RsSOS1 Responding to Salt Stress Might Be Involved in Regulating Salt Tolerance by Maintaining Na+ Homeostasis in Radish (Raphanus sativus L.)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Isolation of RsSOS1 Gene in Radish
2.2. Bioinformatic Analysis of RsSOS1
2.3. Subcellular Localization of RsSOS1 Protein
2.4. Identification of Salt Tolerance of Radish Genotypes
2.5. Expression Profiling of RsSOS1
2.6. Heterologous Expression of RsSOS1 in Arabidopsis and Yeast
2.7. Statistical Analysis
3. Results
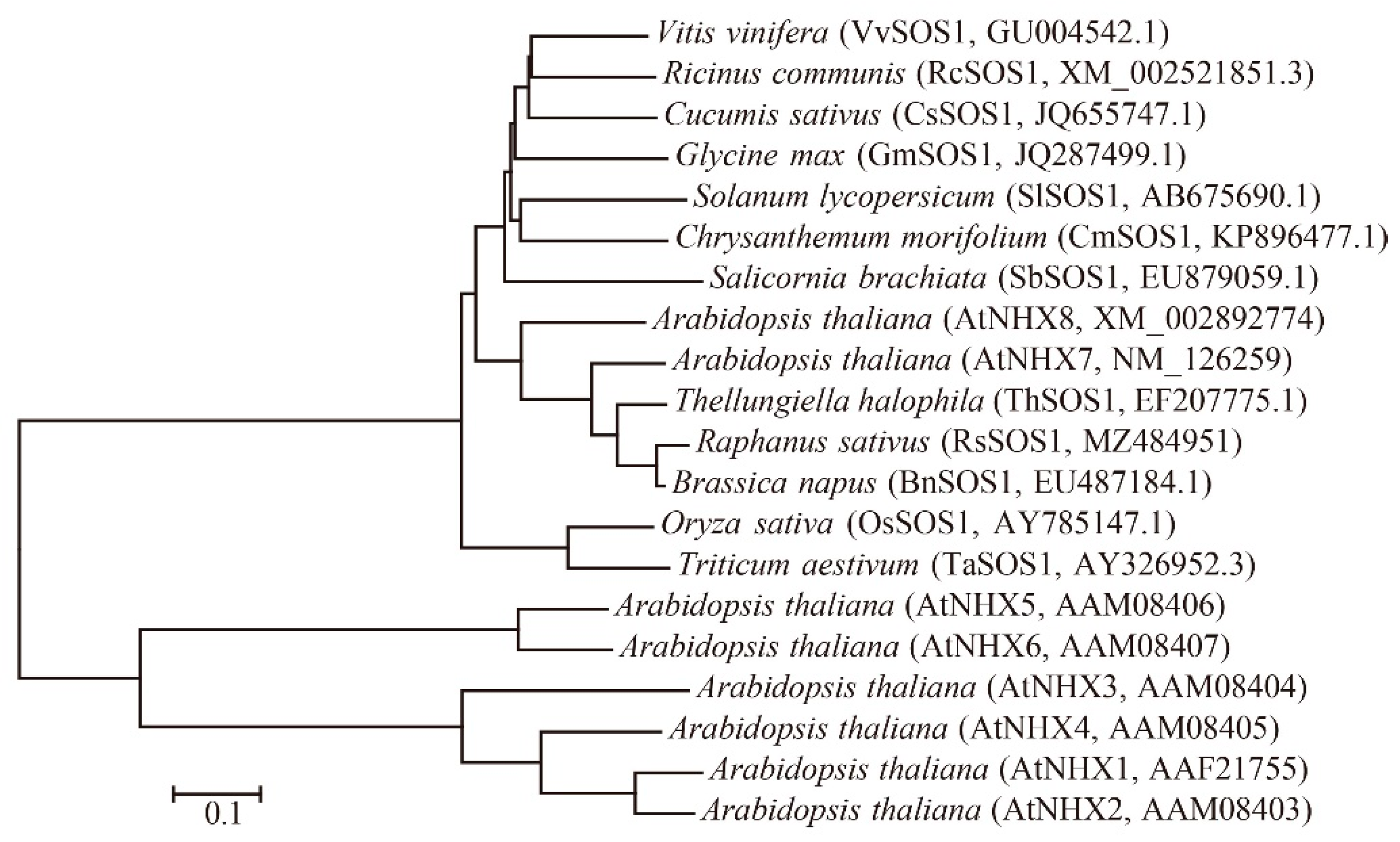
3.1. Isolation and Phylogenetic Analysis of RsSOS1 Gene
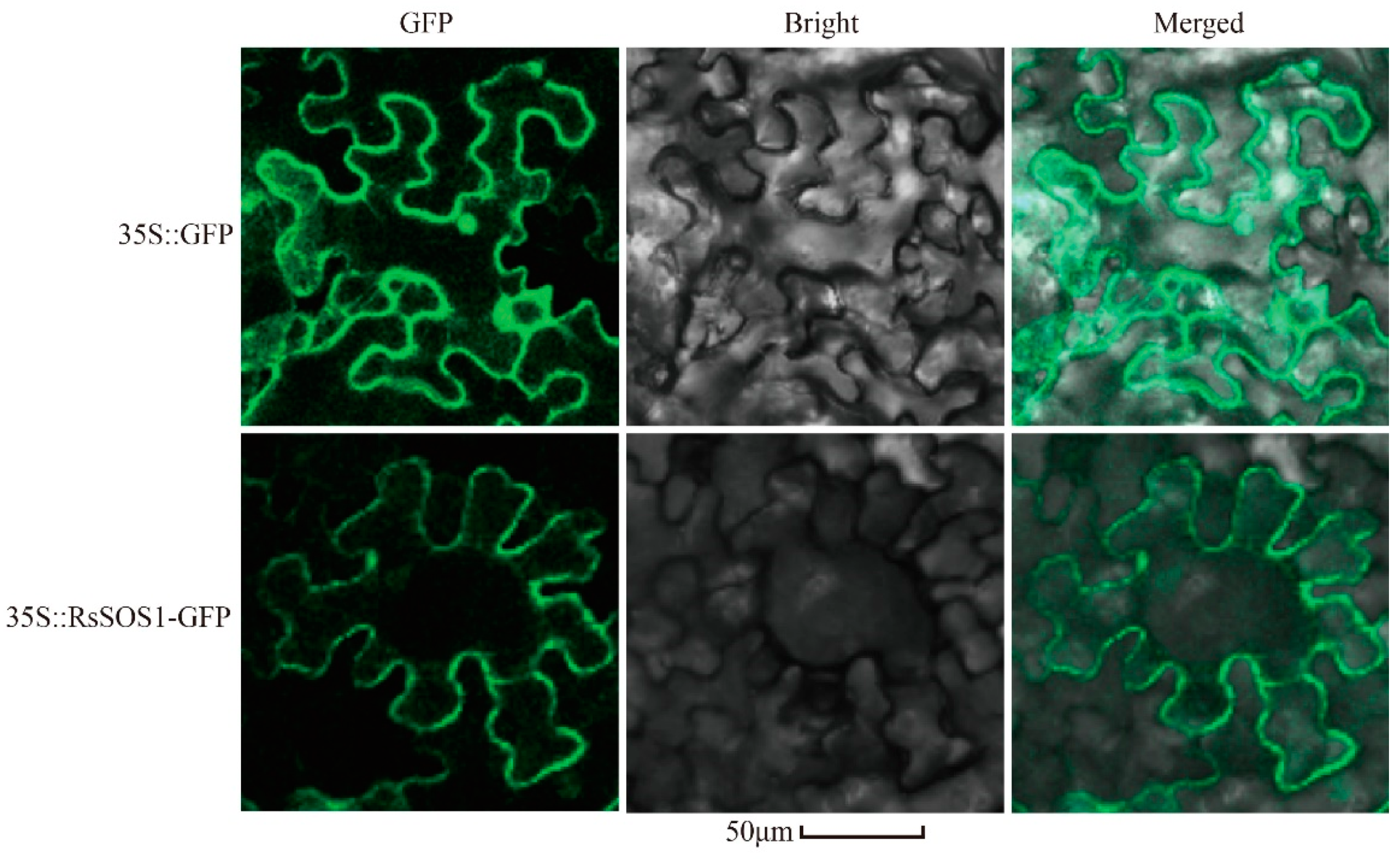
3.2. Subcellular Localization Analysis of RsSOS1
3.3. Identification of Salt Tolerance of Genotype ‘NAU-TR17’ and ‘NAU-TR12’
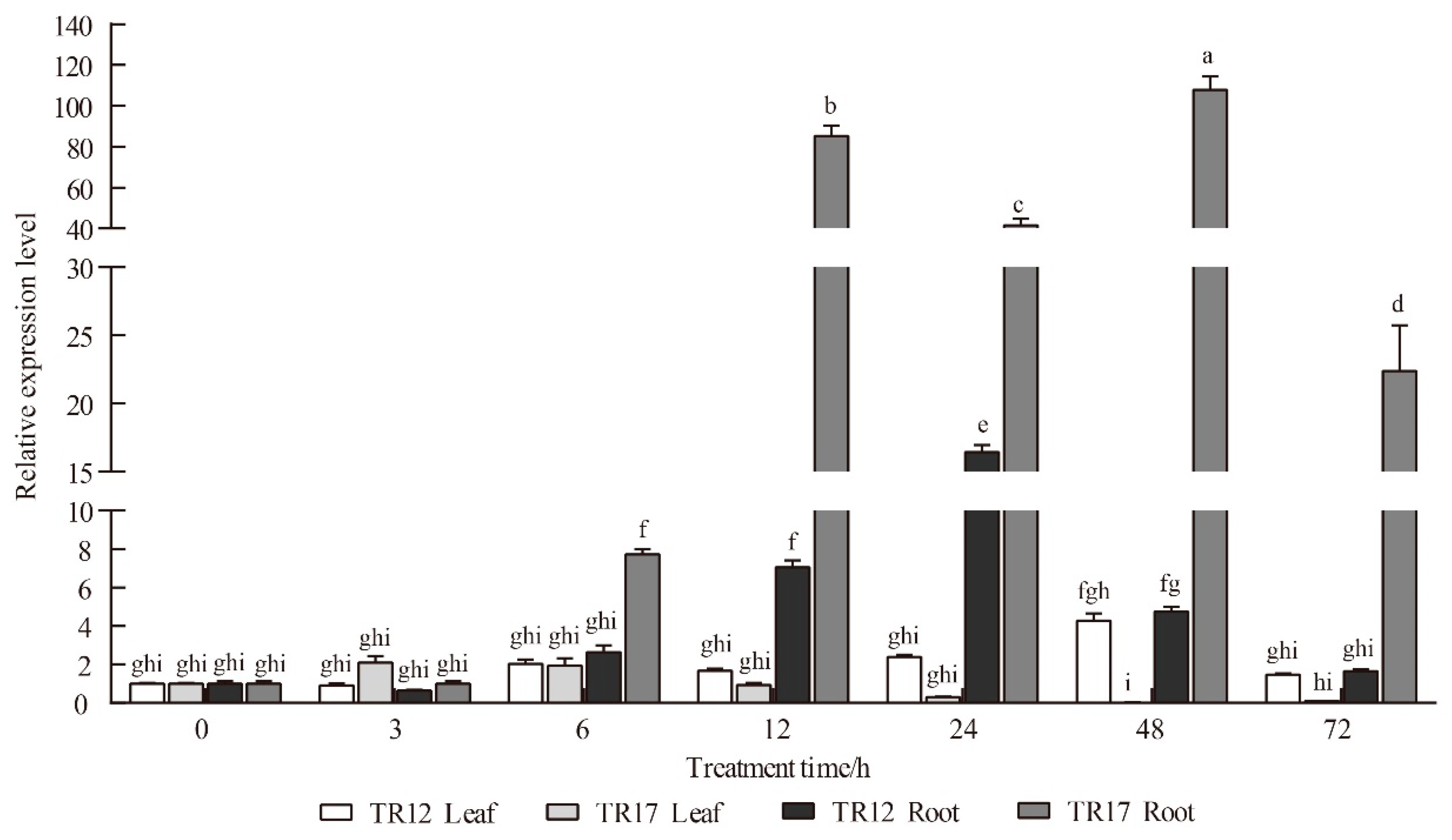
3.4. Expression Profiling of RsSOS1 under Salt Stress
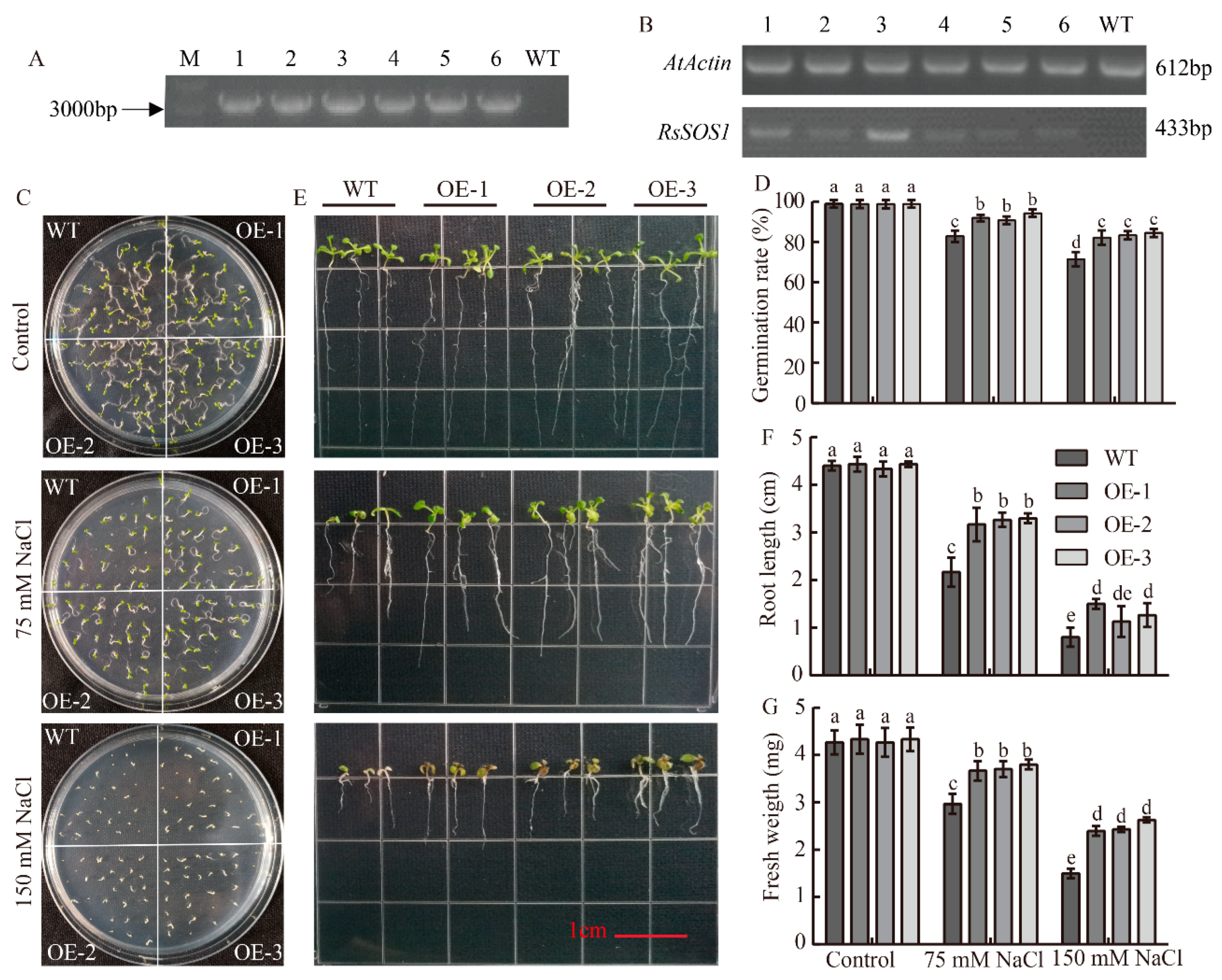
3.5. Overexpression of RsSOS1 in Arabidopsis to Improve Salt Stress Tolerance
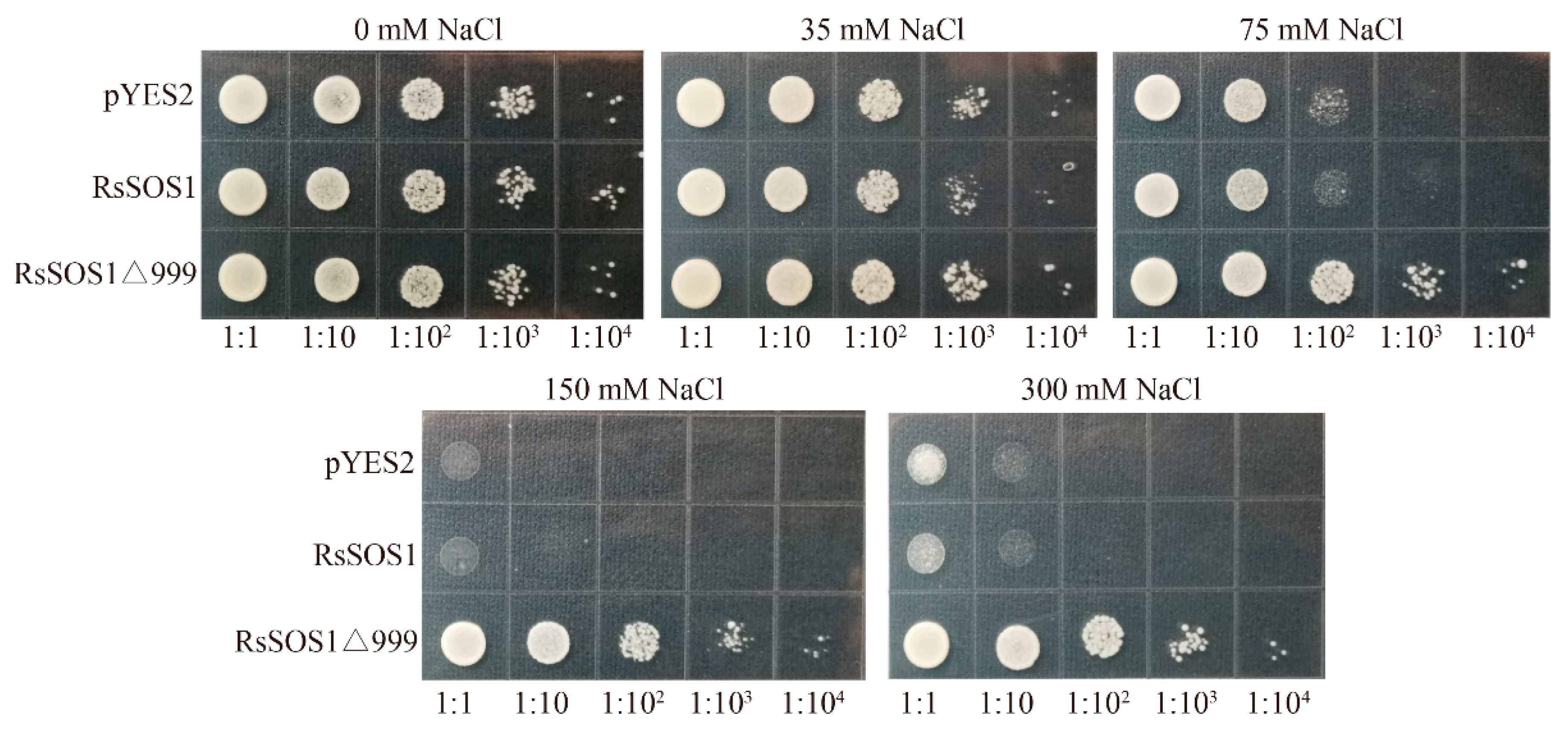
3.6. Functional Complement Analysis of RsSOS1 Protein in AXT3 Yeast
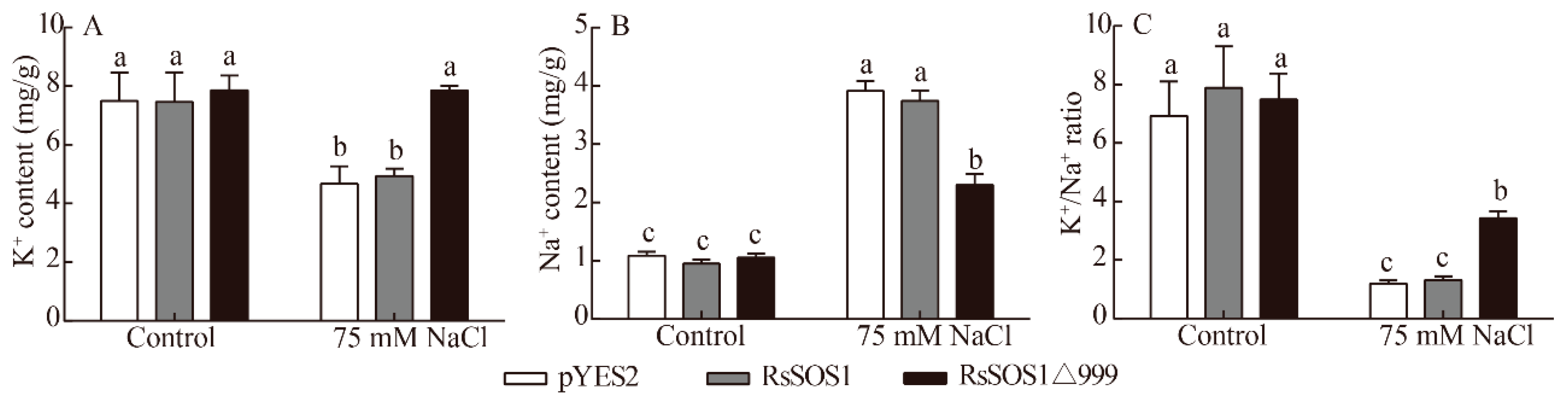
3.7. Determination of Ion Content in Transgenic Yeast
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tang, X.L.; Mu, X.M.; Shao, H.B.; Wang, H.Y.; Brestic, M. Global plant responding mechanisms to salt stress: Physiological and Molecular Levels and Implications in Biotechnology. Crit. Rev. Biotechnol. 2015, 35, 425–437. [Google Scholar] [CrossRef]
- Butcher, K.; Wick, A.F.; Desutter, T.; Chatterjee, A.; Harmon, J. Soil salinity: A threat to global food security. Agron. J. 2016, 108, 2189–2200. [Google Scholar] [CrossRef]
- Parida, A.; Das, A. Salt tolerance and salinity effects on plants: A review. Ecotoxicol. Environ. Saf. 2005, 60, 324–349. [Google Scholar] [CrossRef]
- Huang, L.; Wu, D.; Zhang, G. Advances in studies on ion transporters involved in salt tolerance and breeding crop cultivars with high salt tolerance. J. Zhejiang Univ. Sci. B 2020, 21, 426–441. [Google Scholar] [CrossRef]
- Wang, Y.; Ying, J.L.; Zhang, Y.; Xu, L.; Zhang, W.T.; Ni, M.; Zhu, Y.L.; Liu, L.W. Genome-wide identification and functional characterization of the cation proton antiporter (CPA) family related to salt Stress response in radish (Raphanus sativus L.). Int. J. Mol. Sci. 2020, 21, 8262. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Yin, X.C.; Duan, R.J.; Hao, G.C.; Guo, J.C.; Jiang, X.Y. SpAHA1 and SpSOS1 coordinate in transgenic yeast to improve salt tolerance. PLoS ONE 2015, 10, e0137447. [Google Scholar] [CrossRef]
- Fan, Y.F.; Yin, X.C.; Xie, Q.; Xia, Y.Q.; Wang, Z.Y.; Song, J.; Zhou, Y.; Jiang, X.Y. Co-expression of SpSOS1 and SpAHA1 in transgenic Arabidopsis plants improves salinity tolerance. BMC Plant Biol. 2019, 19, 74. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Y.; Yin, X.C.; Wan, S.M.; Hu, Y.P.; Xie, Q.; Li, R.M.; Zhu, B.B.; Fu, S.P.; Guo, J.C.; Jiang, X.Y. The Sesuvium portulacastrum plasma membrane Na+/H+ antiporter SpSOS1 complemented the salt sensitivity of transgenic Arabidopsis sos1 mutant plants. Plant. Mol. Biol. Rep. 2018, 36, 553–563. [Google Scholar] [CrossRef]
- Mahi, H.; Perez-Hormaeche, J.; Luca, A.; Villalta, I.; Espartero, J.; Gámez-Arjona, F.; Fernández, J.L.; Bundó, M.; Mendoza, I.; Mieulet, D.; et al. A critical role of sodium flux via the plasma membrane Na+/H+ exchanger SOS1 in the salt tolerance of Rice. Plant Physiol. 2019, 180, 1046–1065. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, Q.; Meng, L.; Han, J.W.; Mao, P.C.; Tian, X.X.; Zheng, M.L.; Luis, A.J. SOS1 is a key systemic regulator of salt secretion and K+/Na+ homeostasis in the recretohalophyte Karelinia caspia. Environ. Exp. Bot. 2020, 177, 104098. [Google Scholar] [CrossRef]
- Quintero, F.; Martinez-Atienza, J.; Villalta, I.; Jiang, X.Y.; Kim, W.Y.; Ali, Z.; Fujii, H.; Mendoza, I.; Yun, D.J.; Zhu, J.K.; et al. Activation of the plasma membrane Na+/H+ antiporter SOS1 by phosphorylation of an auto-inhibitory C-terminal domain. Proc. Natl. Acad. Sci. USA 2011, 108, 2611–2616. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, X.C.; Xu, L.; Wang, Y.; Luo, X.B.; Zhu, X.W.; Karanja, B.K.; Nie, S.S.; Feng, H.Y.; Li, C.; Liu, L.W. Transcriptome-based gene expression profiling identifies differentially expressed genes critical for salt stress response in radish (Raphanus sativus L.). Plant Cell Rep. 2016, 35, 329–346. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Zhu, Z.T.; Tao, X.Y.; Wang, F.Q.; Wei, D.Z. RNA-Seq analysis uncovers non-coding small RNA system of Mycobacterium neoaurum in the metabolism of sterols to accumulate steroid intermediates. Microb. Cell Fact. 2016, 15, 64. [Google Scholar] [CrossRef] [Green Version]
- Jeong, Y.M.; Kim, N.; Ahn, B.; Oh, M.J.; Chung, W.H.; Chung, H.; Seongmun, J.; Ki-Byung, L.; Yoon-Jung, H.; Goon-Bo, K.; et al. Elucidating the triplicated ancestral genome structure of radish based on chromosome-level comparison with the Brassica genomes. Theor. Appl. Genet. 2016, 129, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Chinnusamy, V.; Jagendorf, A.; Zhu, J. Understanding and improving salt tolerance in plants. Crop. Sci. 2005, 45, 437–448. [Google Scholar] [CrossRef] [Green Version]
- Bassil, E.; Coku, A.; Blumwald, E. Cellular ion homeostasis: Emerging roles of intracellular NHX Na+/H+ antiporters in plant growth and development. J. Exp. Bot. 2012, 63, 5727–5740. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Fan, L.; Xu, L.; Wang, Y.; Tang, M.J.; Liu, L.W. Genome- and Transcriptome-Wide Characterization of bZIP Gene Family Identifies Potential Members Involved in Abiotic Stress Response and Anthocyanin Biosynthesis in Radish (Raphanus sativus L.). Int. J. Mol. Sci. 2019, 20, 6334. [Google Scholar] [CrossRef] [Green Version]
- Wu, Z.; Li, T.; Liu, X.Y.; Yuan, G.Z.; Hou, H.Z.; Teng, N.J. A novel R2R3-MYB transcription factor LlMYB305 from Lilium longiflorum plays a positive role in thermos tolerance via activating heat-protective genes. Environ. Exp. Bot. 2021, 184, 104399. [Google Scholar] [CrossRef]
- Mei, Y.; Zu, Y.X.; Wu, Y.C.; Zheng, J.Q.; Wang, W.W.; Guo, J. Physiological response of radish seedlings to salt stress. Chin. Agron. Bull. 2015, 31, 49–53. [Google Scholar]
- Xu, Y.Y.; Zhu, X.W.; Gong, Y.Q.; Xu, L.; Wang, Y.; Liu, L.W. Evaluation of reference genes for gene expression studies in radish (Raphanus sativus L.) using quantitative real-time PCR. Biochem. Biophys. Res. Commun. 2012, 424, 398–403. [Google Scholar] [CrossRef]
- Livak, K.; Schmittgen, T. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Martinez-Trujillo, M.; Limones-Briones, V.; Cabrera-Ponce, J.; Herrera-Estrella, L. Improving transformation efficiency of Arabidopsis thaliana by modifying the floral dip method. Plant Mol. Biol. Rep. 2004, 22, 63–70. [Google Scholar] [CrossRef]
- Okada, T.; Yamane, S.; Yamaguchi, M.; Kato, K.; Shinmyo, A.; Tsunemitsu, Y.; Iwasaki, K.; Ueno, D.; Demura, T. Characterization of rice KT/HAK/KUP potassium transporters and K+ uptake by HAK1 from Oryza sativa. Plant Biotechnol. 2018, 35, 101–111. [Google Scholar] [CrossRef] [Green Version]
- Chen, G.; Hu, Q.D.; Luo, L.; Yang, T.Y.; Zhang, S.; Hu, Y.B.; Yu, L.; Xu, G.H. Rice potassium transporter OsHAK1 is essential for maintaining potassium-mediated growth and functions in salt tolerance over low and high potassium concentration ranges. Plant Cell Environ. 2015, 38, 2747–2765. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J. Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol. 2002, 53, 247–273. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, K.; Li, S.; Guo, Q.; Mao, P.; Tian, X.; Meng, L. Cloning and expression analysis of MsSOS1 gene from Melilotus luteus. Biotechnol. Bull. 2017, 33, 120–130. [Google Scholar]
- Martínez-Atienza, J.; Jiang, X.; Garciadeblas, B.; Mendoza, I.; Zhu, J.K.; Pardo, J.M.; Quintero, F.J. Conservation of the salt overly sensitive pathway in rice. Plant Physiol. 2007, 143, 1001–1012. [Google Scholar] [CrossRef] [Green Version]
- Cheng, C.H.; Zhong, Y.M.; Wang, Q.; Cai, Z.M.; Wang, D.D.; Li, C.M. Genome-wide identification and gene expression analysis of SOS family genes in tuber mustard (Brassica juncea var. tumida). PLoS ONE 2019, 14, e0224672. [Google Scholar] [CrossRef]
- Tang, R.J.; Liu, H.; Bao, Y.; Lv, Q.D.; Yang, L.; Zhang, H.X. The woody plant poplar has a functionally conserved salt overly sensitive pathway in response to salinity stress. Plant Mol. Biol. 2010, 74, 367–380. [Google Scholar] [CrossRef] [PubMed]
- Fang, Z.H.; Cui, M.M.; Zhang, Y.; Ren, B.L.; Shi, Y.H.; Liu, J.N.; Wang, Y.Q.; Dong, K.H.; GAO, H.H.; Wu, X.M. Cloning and expression analysis of MsSOS1 gene in Medicago sativa. Grassl. Sci. 2018, 26, 1479–1489. [Google Scholar]
- Shi, P.B.; Hong, L.Z.; Wang, J.; Yue, Y.F.; Wang, W.Y.; Lv, Y.D.; Gu, M.F. Cloning and expression analysis of the Na+/H+ antiporter gene CmaSOS1 from Cucurbita maxima L. J. Nucl. Agric. 2020, 34, 2638–2646. [Google Scholar]
- Wu, Y.X.; Ding, N.; Zhao, X.; Zhao, M.G.; Chang, Z.Q.; Liu, J.Q.; Zhang, L.X. Molecular characterization of PeSOS1: The putative Na+/H+ antiporter of Populus euphratica. Plant Mol. Biol. 2007, 65, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Cosentino, C.; Fischer-Schliebs, E.; Bertl, A.; Thiel, G.; Homann, U. Na+/H+ antiporters are differentially regulated in response to NaCl stress in leaves and roots of Mesembryanthemum crystallinum. New Phytol. 2010, 186, 669–680. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.Z.; Ishitani, M.; Kim, C.; Zhu, J.K. The Arabidopsis thaliana salt tolerance gene SOS1 encodes a putative Na+/H+ antiporter. Proc. Natl. Acad. Sci. USA 2000, 97, 6896–6901. [Google Scholar] [CrossRef] [Green Version]
- Cheng, C.; Liu, Y.; Liu, Y.M.; Liu, X.; An, J.; Jiang, L.; Yu, B.J. Recretohalophyte Tamarix TrSOS1 confers higher salt tolerance to transgenic plants and yeast than glycophyte soybean GmSOS1. Environ. Exp. Bot. 2019, 165, 196–207. [Google Scholar] [CrossRef]
- Chen, X.G.; Lu, X.K.; Shu, N.; Wang, D.L.; Wang, S.; Wang, J.J.; Guo, L.X.; Guo, X.N.; Fan, W.L.; Lin, Z.X.; et al. GhSOS1, a plasma membrane Na+/H+ antiporter gene from upland cotton, enhances salt tolerance in transgenic Arabidopsis thaliana. PLoS ONE 2017, 12, e0181450. [Google Scholar] [CrossRef]
- Zhao, X.F.; Wei, P.P.; Liu, Z.; Yu, B.J.; Shi, H.Z. Soybean Na+/H+ antiporter GmsSOS1 enhances antioxidant enzyme activity and reduces Na+ accumulation in Arabidopsis and yeast cells under salt stress. Acta Physiol. Plant. 2017, 39, 19. [Google Scholar] [CrossRef]
- Fan, Y.F.; Zhou, Y.; Wan, S.M.; Mo, C.Y.; Jiang, X.Y. Over-expression of a Sesuvium portulacastrum salt overly sensitive gene (SpSOS1) enhances salt tolerance of transgenic Arabidopsis. Mol. Plant Breed. 2017, 15, 1347–1353. [Google Scholar]
- Che, B.N.; Cheng, C.; Fang, J.J.; Liu, Y.M.; Jiang, L.; Yu, B.Y. The Recretohalophyte Tamarix TrSOS1 Gene Confers Enhanced Salt Tolerance to Transgenic Hairy Root Composite Cotton Seedlings Exhibiting Virus-Induced Gene Silencing of GhSOS1. Int. J. Mol. Sci. 2019, 20, 2930. [Google Scholar] [CrossRef] [Green Version]
- Guo, X.Y.; Fan, Y.F.; Zhou, Y.; Yang, Y.; Jiang, X.Y. Isolation and functional analysis of super active mutant gene BgSOS1-3000 in Bruguiera gymnorrhiza. Mol. Plant Breed. 2016, 14, 851–857. [Google Scholar]
- Feki, K.; Quintero, F.J.; Pardo, J.M.; Masmoudi, K. Regulation of durum wheat Na+/H+ exchanger TdSOS1 by phosphorylation. Plant Mol. Biol. 2011, 76, 545–556. [Google Scholar] [CrossRef] [PubMed] [Green Version]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, W.; Li, J.; Dong, J.; Wang, Y.; Xu, L.; Li, K.; Yi, X.; Zhu, Y.; Liu, L. RsSOS1 Responding to Salt Stress Might Be Involved in Regulating Salt Tolerance by Maintaining Na+ Homeostasis in Radish (Raphanus sativus L.). Horticulturae 2021, 7, 458. https://doi.org/10.3390/horticulturae7110458
Zhang W, Li J, Dong J, Wang Y, Xu L, Li K, Yi X, Zhu Y, Liu L. RsSOS1 Responding to Salt Stress Might Be Involved in Regulating Salt Tolerance by Maintaining Na+ Homeostasis in Radish (Raphanus sativus L.). Horticulturae. 2021; 7(11):458. https://doi.org/10.3390/horticulturae7110458
Chicago/Turabian StyleZhang, Wanting, Jingxue Li, Junhui Dong, Yan Wang, Liang Xu, Kexin Li, Xiaofang Yi, Yuelin Zhu, and Liwang Liu. 2021. "RsSOS1 Responding to Salt Stress Might Be Involved in Regulating Salt Tolerance by Maintaining Na+ Homeostasis in Radish (Raphanus sativus L.)" Horticulturae 7, no. 11: 458. https://doi.org/10.3390/horticulturae7110458
APA StyleZhang, W., Li, J., Dong, J., Wang, Y., Xu, L., Li, K., Yi, X., Zhu, Y., & Liu, L. (2021). RsSOS1 Responding to Salt Stress Might Be Involved in Regulating Salt Tolerance by Maintaining Na+ Homeostasis in Radish (Raphanus sativus L.). Horticulturae, 7(11), 458. https://doi.org/10.3390/horticulturae7110458








