Phylogenetic Analysis and Molecular Diversity of Capsicum Based on rDNA-ITS Region
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. DNA Sequence of rDNA-ITS Region
2.3. Sequence Alignment and Phylogenetic Analysis
2.4. Evaluation of Genetic Diversity
3. Results
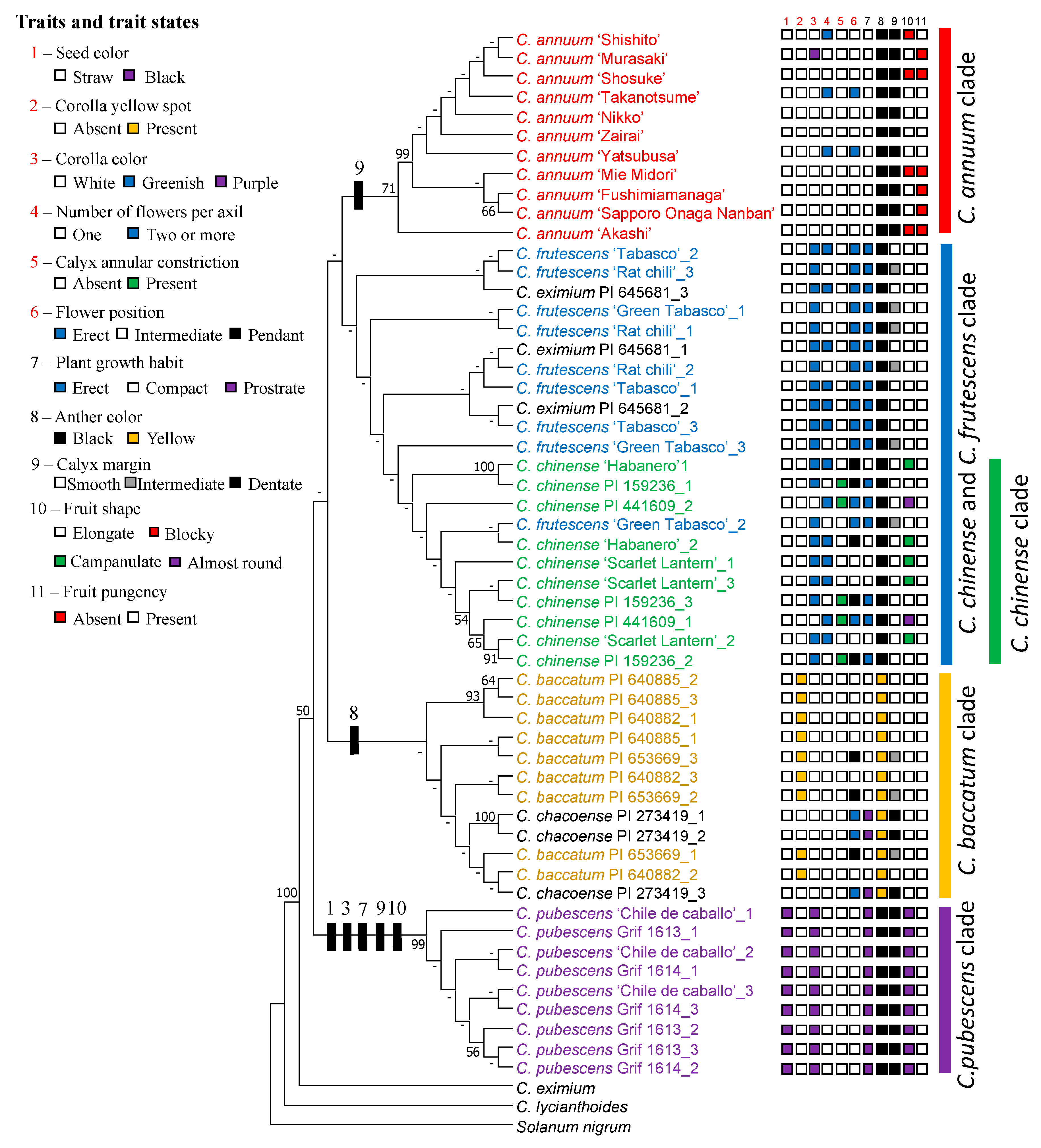
3.1. Morphology Characters of Each Capsicum Species
3.2. Variations of Sequence Length and GC Content, and Genetic Diversity in rDNA-ITS
3.3. Phylogenetic Relationship Between Capsicum Species Based on rDNA-ITS
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Perry, L.; Dickau, R.; Zarrillo, S.; Holst, I.; Pearsall, D.M.; Piperno, D.R.; Berman, M.J.; Cooke, R.G.; Rademaker, K.; Ranere, A.J.; et al. Starch fossils and the domestication and dispersal of chili peppers (Capsicum spp. L.) in the Americas. Science 2007, 315, 986–988. [Google Scholar] [CrossRef] [PubMed]
- FAO. Available online: http://faostat.fao.org/site/567/default.aspx#ancor (accessed on 5 April 2020).
- Kobayashi, A.; Osaka, T.; Namba, Y.; Inoue, S.; Lee, T.H.; Kimura, S. Capsaicin activates heat loss and heat production simultaneously and independently in rats. Am. J. Physiol. Regul. Integr. Comp. Physiol. 1998, 275, R92–R98. [Google Scholar] [CrossRef]
- Bach, F.W.; Yaksh, T.L. Release of β-endorphin immunoreactivity into ventriculo-cisternal perfusate by lumbar intrathecal capsaicin in the rat. Brain Res. 1995, 701, 192–200. [Google Scholar] [CrossRef]
- Watanabe, T.; Kawada, T.; Kato, T.; Harada, T.; Iwai, K. Effects of capsaicin analogs on adrenal catecholamine secretion in rats. Life Sci. 1994, 54, 369–374. [Google Scholar] [CrossRef]
- Carrizo Garcia, C.; Sterpetti, M.; Volpi, P.; Ummarino, M.; Saccardo, F. Wild Capsicums: Identification and in situ analysis of Brazilian species. In Proceedings of the Breakthroughs in the Genetics and Breeding of Capsicum and Eggplant, Torino, Italy, 2–4 September 2013; pp. 205–213. [Google Scholar]
- Heiser, C.B.; Pickersgill, B. Names for the cultivated Capsicum species (Solanaceae). Taxon 1969, 18, 277–283. [Google Scholar] [CrossRef]
- Carrizo García, C.; Barfuss, M.H.J.; Sehr, E.M.; Barboza, G.E.; Samuel, R.; Moscone, E.A.; Ehrendorfer, F. Phylogenetic relationships, diversification and expansion of chili peppers (Capsicum, Solanaceae). Ann. Bot. 2016, 118, 35–51. [Google Scholar] [CrossRef] [PubMed]
- Pickersgill, B. Relationships between weedy and cultivated forms in some species of chili peppers (Genus Capsicum). Evolution 1971, 25, 683–691. [Google Scholar]
- Kraft, K.H.; Brown, C.H.; Nabhan, G.P.; Luedeling, E.; Luna Ruiz, J.d.J.; Coppens d’Eeckenbrugge, G.; Hijmans, R.J.; Gepts, P. Multiple lines of evidence for the origin of domesticated chili pepper, Capsicum annuum in Mexico. Proc. Natl. Acad. Sci. USA 2014, 111, 6165–6170. [Google Scholar] [CrossRef]
- IBPGR. Genetic Resources of Capsicum: A global Plan of Action; IBPGR Executive Secretariat: Rome, Italy, 1983. [Google Scholar]
- Moscone, E.A.; Scaldaferro, M.A.; Grabiele, M.; Cecchini, N.M.; Sánchez García, Y.; Jarret, R.; Daviña, J.R.; Ducasse, D.A.; Barboza, G.E.; Ehrendorfer, F. The evolution of chili peppers (Capsicum—Solanaceae): A cytogenetic perspective. Acta Hortic. 2007, 745, 137–170. [Google Scholar] [CrossRef]
- Tong, N.; Bosland, P.W. Capsicum tovarii, a new member of the Capsicum baccatum complex. Euphytica 1999, 109, 71–77. [Google Scholar] [CrossRef]
- Pickersgill, B. The genus Capsicum—A multidisciplinary approach to the taxonomy of cultivated and wild plants. Biol. Zentralbl. 1988, 107, 381–389. [Google Scholar]
- Ince, A.G.; Karaca, M.; Onus, A.N. Genetic relationships within and between Capsicum species. Biochem. Genet. 2010, 48, 83–95. [Google Scholar] [CrossRef] [PubMed]
- McLeod, M.J.; Guttman, S.I.; Eshbaugh, W.H.; Rayle, R.E. An electrophoretic study of evolution in Capsicum (Solanaceae). Evolution 1983, 37, 562–574. [Google Scholar] [CrossRef] [PubMed]
- Paran, I.; Aftergoot, E.; Shifriss, C. Variation in Capsicum annuum revealed by RAPD and AFLP markers. Euphytica 1998, 99, 167–173. [Google Scholar] [CrossRef]
- Silvar, C.; García González, C.A. Deciphering genetic diversity in the origins of pepper (Capsicum spp.) and comparison with worldwide variability. Crop Sci. 2016, 56, 3100–3111. [Google Scholar] [CrossRef]
- Nicolaï, M.; Cantet, M.; Lefebvre, V.; Sage-Palloix, A.M.; Palloix, A. Genotyping a large collection of pepper (Capsicum spp.) with SSR loci brings new evidence for the wild origin of cultivated C. annuum and the structuring of genetic diversity by human selection of cultivar types. Genet. Resour. Crop Evol. 2013, 60, 2375–2390. [Google Scholar] [CrossRef]
- Jeong, H.J.; Jo, Y.D.; Park, S.W.; Kang, B.C. Identification of Capsicum species using SNP markers based on high resolution melting analysis. Genome 2010, 53, 1029–1040. [Google Scholar] [CrossRef]
- Hollingsworth, P.M.; Graham, S.W.; Little, D.P. Choosing and using a plant DNA barcode. PLoS ONE 2011, 6, e19254. [Google Scholar] [CrossRef]
- Purkayastha, J.; Alam, S.I.; Gogoi, H.K.; Singh, L.; Veer, V. Molecular characterization of ‘Bhut Jolokia’ the hottest chilli. J. Biosci. 2012, 37, 757–768. [Google Scholar] [CrossRef]
- Sun, Y.L.; Choi, I.L.; Lee, Y.B.; Choi, K.Y.; Hong, S.K.; Kang, H.M. Molecular diversity and phylogentic analysis of Capsicum annuum varieties using the nrDNA ITS region. Sci. Hortic. 2014, 165, 336–343. [Google Scholar] [CrossRef]
- Kehie, M.; Kumaria, S.; Devi, K.S.; Tandon, P. Genetic diversity and molecular evolution of Naga King Chili inferred from internal transcribed spacer sequence of nuclear ribosomal DNA. Meta Gene 2016, 7, 56–63. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.W.; Li, Y.; Wang, H.M.; Xu, X.; Li, T.; Luo, S. Analysis on the internal transcribed spacers (ITS) sequences and phylogenetic of pepper. Acta Hortic. Sin. 2014, 41, 881–888. [Google Scholar]
- IPGRI; AVRDC. Descriptors for Capsicum (Capsicum spp.); International Plant Genetic Resources Institute: Washington, DC, USA, 1995; p. 100. [Google Scholar]
- Whitson, M.; Manos, P.S. Untangling Physalis (Solanaceae) from the Physaloids: A two-gene phylogeny of the Physalinae. Syst. Bot. 2005, 30, 216–230. [Google Scholar] [CrossRef]
- Smith, S.D.; Baum, D.A. Phylogenetics of the florally diverse Andean clade Iochrominae (Solanaceae). Am. J. Bot. 2006, 93, 1140–1153. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.L.; Han, J.P. Identification of medicinal plants using DNA barcoding technique. Wiley Online Libr. 2009, (unpublished). [Google Scholar]
- Murray, M.G.; Thompson, W.F. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 1980, 8, 4321–4326. [Google Scholar] [CrossRef] [PubMed]
- Sui, Y.H.; Hui, N.B. Acquisition, identification and analysis of an interspecific Capsicum hybrid (C. annuum × C. chinense). J. Hortic. Sci. Biotechnol. 2015, 90, 31–38. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef]
- Nei, M.; Kumar, S. Molecular Evolution and Phylogenetics; Oxford University Press: Oxford, UK, 2000. [Google Scholar]
- Rozas, J.; Ferrer-Mata, A.; Sánchez-DelBarrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sánchez-Gracia, A. DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef]
- Eshbaugh, W.H. The taxonomy of the genus capsicum. In Peppers: Botany, Production and Uses; Russo, V.M., Ed.; CAB International: Oxford, MS, USA, 2012; pp. 14–28. [Google Scholar]
- Hunziker, A.T. Estudios sobre Solanaceae, I. Sinopsis de las species silvestres de Capsicum de Argentina y Paraguay. Darwiniana 1950, 9, 225–247. [Google Scholar]
- Watterson, G.A. On the number of segregating sites in genetical models without recombination. Theor. Popul. Biol. 1975, 7, 256–276. [Google Scholar] [CrossRef]
- Nei, M. Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics 1978, 89, 583–590. [Google Scholar] [PubMed]
- Nelson, J.O.; Watase, G.J.; Warsinger-Pepe, N.; Yamashita, Y.M. Mechanisms of rDNA Copy Number Maintenance. Trends Genet. 2019, 35, 734–742. [Google Scholar] [CrossRef] [PubMed]
- Elder, J.F.; Turner, B.J. Concerted evolution of repetitive DNA sequences in Eukaryotes. Q. Rev. Biol. 1995, 70, 297–320. [Google Scholar] [CrossRef] [PubMed]
- Wei, X.X.; Wang, X.Q.; Hong, D.Y. Marked intragenomic heterogeneity and gographical differentiation of nrDNA ITS in Larix potaninii (Pinaceae). J. Mol. Evol. 2003, 57, 623–635. [Google Scholar] [CrossRef]
- Won, H.; Renner, S.S. The internal transcribed spacer of nuclear ribosomal DNA in the gymnosperm Gnetum. Mol. Phylogenet. Evol. 2005, 36, 581–597. [Google Scholar] [CrossRef]
- Muir, G.; Fleming, C.C.; Schlötterer, C. Three divergent rDNA clusters predate the species divergence in Quercus petraea (Matt.) Liebl. and Quercus robur L. Mol. Biol. Evol. 2001, 18, 112–119. [Google Scholar] [CrossRef]
- Wei, X.X.; Wang, X.Q. Recolonization and radiation in Larix (Pinaceae): Evidence from nuclear ribosomal DNA paralogues. Mol. Ecol. 2004, 13, 3115–3123. [Google Scholar] [CrossRef]
- Colonna, V.; D’Agostino, N.; Garrison, E.; Albrechtsen, A.; Meisner, J.; Facchiano, A.; Cardi, T.; Tripodi, P. Genomic diversity and novel genome-wide association with fruit morphology in Capsicum, from 746k polymorphic sites. Sci. Rep. 2019, 9, 10067. [Google Scholar] [CrossRef]
- Csilléry, G. Pepper taxonomy and the botanical description of the species. Acta. Agron. Hung. 2006, 54, 151–166. [Google Scholar] [CrossRef]
- Carvalho, S.; Ragassi, C.; Bianchetti, L.; Reifschneider, F.; Buso, G.; Faleiro, F. Morphological and genetic relationships between wild and domesticated forms of peppers (Capsicum frutescens L. and C. chinense Jacquin). Genet. Mol. Res. 2014, 13, 7447–7464. [Google Scholar] [CrossRef] [PubMed]
- Heiser, C.B.; Smith, P.G. New species of Capsicum from South America. Brittonia 1958, 10, 194–201. [Google Scholar] [CrossRef]
- Datta, S.; Das, L. Characterization and genetic variability analysis in Capsicum annuum L. germplasm. SAARC J. Agric. 2014, 11, 91–103. [Google Scholar] [CrossRef]
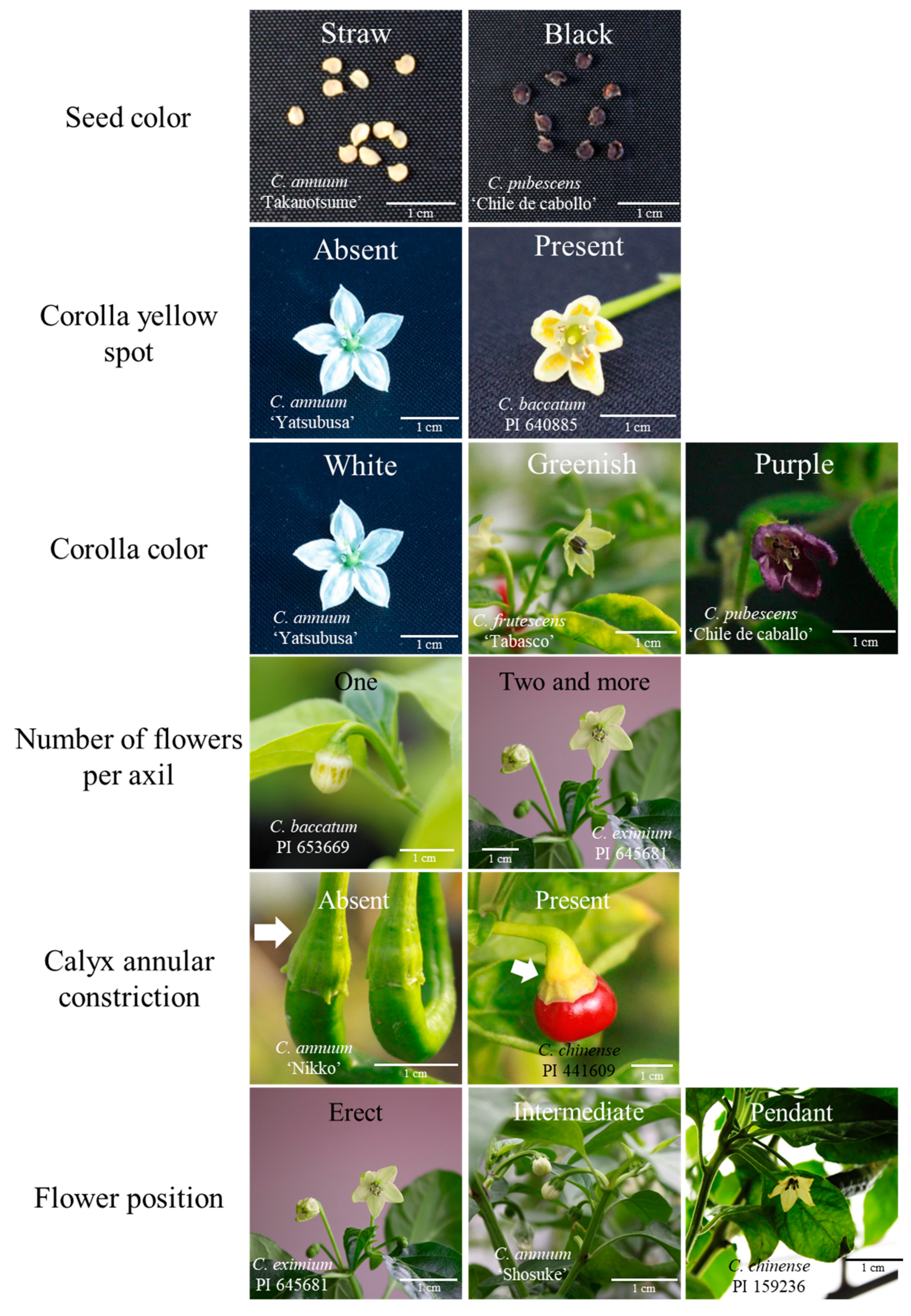

| Species | Accession No. | Cultivar and Line Name | Origin | Sources | rDNA-ITS Accession No. | Reference of rDNA-ITS |
|---|---|---|---|---|---|---|
| C. annuum | JP 32511 | Sapporo Onaga Nanban | Japan | NARO a | LC510550 | This study |
| JP 32520 | Mie Midori | Japan | NARO a | LC510551 | This study | |
| JP 32523 | Akashi | Japan | NARO a | LC510552 | This study | |
| JP 32549 | Yatsubusa | Japan | NARO a | LC510553 | This study | |
| JP 32555 | Zairai | Japan | NARO a | LC510554 | This study | |
| JP 32562 | Nikko | Japan | NARO a | LC510555 | This study | |
| JP 32566 | Fushimiamanaga | Japan | NARO a | LC510556 | This study | |
| JP 82498 | Takanotsume | Japan | NARO a | LC510557 | This study | |
| JP 123787 | Shosuke | Japan | NARO a | LC510558 | This study | |
| JP 124339 | Murasaki | Japan | NARO a | LC510559 | This study | |
| PI 640723 | Shishito | Japan | NARO a | LC510560 | This study | |
| C. chinense | PI 159236 | USA | USDA b | LC510561-510563 | This study | |
| PI 315008 | Scarlet Lantern | Peru | USDA b | LC510564-510566 | This study | |
| PI 438614 | Habanero | Mexico | USDA b | LC510567-510568 | This study | |
| PI 441609 | Brazil | USDA b | LC510569-510570 | This study | ||
| C. frutescens | PI 439512 | Rat chili | Mexico | USDA b | LC510571-510573 | This study |
| PI 586675 | Tabasco | USA | USDA b | LC510574-510576 | This study | |
| PI 634826 | Greenleaf Tabasco | USA | USDA b | LC510577-510579 | This study | |
| C. baccatum | PI 640882 | Peru | USDA b | LC510580-510582 | This study | |
| PI 640885 | India | USDA b | LC510583-510585 | This study | ||
| PI 653669 | Colombia | USDA b | LC510586-510588 | This study | ||
| C. pubescens | PI 593624 | Chile de caballo | Guatemala | USDA b | LC510589-510591 | This study |
| Grif 1613 | Unknown | USDA b | LC510592-510594 | This study | ||
| Grif 1614 | Mexico | USDA b | LC510595-510597 | This study | ||
| C. chacoense | PI 273419 | Argentina | USDA b | LC510598-510600 | This study | |
| C. eximium | PI 645681 | Australia | USDA b | LC510601-510603 | This study | |
| Mexico | AY665841 | [27] | ||||
| C. lycianthoides | USA | DQ314158 | [28] | |||
| Solanum nigrum | China | FJ980391 | [29] |
| Species | Number of Plant Materials | Number of Sequences a | Number of Haplotypes | Haplotype Diversity | Number of Mutation Nucleotides | Nucleotide Polymorphism (θw) | Average Number of Nucleotide Differences | Nucleotide Diversity (π) |
|---|---|---|---|---|---|---|---|---|
| C. annuum | 11 | 11 | 4 | 0.49091 | 9 | 0.00808 | 1.78182 | 0.00476 |
| C. chinense | 4 | 10 | 9 | 0.95556 | 46 | 0.03661 | 14.55556 | 0.03239 |
| C. frutescens | 3 | 9 | 9 | 1 | 150 | 0.13385 | 40.47222 | 0.09922 |
| C. baccatum | 3 | 9 | 9 | 1 | 110 | 0.10795 | 34.11111 | 0.09187 |
| C. pubescens | 3 | 9 | 9 | 1 | 74 | 0.06188 | 18.52778 | 0.04449 |
| C. chacoense | 1 | 3 | 3 | 1 | 84 | n.d. | 56 | n.d. |
| C. eximium | 1 | 3 | 3 | 1 | 49 | n.d. | 32.66667 | n.d. |
| Total | 26 | 57 | 48 | 0.981 | 279 | 0.14439 | 34.323 | 0.08192 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shiragaki, K.; Yokoi, S.; Tezuka, T. Phylogenetic Analysis and Molecular Diversity of Capsicum Based on rDNA-ITS Region. Horticulturae 2020, 6, 87. https://doi.org/10.3390/horticulturae6040087
Shiragaki K, Yokoi S, Tezuka T. Phylogenetic Analysis and Molecular Diversity of Capsicum Based on rDNA-ITS Region. Horticulturae. 2020; 6(4):87. https://doi.org/10.3390/horticulturae6040087
Chicago/Turabian StyleShiragaki, Kumpei, Shuji Yokoi, and Takahiro Tezuka. 2020. "Phylogenetic Analysis and Molecular Diversity of Capsicum Based on rDNA-ITS Region" Horticulturae 6, no. 4: 87. https://doi.org/10.3390/horticulturae6040087
APA StyleShiragaki, K., Yokoi, S., & Tezuka, T. (2020). Phylogenetic Analysis and Molecular Diversity of Capsicum Based on rDNA-ITS Region. Horticulturae, 6(4), 87. https://doi.org/10.3390/horticulturae6040087






