, M.C.; Macedo, D.; Vasconcelos, S.M. Cocos nucifera (L.) (Arecaceae): A phytochemical and pharmacological review. Braz. J. Med. Biol. Res. 2015, 48, 953–964. [Google Scholar] [CrossRef] [Green Version]
- Durand-Gasselin, T.; Asmady, H.; Flori, A.; Jacquemard, J.C.; Hayun, Z.; Breton, F.; de Franqueville, H. Possible sources of genetic resistance in oil palm (Elaeis guineensis Jacq.) to basal stem rot caused by Ganoderma boninense--prospects for future breeding. Mycopathologia 2005, 159, 93–100. [Google Scholar] [CrossRef]
- Hanold, D.; Randles, J.W. Coconut Cadang-Cadang Disease and Its Viroid Agent. Plant Dis. 1991, 75, 330–335. [Google Scholar] [CrossRef] [Green Version]
- Ameline-Torregrosa, C.; Wang, B.-B.; O’Bleness, M.S.; Deshpande, S.; Zhu, H.; Roe, B.; Young, N.D.; Cannon, S.B. Identification and characterization of nucleotide-binding site-leucine-rich repeat genes in the model plant Medicago truncatula. Plant Physiol. 2008, 146, 5–21. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, L.T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef] [Green Version]
- Minh, B.Q.; Nguyen, M.A.; von Haeseler, A. Ultrafast approximation for phylogenetic bootstrap. Mol. Biol. Evol. 2013, 30, 1188–1195. [Google Scholar] [CrossRef]
- Chen, K.; Durand, D.; Farach-Colton, M. NOTUNG: A program for dating gene duplications and optimizing gene family trees. J. Comput. Biol. A J. Comput. Mol. Cell Biol. 2000, 7, 429–447. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Tang, H.; Debarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heller, J.; Clave, C.; Gladieux, P.; Saupe, S.J.; Glass, N.L. NLR surveillance of essential SEC-9 SNARE proteins induces programmed cell death upon allorecognition in filamentous fungi. Proc. Natl. Acad. Sci. USA 2018, 115, E2292–E2301. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dardick, C.; Schwessinger, B.; Ronald, P. Non-arginine-aspartate (non-RD) kinases are associated with innate immune receptors that recognize conserved microbial signatures. Curr. Opin. Plant. Biol. 2012, 15, 358–366. [Google Scholar] [CrossRef]
- Leister, D. Tandem and segmental gene duplication and recombination in the evolution of plant disease resistance gene. Trends Genet 2004, 20, 116–122. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stolzer, M.; Lai, H.; Xu, M.; Sathaye, D.; Vernot, B.; Durand, D. Inferring duplications, losses, transfers and incomplete lineage sorting with nonbinary species trees. Bioinformatics 2012, 28, i409–i415. [Google Scholar] [CrossRef] [Green Version]
- Bergelson, J.; Kreitman, M.; Stahl, E.A.; Tian, D. Evolutionary dynamics of plant R-genes. Science 2001, 292, 2281–2285. [Google Scholar] [CrossRef] [Green Version]
- Botella, M.A.; Parker, J.E.; Frost, L.N.; Bittner-Eddy, P.D.; Beynon, J.L.; Daniels, M.J.; Holub, E.B.; Jones, J.D. Three genes of the Arabidopsis RPP1 complex resistance locus recognize distinct Peronospora parasitica avirulence determinants. Plant Cell 1998, 10, 1847–1860. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baggs, E.L.; Monroe, J.G.; Thanki, A.S.; O’Grady, R.; Schudoma, C.; Haerty, W.; Krasileva, K.V. Convergent Loss of an EDS1/PAD4 Signaling Pathway in Several Plant Lineages Reveals Coevolved Components of Plant Immunity and Drought Response. Plant Cell 2020, 32, 2158–2177. [Google Scholar] [CrossRef]
- Wang, J.; Hu, M.; Wang, J.; Qi, J.; Han, Z.; Wang, G.; Qi, Y.; Wang, H.W.; Zhou, J.M.; Chai, J. Reconstitution and structure of a plant NLR resistosome conferring immunity. Science 2019, 364, eaav5870. [Google Scholar] [CrossRef]
- Qian, L.H.; Zhou, G.C.; Sun, X.Q.; Lei, Z.; Zhang, Y.M.; Xue, J.Y.; Han, Y.Y. Distinct Patterns of Gene Gain and Loss: Diverse Evolutionary Modes of NBS-Encoding Genes in Three Solanaceae Crop Species. G3-Genes Genomes Genetics 2017, 7, 1577–1585. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, X.; Feng, Y.; Cheng, H.; Tian, D.; Yang, S.; Chen, J.Q. Relative evolutionary rates of NBS-encoding genes revealed by soybean segmental duplication. Mol. Genet. Genom. 2011, 285, 79–90. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Gu, T.; Pan, C.; Feng, Z.; Ding, J.; Hang, Y.; Chen, J.Q.; Tian, D. Genetic variation of NBS-LRR class resistance genes in rice lines. Theor Appl Genet. 2008, 116, 165–177. [Google Scholar] [CrossRef]
- Kato, H.; Saito, T.; Ito, H.; Komeda, Y.; Kato, A. Overexpression of the TIR-X gene results in a dwarf phenotype and activation of defense-related gene expression in Arabidopsis thaliana. J. Plant Physiol. 2014, 171, 382–388. [Google Scholar] [CrossRef] [PubMed]
- Nandety, R.S.; Caplan, J.L.; Cavanaugh, K.A.; Perroud, B.; Wroblewski, T.; Michelmore, R.W.; Meyers, B.C. The role of TIR-NBS and TIR-X proteins in plant basal defense responses. Plant Physiol. 2013, 162, 1459–1472. [Google Scholar] [CrossRef] [Green Version]
- Cai, H.; Wang, W.; Rui, L.; Han, L.; Luo, M.; Liu, N.; Tang, D. The TIR-NBS protein TN13 associates with the CC-NBS-LRR resistance protein RPS5 and contributes to RPS5-triggered immunity in Arabidopsis. Plant J. Cell Mol. Biol. 2021, 107, 775–786. [Google Scholar] [CrossRef]
- Seong, K.; Seo, E.; Witek, K.; Li, M.; Staskawicz, B. Evolution of NLR resistance genes with noncanonical N-terminal domains in wild tomato species. New Phytol. 2020, 227, 1530–1543. [Google Scholar] [CrossRef]
- Kanzaki, H.; Yoshida, K.; Saitoh, H.; Fujisaki, K.; Hirabuchi, A.; Alaux, L.; Fournier, E.; Tharreau, D.; Terauchi, R. Arms race co-evolution of Magnaporthe oryzae AVR-Pik and rice Pik genes driven by their physical interactions. Plant J. 2012, 72, 894–907. [Google Scholar] [CrossRef] [PubMed]
- Cesari, S.; Thilliez, G.; Ribot, C.; Chalvon, V.; Michel, C.; Jauneau, A.; Rivas, S.; Alaux, L.; Kanzaki, H.; Okuyama, Y.; et al. The rice resistance protein pair RGA4/RGA5 recognizes the Magnaporthe oryzae effectors AVR-Pia and AVR1-CO39 by direct binding. Plant Cell 2013, 25, 1463–1481. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sarris, P.F.; Duxbury, Z.; Huh, S.U.; Ma, Y.; Segonzac, C.; Sklenar, J.; Derbyshire, P.; Cevik, V.; Rallapalli, G.; Saucet, S.B.; et al. A Plant Immune Receptor Detects Pathogen Effectors that Target WRKY Transcription Factors. Cell 2015, 161, 1089–1100. [Google Scholar] [CrossRef] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).