Identification of Long Non-Coding RNAs Associated with Tomato Fruit Expansion and Ripening by Strand-Specific Paired-End RNA Sequencing
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Growth Conditions
2.2. RNA Isolation and Quality Control
2.3. Library Preparation and Sequencing
2.4. Data Analysis
2.5. LncRNAs Calling
2.6. Differential Expression Analysis
3. Results
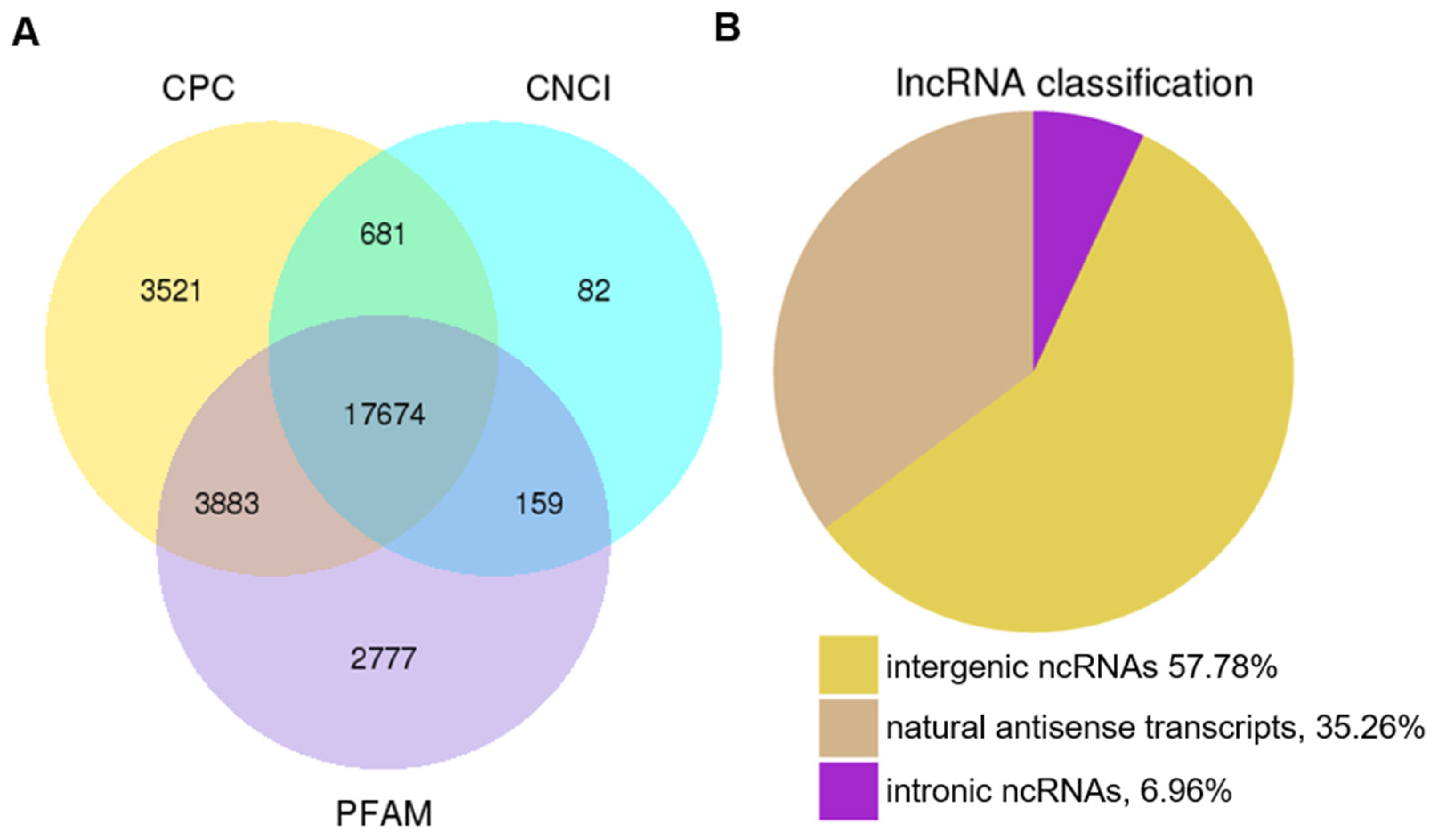
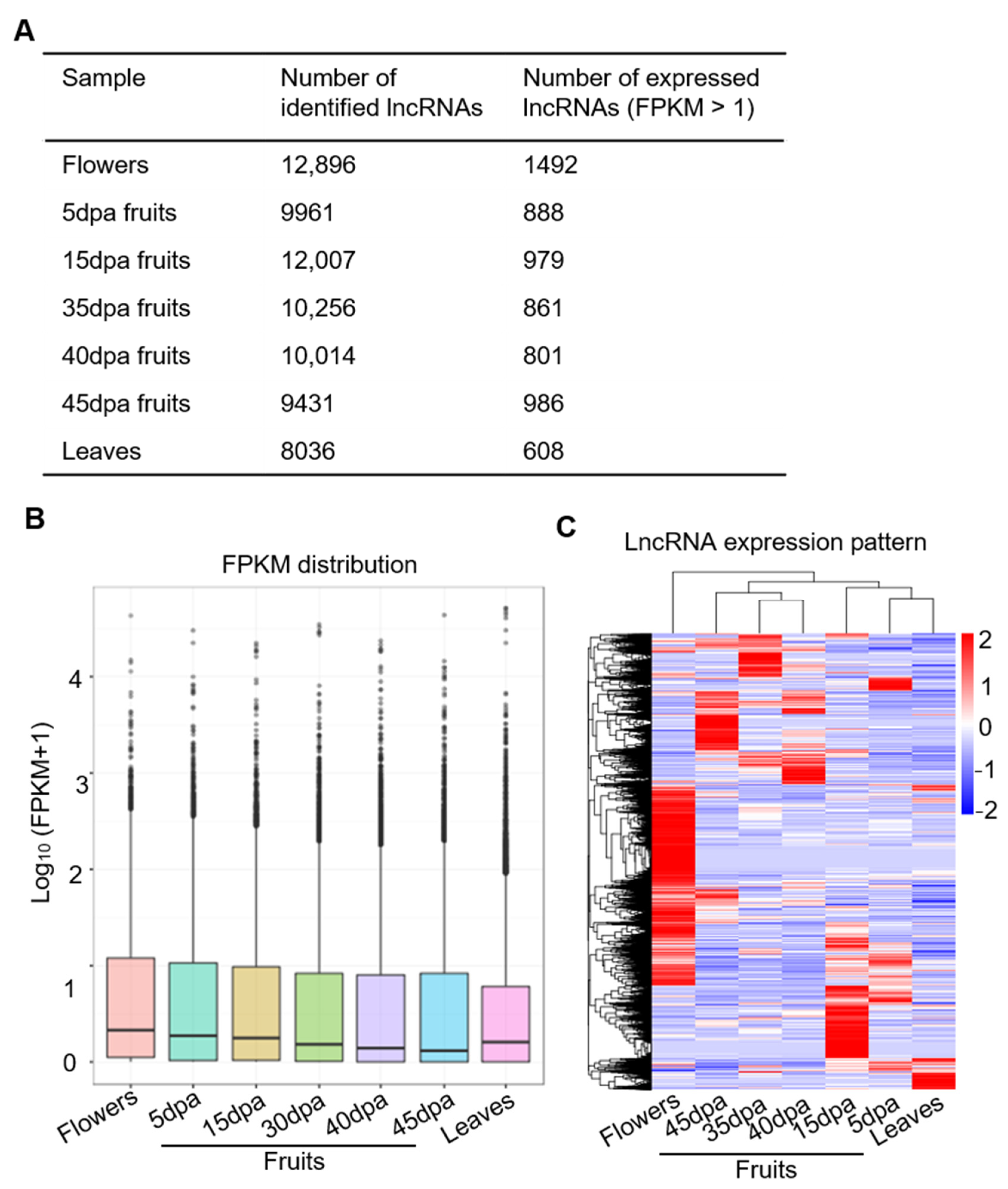
3.1. Genome-Wide Identification of lncRNAs in Tomato Fruit at Different Developmental Stages
3.2. Genome-Wide lncRNAs Expression Pattern and Their Distribution on Tomato Chromosomes
3.3. Characterization of lncRNA Identified in the Tomato Genome
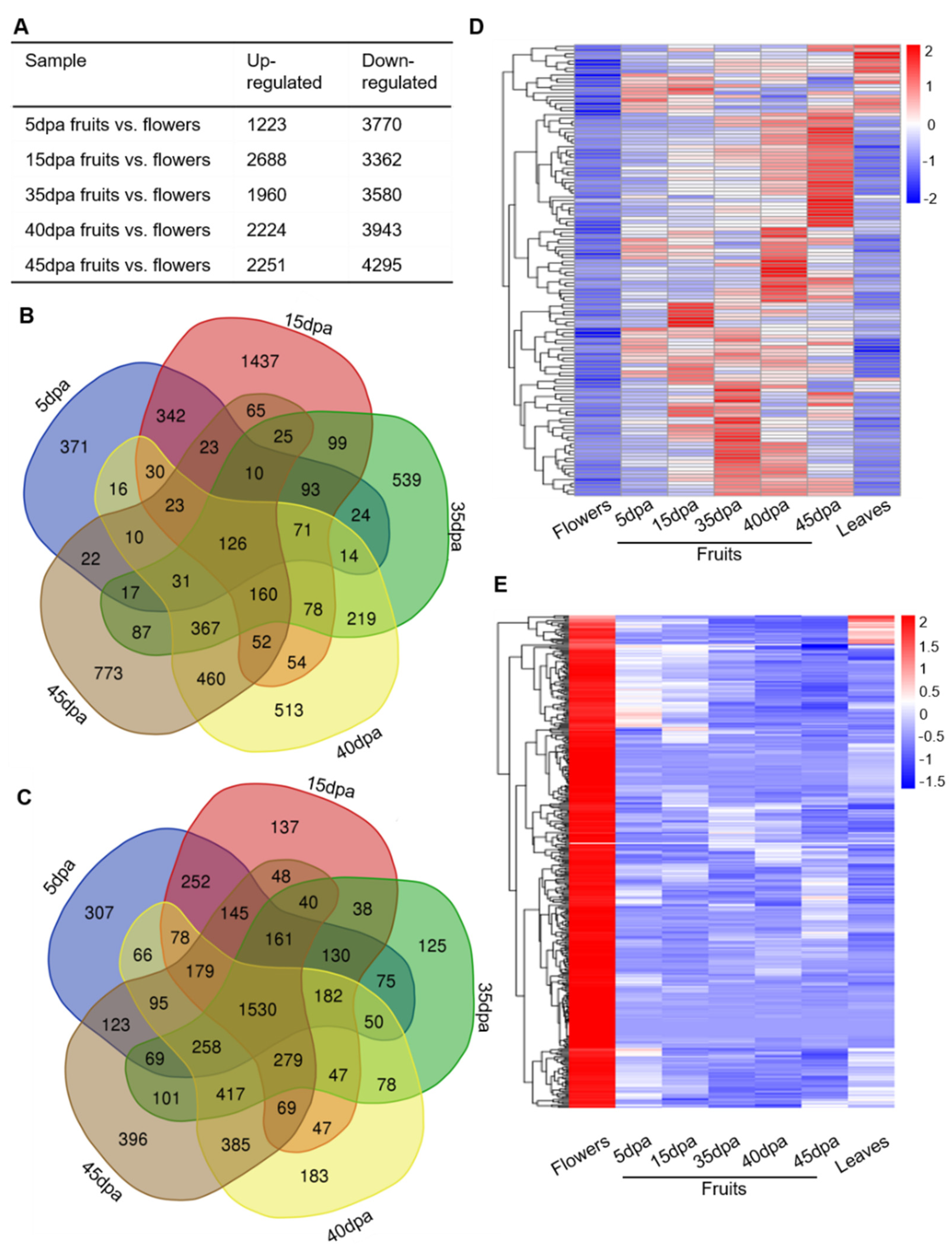
3.4. Analysis of Tomato Tissue- and Stage-Dependent lncRNAs during Fruit Expansion and Ripening
3.5. Specific lncRNA Candidates in Regulating Tomato Fruit Expansion and Ripening
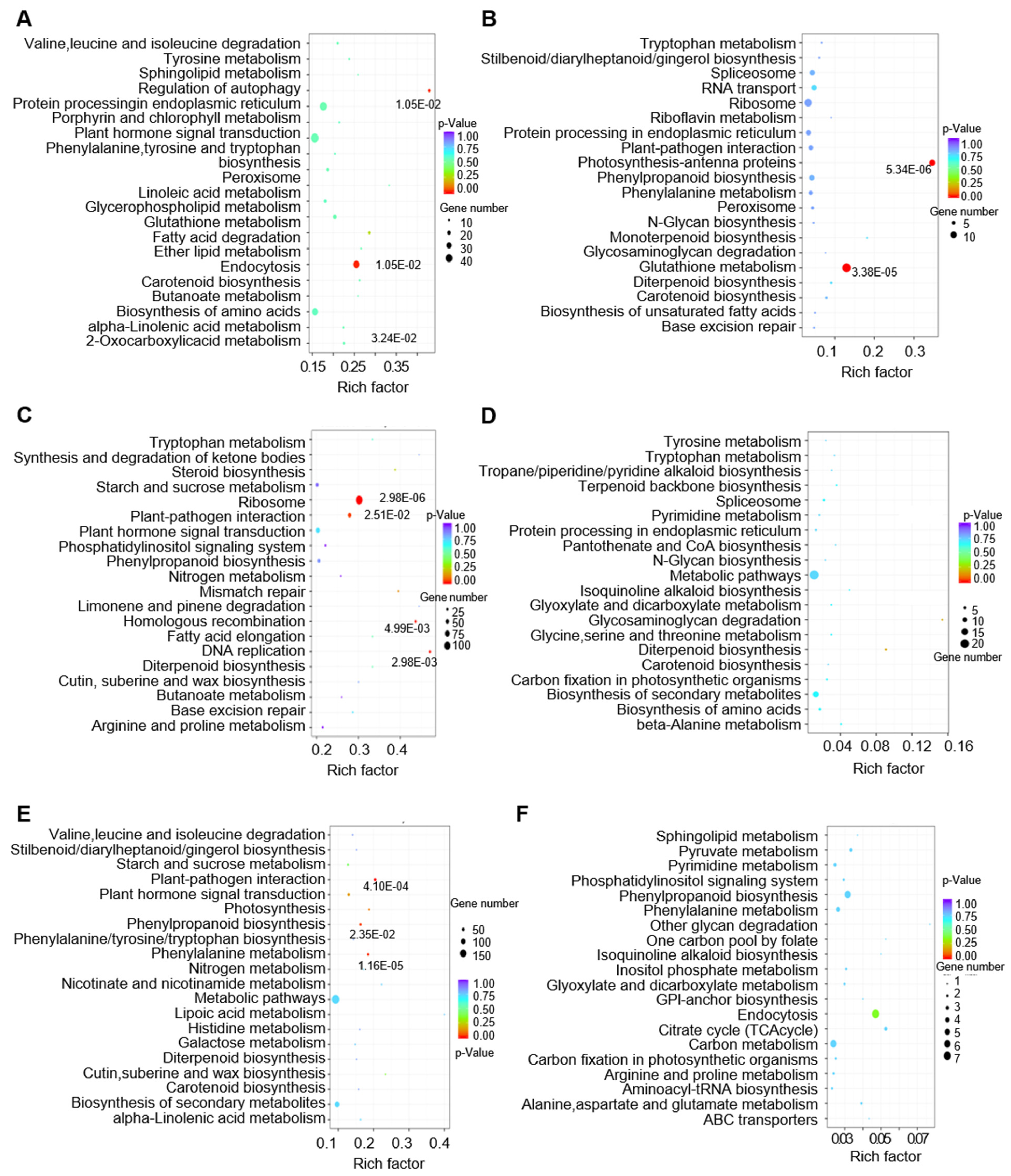
3.6. Biological Function Analysis of lncRNAs in Regulating Tomato Fruit Expansion and Ripening
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Cech, T.R.; Steitz, J.A. The Noncoding RNA Revolution—Trashing Old Rules to Forge New Ones. Cell 2014, 157, 77–94. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rymarquis, L.A.; Kastenmayer, J.P.; Hüttenhofer, A.G.; Green, P.J. Diamonds in the rough: mRNA-like non-coding RNAs. Trends Plant Sci. 2008, 13, 329–334. [Google Scholar] [CrossRef] [PubMed]
- Studniarek, C.; Egloff, S.; Murphy, S. Noncoding RNAs Set the Stage for RNA Polymerase II Transcription. Trends Genet. 2021, 37, 279–291. [Google Scholar] [CrossRef]
- Hombach, S.; Kretz, M. Non-coding RNAs: Classification, Biology and Functioning. Adv. Exp. Med. Biol. 2016, 937, 3–17. [Google Scholar] [CrossRef] [PubMed]
- Kapranov, P.; Cheng, J.; Dike, S.; Nix, D.A.; Duttagupta, R.; Willingham, A.T.; Stadler, P.F.; Hertel, J.; Hackermüller, J.; Hofacker, I.L.; et al. RNA Maps Reveal New RNA Classes and a Possible Function for Pervasive Transcription. Science 2007, 316, 1484–1488. [Google Scholar] [CrossRef] [Green Version]
- Zhang, P.; Wu, W.; Chen, Q.; Chen, M. Non-Coding RNAs and their Integrated Networks. J. Integr. Bioinform. 2019, 16, 20190027. [Google Scholar] [CrossRef]
- Kowalski, M.P.; Krude, T. Functional roles of non-coding Y RNAs. Int. J. Biochem. Cell Biol. 2015, 66, 20–29. [Google Scholar] [CrossRef] [Green Version]
- Ponjavic, J.; Ponting, C.P.; Lunter, G. Functionality or transcriptional noise? Evidence for selection within long noncoding RNAs. Genome Res. 2007, 17, 556–565. [Google Scholar] [CrossRef] [Green Version]
- Yang, X.; Liu, Y.; Zhang, H.; Wang, J.; Zinta, G.; Xie, S.; Zhu, W.; Nie, W.-F. Genome-Wide Identification of Circular RNAs in Response to Low-Temperature Stress in Tomato Leaves. Front. Genet. 2020, 11, 591806. [Google Scholar] [CrossRef] [PubMed]
- Bhatia, G.; Sharma, S.; Upadhyay, S.K.; Singh, K. Long Non-coding RNAs Coordinate Developmental Transitions and Other Key Biological Processes in Grapevine. Sci. Rep. 2019, 9, 3552. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deng, N.; Hou, C.; He, B.; Ma, F.; Song, Q.; Shi, S.; Liu, C.; Tian, Y. A full-length transcriptome and gene expression analysis reveal genes and molecular elements expressed during seed development in Gnetum luofuense. BMC Plant Biol. 2020, 20, 531. [Google Scholar] [CrossRef]
- Jiang, N.; Cui, J.; Shi, Y.; Yang, G.; Zhou, X.; Hou, X.; Meng, J.; Luan, Y. Tomato lncRNA23468 functions as a competing endogenous RNA to modulate NBS-LRR genes by decoying miR482b in the tomato-Phytophthora infestans interaction. Hortic. Res. 2019, 6, 28. [Google Scholar] [CrossRef] [Green Version]
- Yang, F.; Zhao, D.; Fan, H.; Zhu, X.; Wang, Y.; Liu, X.; Duan, Y.; Xuan, Y.; Chen, L. Functional Analysis of Long Non-Coding RNAs Reveal Their Novel Roles in Biocontrol of Bacteria-Induced Tomato Resistance to Meloidogyne incognita. Int. J. Mol. Sci. 2020, 21, 911. [Google Scholar] [CrossRef] [Green Version]
- Gai, Y.P.; Yuan, S.S.; Zhao, Y.N.; Zhao, H.N.; Zhang, H.L.; Ji, X.L. A Novel LncRNA, MuLnc1, Associated with Environmental Stress in Mulberry (Morus multicaulis). Front. Plant Sci. 2018, 9, 669. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, N.; Cui, J.; Hou, X.; Yang, G.; Xiao, Y.; Han, L.; Meng, J.; Luan, Y. Sl-lncRNA15492 interacts with Sl-miR482a and affects Solanum lycopersicum immunity against Phytophthora infestans. Plant J. 2020, 103, 1561–1574. [Google Scholar] [CrossRef]
- Moh, N.M.M.; Zhang, P.; Chen, Y.; Chen, M. Computational Identification of miRNAs and Temperature-Responsive lncRNAs From Mango (Mangifera indica L.). Front. Genet. 2021, 12, 607248. [Google Scholar] [CrossRef] [PubMed]
- Bai, Y.; Dai, X.; Li, Y.; Wang, L.; Li, W.; Liu, Y.; Cheng, Y.; Qin, Y. Identification and characterization of pineapple leaf lncRNAs in crassulacean acid metabolism (CAM) photosynthesis pathway. Sci. Rep. 2019, 9, 6658. [Google Scholar] [CrossRef] [PubMed]
- Wong, D.C.J.; Matus, J.T. Constructing Integrated Networks for Identifying New Secondary Metabolic Pathway Regulators in Grapevine: Recent Applications and Future Opportunities. Front. Plant Sci. 2017, 8, 505. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yao, S.; Wang, Z.; Cao, Q.; Xie, J.; Wang, X.; Zhang, R.; Deng, L.; Ming, J.; Zeng, K. Molecular basis of postharvest granulation in orange fruit revealed by metabolite, transcriptome and methylome profiling. Postharvest Biol. Technol. 2020, 166, 111205. [Google Scholar] [CrossRef]
- Zuo, J.; Grierson, D.; Courtney, L.T.; Wang, Y.; Gao, L.; Zhao, X.; Zhu, B.; Luo, Y.; Wang, Q.; Giovannoni, J.J. Relationships between genome methylation, levels of non-coding RNAs, mRNAs and metabolites in ripening tomato fruit. Plant J. 2020, 103, 980–994. [Google Scholar] [CrossRef]
- Chen, Y.; Cheng, C.; Feng, X.; Lai, R.; Gao, M.; Chen, W.; Wu, R. Integrated analysis of lncRNA and mRNA transcriptomes reveals the potential regulatory role of lncRNA in kiwifruit ripening and softening. Sci. Rep. 2021, 11, 1671. [Google Scholar] [CrossRef]
- Li, R.; Fu, D.; Zhu, B.; Luo, Y.; Zhu, H. CRISPR/Cas9--mediated mutagenesis of lncRNA1459 alters tomato fruit ripening. Plant J. 2018, 94, 513–524. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, Y.; Qu, Z.; Lei, J.; He, R.; Adelson, D.L.; Zhu, Y.; Yang, Z.; Wang, D. The long noncoding RNA FRILAIR regulates strawberry fruit ripening by functioning as a noncanonical target mimic. PLoS Genet. 2021, 17, e1009461. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Wang, H.; Chua, N.H. Long noncoding RNA transcriptome of plants. Plant Biotechnol. J. 2015, 13, 319–328. [Google Scholar] [CrossRef] [PubMed]
- Cho, J.; Koo, D.H.; Nam, Y.W.; Han, C.T.; Lim, H.T.; Bang, J.W.; Hur, Y. Isolation and characterization of cDNA clones expressed under male sex expression conditions in a monoecious cucumber plant (Cucumis sativus L. cv. Winter Long). Euphytica 2006, 146, 271–281. [Google Scholar] [CrossRef]
- Ma, J.; Yan, B.; Qu, Y.; Qin, F.; Yang, Y.; Hao, X.; Yu, J.; Zhao, Q.; Zhu, D.; Ao, G. Zm401, a short-open reading-frame mRNA or noncoding RNA, is essential for tapetum and microspore development and can regulate the floret formation in maize. J. Cell. Biochem. 2008, 105, 136–146. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Jung, C.; Xu, J.; Wang, H.; Deng, S.; Bernad, L.; Arenas-Huertero, C.; Chua, N.-H. Data from: Genome-wide analysis uncovers regulation of long intergenic noncoding RNAs in Arabidopsis. Plant Cell 2012, 24, 4333–4345. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, H.; Chung, P.J.; Liu, J.; Jang, I.C.; Kean, M.J.; Xu, J.; Chua, N.-H. Genome-wide identification of long noncoding natural antisense transcripts and their responses to light in Arabidopsis. Genome Res. 2014, 24, 444–453. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.; Niu, Q.W.; Wu, H.W.; Liu, J.; Ye, J.; Yu, N.; Chua, N.H. Analysis of non-coding transcriptome in rice and maize uncovers roles of conserved lncRNAs associated with agriculture traits. Plant J. 2015, 84, 404–416. [Google Scholar] [CrossRef]
- Lv, Y.; Liang, Z.; Ge, M.; Qi, W.; Zhang, T.; Lin, F.; Peng, Z.; Zhaohua, P. Genome-wide identification and functional prediction of nitrogen-responsive intergenic and intronic long non-coding RNAs in maize (Zea mays L.). BMC Genom. 2016, 17, 350. [Google Scholar] [CrossRef] [Green Version]
- Zhu, B.; Yang, Y.; Li, R.; Fu, D.; Wen, L.; Luo, Y.; Zhu, H. RNA sequencing and functional analysis implicate the regulatory role of long non-coding RNAs in tomato fruit ripening. J. Exp. Bot. 2015, 66, 4483–4495. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Chen, X.; Wang, C.; Xu, Z.; Wang, Y.; Liu, X.; Kang, Z.; Ji, W. Long non-coding genes implicated in response to stripe rust pathogen stress in wheat (Triticum aestivum L.). Mol. Biol. Rep. 2013, 40, 6245–6253. [Google Scholar] [CrossRef]
- Golicz, A.A.; Singh, M.B.; Bhalla, P.L. The Long Intergenic Noncoding RNA (LincRNA) Landscape of the Soybean Genome. Plant Physiol. 2018, 176, 2133–2147. [Google Scholar] [CrossRef] [Green Version]
- Hou, X.; Du, Y.; Liu, X.; Zhang, H.; Liu, Y.; Yan, N.; Zhang, Z. Genome-Wide Analysis of Long Non-Coding RNAs in Potato and Their Potential Role in Tuber Sprouting Process. Int. J. Mol. Sci. 2017, 19, 101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, T.-Z.; Liu, M.; Zhao, M.-G.; Chen, R.; Zhang, W.-H. Identification and characterization of long non-coding RNAs involved in osmotic and salt stress in Medicago truncatula using genome-wide high-throughput sequencing. BMC Plant Biol. 2015, 15, 131. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hao, Z.; Fan, C.; Cheng, T.; Su, Y.; Wei, Q.; Li, G. Genome-Wide Identification, Characterization and Evolutionary Analysis of Long Intergenic Noncoding RNAs in Cucumber. PLoS ONE 2015, 10, e0121800. [Google Scholar] [CrossRef]
- Zhang, Y.C.; Liao, J.Y.; Li, Z.Y.; Yu, Y.; Zhang, J.P.; Li, Q.F.; Qu, L.H.; Shu, W.S.; Chen, Y.Q. Genome-wide screening and functional analysis identify a large number of long noncoding RNAs involved in the sexual reproduction of rice. Genome Biol. 2014, 15, 512. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharma, S.; Taneja, M.; Tyagi, S.; Singh, K.; Upadhyay, S.K. Survey of High Throughput RNA-Seq Data Reveals Potential Roles for lncRNAs during Development and Stress Response in Bread Wheat. Front. Plant Sci. 2017, 8, 1019. [Google Scholar]
- Qi, X.; Xie, S.; Liu, Y.; Yi, F.; Yu, J. Genome-wide annotation of genes and noncoding RNAs of foxtail millet in response to simulated drought stress by deep sequencing. Plant Mol. Biol. 2013, 83, 459–473. [Google Scholar] [CrossRef]
- Ding, J.; Lu, Q.; Ouyang, Y.; Mao, H.; Zhang, P.; Yao, J.; Xu, C.; Li, X.; Xiao, J.; Zhang, Q. A long noncoding RNA regulates photoperiod-sensitive male sterility, an essential component of hybrid rice. Proc. Natl. Acad. Sci. USA 2012, 109, 2654–2659. [Google Scholar] [CrossRef] [Green Version]
- Heo, J.B.; Sung, S. Vernalization-Mediated Epigenetic Silencing by a Long Intronic Noncoding RNA. Science 2011, 331, 76–79. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, Q.; Csorba, T.; Skourti-Stathaki, K.; Proudfoot, N.J.; Dean, C. R-Loop Stabilization Represses Antisense Transcription at the Arabidopsis FLC Locus. Science 2013, 340, 619–621. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Csorba, T.; Questa, J.I.; Sun, Q.; Dean, C. Antisense COOLAIR mediates the coordinated switching of chromatin states at FLC during vernalization. Proc. Natl. Acad. Sci. USA 2014, 111, 16160–16165. [Google Scholar] [CrossRef] [Green Version]
- Xu, X.-W.; Zhou, X.; Wang, R.-R.; Peng, W.-L.; An, Y.; Chen, L.-L. Functional analysis of long intergenic non-coding RNAs in phosphate-starved rice using competing endogenous RNA network. Sci. Rep. 2016, 6, 20715. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yuan, J.; Zhang, Y.; Dong, J.; Sun, Y.; Lim, B.L.; Liu, D.; Lu, Z.J. Systematic characterization of novel lncRNAs responding to phosphate starvation in Arabidopsis thaliana. BMC Genom. 2016, 17, 655. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tomato Genome Consortium. The tomato genome sequence provides insights into fleshy fruit evolution. Nature 2012, 485, 635–641. [Google Scholar] [CrossRef] [Green Version]
- Nie, W.F. DNA methylation: From model plants to vegetable crops. Biochem. Soc. Trans. 2021, 49, 1479–1487. [Google Scholar] [CrossRef] [PubMed]
- Tang, D.; Gallusci, P.; Lang, Z. Fruit development and epigenetic modifications. New Phytol. 2020, 228, 839–844. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Yang, Y.; Jin, L.; Ling, X.; Liu, T.; Chen, T.; Ji, Y.; Yu, W.; Zhang, B. Re-analysis of long non-coding RNAs and prediction of circRNAs reveal their novel roles in susceptible tomato following TYLCV infection. BMC Plant Biol. 2018, 18, 104. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Liu, T.; Shen, D.; Wang, J.; Ling, X.; Hu, Z.; Chen, T.; Hu, J.; Huang, J.; Yu, W.; et al. Tomato yellow leaf curl virus intergenic siRNAs target a host long noncoding RNA to modulate disease symptoms. PLOS Pathog. 2019, 15, e1007534. [Google Scholar] [CrossRef]
- Yang, Z.; Yang, C.; Wang, Z.; Yang, Z.; Chen, D.; Wu, Y. LncRNA expression profile and ceRNA analysis in tomato during flowering. PLoS ONE 2019, 14, e0210650. [Google Scholar] [CrossRef]
- Wang, X.; Ai, G.; Zhang, C.; Cui, L.; Wang, J.; Li, H.; Zhang, J.; Ye, Z. Expression and diversification analysis reveals transposable elements play important roles in the origin of Lycopersicon-specific lncRNAs in tomato. New Phytol. 2016, 209, 1442–1455. [Google Scholar] [CrossRef]
- Zhao, T.; Mei, H.; Cao, Z.; Wang, L.; Tao, X.; Feng, S.; Fang, L.; Guan, X. Absence of CG methylation alters the long noncoding transcriptome landscape in multiple species. FEBS Lett. 2021, 595, 1734–1747. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Gao, L.; Li, J.; Zhu, B.; Zhu, H.; Luo, Y.; Wang, Q.; Zuo, J. Analysis of long-non-coding RNAs associated with ethylene in tomato. Gene 2018, 674, 151–160. [Google Scholar] [CrossRef] [PubMed]
- Yu, T.; Tzeng, D.T.W.; Li, R.; Chen, J.; Zhong, S.; Fu, D.; Zhu, B.; Luo, Y.; Zhu, H. Genome-wide identification of long non-coding RNA targets of the tomato MADS box transcription factor RIN and function analysis. Ann. Bot. 2018, 123, 469–482. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.; Kang, B.; Li, M.; Xiao, L.; Xiao, H.; Shen, H.; Yang, W. Transcription of lncRNA ACoS-AS1 is essential to trans-splicing between SlPsy1 and ACoS-AS1 that causes yellow fruit in tomato. RNA Biol. 2020, 17, 596–607. [Google Scholar] [CrossRef] [PubMed]
- Xue, L.; Sun, M.; Wu, Z.; Yu, L.; Yu, Q.; Tang, Y.; Jiang, F. LncRNA regulates tomato fruit cracking by coordinating gene expression via a hormone-redox-cell wall network. BMC Plant Biol. 2020, 20, 162. [Google Scholar] [CrossRef] [Green Version]
- Zhou, C.; Zhu, J.; Qian, N.; Guo, J.; Yan, C. Bacillus subtilis SL18r Induces Tomato Resistance Against Botrytis cinerea, Involving Activation of Long Non-coding RNA, MSTRG18363, to Decoy miR1918. Front. Plant Sci. 2021, 11, 634819. [Google Scholar] [CrossRef]
- Cui, J.; Jiang, N.; Hou, X.; Wu, S.; Zhang, Q.; Meng, J.; Luan, Y. Genome-Wide Identification of lncRNAs and Analysis of ceRNA Networks During Tomato Resistance to Phytophthora infestans. Phytopathol. 2020, 110, 456–464. [Google Scholar] [CrossRef] [PubMed]
- Cui, J.; Jiang, N.; Meng, J.; Yang, G.; Liu, W.; Zhou, X.; Ma, N.; Hou, X.; Luan, Y. LncRNA33732-respiratory burst oxidase module associated with WRKY1 in tomato- Phytophthora infestans interactions. Plant J. 2018, 97, 933–946. [Google Scholar] [CrossRef]
- Cui, J.; Luan, Y.; Jiang, N.; Bao, H.; Meng, J. Comparative transcriptome analysis between resistant and susceptible tomato allows the identification of lncRNA16397 conferring resistance to Phytophthora infestans by co-expressing glutaredoxin. Plant J. 2017, 89, 577–589. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hou, X.; Cui, J.; Liu, W.; Jiang, N.; Zhou, X.; Qi, H.; Meng, J.; Luan, Y. LncRNA39026 Enhances Tomato Resistance to Phytophthora infestans by Decoying miR168a and Inducing PR Gene Expression. Phytopathology 2020, 110, 873–880. [Google Scholar] [CrossRef] [PubMed]
- Liao, X.; Wang, J.; Zhu, S.; Xie, Q.; Wang, L.; Yu, H.; Ye, Z.; Yang, C. Transcriptomic and functional analyses uncover the regulatory role of lncRNA000170 in tomato multicellular trichome formation. Plant J. 2020, 104, 18–29. [Google Scholar] [CrossRef]
- Eom, S.H.; Lee, H.J.; Lee, J.H.; Wi, S.H.; Kim, S.K.; Hyun, T.K. Identification and Functional Prediction of Drought-Responsive Long Non-Coding RNA in Tomato. Agronomy 2019, 9, 629. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Gao, L.; Zhu, B.; Zhu, H.; Luo, Y.; Wang, Q.; Zuo, J. Integrative analysis of long non-coding RNA acting as ceRNAs involved in chilling injury in tomato fruit. Gene 2018, 667, 25–33. [Google Scholar] [CrossRef]
- Lin, Y.; Jiang, L.; Chen, Q.; Li, Y.; Zhang, Y.; Luo, Y.; Zhang, Y.; Sun, B.; Wang, X.; Tang, H. Comparative Transcriptome Profiling Analysis of Red- and White-Fleshed Strawberry (Fragaria x ananassa) Provides New Insight into the Regulation of the Anthocyanin Pathway. Plant Cell Physiol. 2018, 59, 1844–1859. [Google Scholar] [CrossRef] [PubMed]
- Feng, S.; Fang, H.; Liu, X.; Dong, Y.; Wang, Q.; Yang, K.Q. Genome-wide identification and characterization of long non-coding RNAs conferring resistance to Colletotrichum gloeosporioides in walnut (Juglans regia). BMC Genom. 2021, 22, 15. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Yang, T.; Tang, Y.; Aisimutuola, P.; Zhang, G.; Wang, B.; Li, N.; Wang, J.; Yu, Q. Transcriptomic profile analysis of non-coding RNAs involved in Capsicum chinense Jacq. fruit ripening. Sci. Hortic. 2020, 264, 109158. [Google Scholar] [CrossRef]
- Ma, H.; Yang, T.; Li, Y.; Zhang, J.; Wu, T.; Song, T.; Yao, Y.; Tian, J. The long noncoding RNA MdLNC499 bridges MdWRKY1 and MdERF109 function to regulate early-stage light-induced anthocyanin accumulation in apple fruit. Plant Cell 2021, 33, 3309–3330. [Google Scholar] [CrossRef] [PubMed]
- Yang, T.; Ma, H.; Zhang, J.; Wu, T.; Song, T.; Tian, J.; Yao, Y. Systematic identification of long noncoding RNA s expressed during light—Induced anthocyanin accumulation in apple fruit. Plant J. 2019, 100, 572–590. [Google Scholar] [CrossRef]
- Li, Y.; Wu, C.; Liu, C.; Yu, J.; Duan, X.; Fan, W.; Wang, J.; Zhang, X.; Yan, G.; Li, T.; et al. Functional identification of lncRNAs in sweet cherry (Prunus avium) pollen tubes via transcriptome analysis using single-molecule long-read sequencing. Hortic. Res. 2019, 6, 135. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, G.; Chen, D.; Zhang, T.; Duan, A.; Zhang, J.; He, C. Transcriptomic and functional analyses unveil the role of long non-coding RNAs in anthocyanin biosynthesis during sea buckthorn fruit ripening. DNA Res. 2018, 25, 465–476. [Google Scholar] [CrossRef]
- Zhang, G.; Duan, A.; Zhang, J.; He, C. Genome-wide analysis of long non-coding RNAs at the mature stage of sea buckthorn (Hippophae rhamnoides Linn) fruit. Gene 2017, 596, 130–136. [Google Scholar] [CrossRef]
- Liu, H.; Lu, Y.; Wang, J.; Hu, J.; Wuyun, T. Genome-wide screening of long non-coding RNAs involved in rubber biosynthesis in Eucommia ulmoides. J. Integr. Plant Biol. 2018, 60, 1070–1082. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, W.; Li, C.; Li, S.; Peng, M. Long noncoding RNAs that respond to Fusarium oxysporum infection in ‘Cavendish’ banana (Musa acuminata). Sci. Rep. 2017, 7, 16939. [Google Scholar] [CrossRef] [Green Version]
- Tian, Y.; Bai, S.; Dang, Z.; Hao, J.; Zhang, J.; Hasi, A. Genome-wide identification and characterization of long non-coding RNAs involved in fruit ripening and the climacteric in Cucumis melo. BMC Plant Biol. 2019, 19, 369. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parkhomchuk, D.; Borodina, T.; Amstislavskiy, V.; Banaru, M.; Hallen, L.; Krobitsch, S.; Lehrach, H.; Soldatov, A. Transcriptome analysis by strand-specific sequencing of complementary DNA. Nucleic Acids Res. 2009, 37, e123. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.-C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garber, M.; Grabherr, M.G.; Guttman, M.; Trapnell, C. Computational methods for transcriptome annotation and quantification using RNA-seq. Nat. Methods 2011, 8, 469–477. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, X.; Zhang, X.; Yang, Y.; Zhang, H.; Zhu, W.; Nie, W.F. The histone variant Sl_H2A.Z regulates carotenoid biosynthesis and gene expression during tomato fruit ripening. Hortic. Res. 2021, 8, 85. [Google Scholar] [CrossRef] [PubMed]
- Pauli, A.; Valen, E.; Lin, M.F.; Garber, M.; Vastenhouw, N.L.; Levin, J.; Fan, L.; Sandelin, A.; Rinn, J.; Regev, A.; et al. Systematic identification of long noncoding RNAs expressed during zebrafish embryogenesis. Genome Res. 2011, 22, 577–591. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shuai, P.; Liang, D.; Tang, S.; Zhang, Z.; Ye, C.-Y.; Su, Y.; Xia, X.; Yin, W. Genome-wide identification and functional prediction of novel and drought-responsive lincRNAs in Populus trichocarpa. J. Exp. Bot. 2014, 65, 4975–4983. [Google Scholar] [CrossRef] [PubMed]


| Num. | Biological Function/Main Finding | References |
|---|---|---|
| 1 | Identification of lncRNAs by strand-specific paired-end RNA sequencing of tomato leaves, flowers, and roots | [51] |
| 2 | Integration and relationships between DNA methylation, lncRNAs, mRNAs, and metabolites in ripening tomato fruit | [20] |
| 3 | The connection between lncRNAs and transposable elements, and between lncRNAs and DNA CG methylation | [52,53] |
| 4 | Regulating tomato fruit ripening | [20,22,31,54,55] |
| 5 | Function in trans-splicing event SlPsy1-ACoS-AS1, and generation of yellow fruit in tomatoes | [56] |
| 6 | LncRNA regulates tomato fruit cracking | [57] |
| 7 | LncRNA is involved in rhizobacterial strain-induced systemic resistance (ISR) to the foliar pathogen Botrytis cinerea in leaves | [58] |
| 8 | LncRNAs regulate rhizosphere bacteria-induced tomato resistance to Meloidogyne incognita in roots | [13] |
| 9 | Function in regulating the resistance to Phytophthora infestans in tomato plants | [12,15,59,60,61,62] |
| 10 | In response to tomato yellow leaf curl virus (TYLCV) interaction | [49,50] |
| 11 | Regulation in tomato multicellular trichome formation | [63] |
| 12 | In response to drought stress in tomato leaves | [64] |
| 13 | In response to chilling injury in tomato fruits | [65] |
| Num. | Species | Biological Function | References |
|---|---|---|---|
| 1 | Strawberry | Fruit ripening; anthocyanin accumulation | [23,66] |
| 2 | Mango | Abiotic stress | [16] |
| 3 | Walnut | Biotic stress | [67] |
| 4 | Kiwifruit | Fruit ripening | [21] |
| 5 | Orange | Fruit granulation | [19] |
| 6 | Pepper | Fruit ripening | [68] |
| 7 | Gnetum luofuense | Seed development | [11] |
| 8 | Apple | Fruit anthocyanin accumulation | [69,70] |
| 9 | Sweet cherry | Pollen development | [71] |
| 10 | Grapevine | Different developmental stages of leaf, inflorescence, and berry tissues | [10] |
| 11 | Pineapple | Crassulacean acid metabolism photosynthesis pathway in leaves | [17] |
| 12 | Sea buckthorn | Fruit ripening | [72,73] |
| 13 | Eucommia ulmoides | Rubber biosynthesis | [74] |
| 14 | Mulberry | Abiotic stress | [14] |
| 15 | Banana | Biotic stress | [75] |
| 16 | Cucumis melo | Fruit ripening | [76] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, J.; Feng, Y.; Ding, X.; Huo, J.; Nie, W.-F. Identification of Long Non-Coding RNAs Associated with Tomato Fruit Expansion and Ripening by Strand-Specific Paired-End RNA Sequencing. Horticulturae 2021, 7, 522. https://doi.org/10.3390/horticulturae7120522
Wang J, Feng Y, Ding X, Huo J, Nie W-F. Identification of Long Non-Coding RNAs Associated with Tomato Fruit Expansion and Ripening by Strand-Specific Paired-End RNA Sequencing. Horticulturae. 2021; 7(12):522. https://doi.org/10.3390/horticulturae7120522
Chicago/Turabian StyleWang, Jinyu, Yan Feng, Xiaotao Ding, Jingtian Huo, and Wen-Feng Nie. 2021. "Identification of Long Non-Coding RNAs Associated with Tomato Fruit Expansion and Ripening by Strand-Specific Paired-End RNA Sequencing" Horticulturae 7, no. 12: 522. https://doi.org/10.3390/horticulturae7120522
APA StyleWang, J., Feng, Y., Ding, X., Huo, J., & Nie, W.-F. (2021). Identification of Long Non-Coding RNAs Associated with Tomato Fruit Expansion and Ripening by Strand-Specific Paired-End RNA Sequencing. Horticulturae, 7(12), 522. https://doi.org/10.3390/horticulturae7120522







