Genome-Wide Characterization of Cucumber (Cucumis sativus L.) GRAS Genes and Their Response to Various Abiotic Stresses
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Identification of the CsGRAS Genes
2.3. Phylogenetic Analysis of CsGRAS Genes
2.4. Protein Property, Gene Structure, and Conserved Motif Analysis
2.5. Chromosomal Mapping and Synteny Analysis
2.6. Promoter Cis-Elements Analysis
2.7. Prediction of the CsGRAS Protein–Protein Interaction Network
2.8. Expression Profile Analysis of CsGRAS Genes via RNA-Seq Data
2.9. qRT-PCR Gene Expression Analysis
2.10. RNA Extraction and Quantitative Real-Time PCR Analysis
3. Results
3.1. Syntenic Relations of the GRAS Gene Family in Cucumber, Arabidopsis, and Tomato
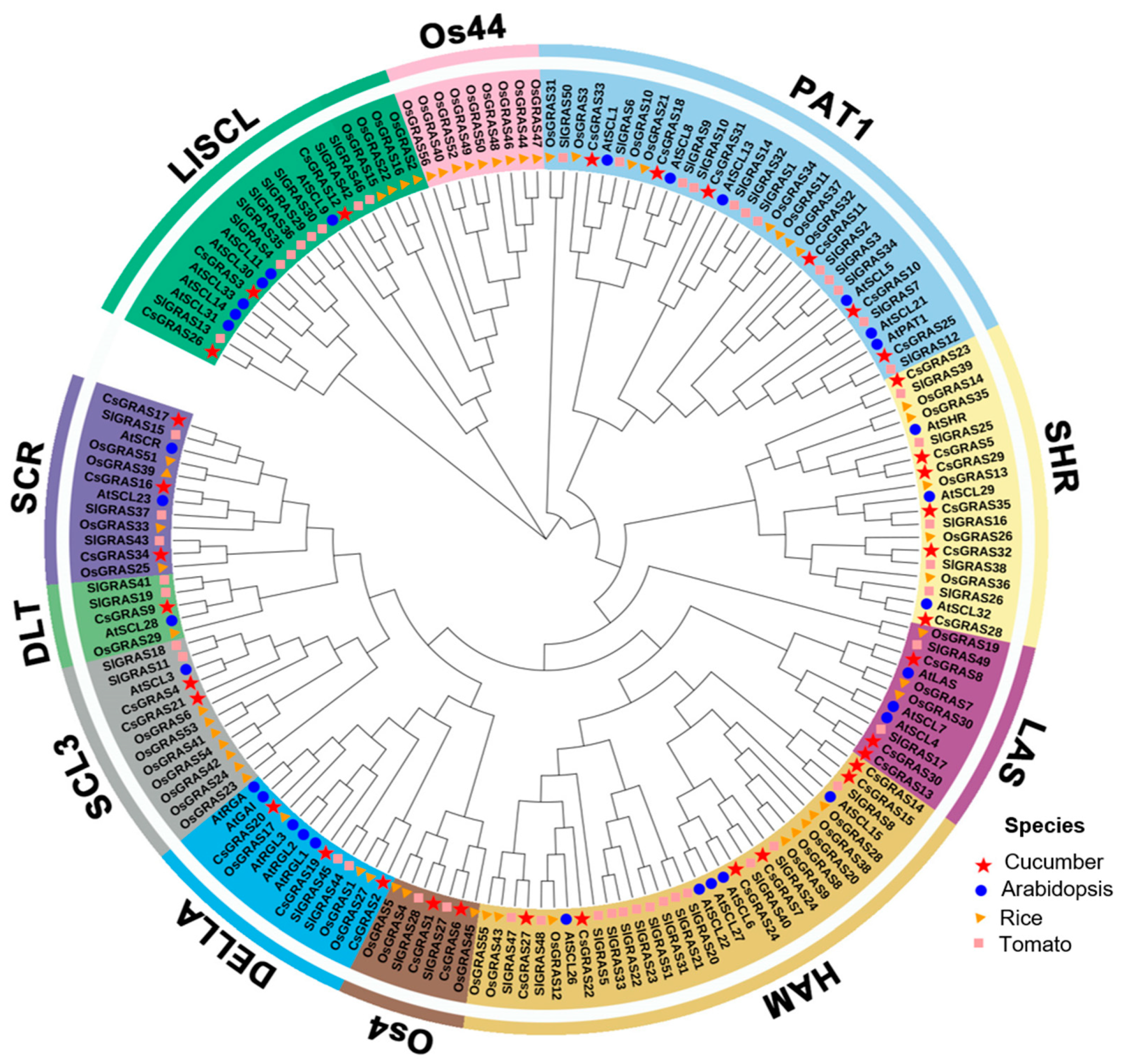
3.2. Phylogenetic and Structural Analysis of the Cucumber GRAS Genes
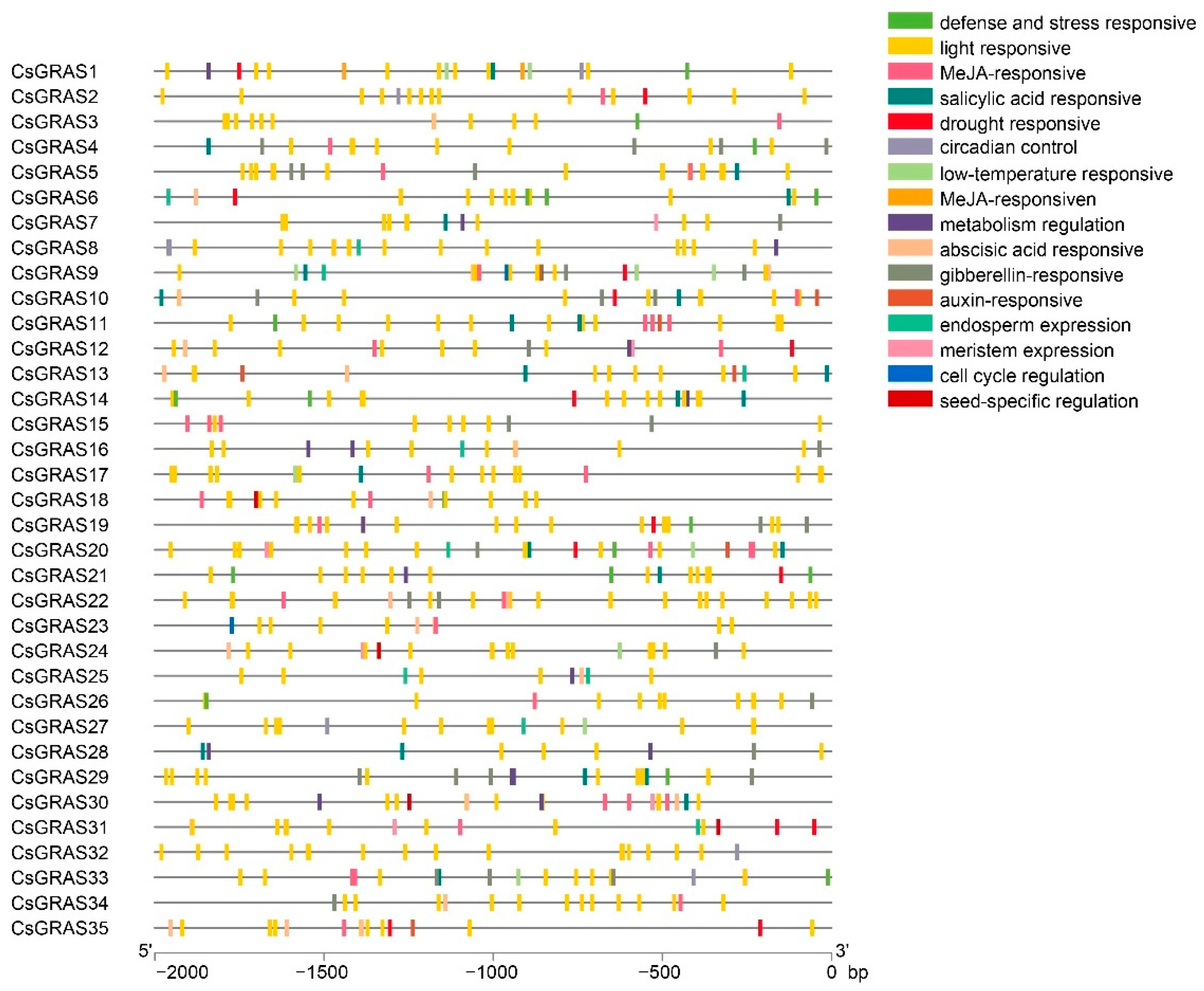
3.3. Analysis of Cis-Elements in Promoters and Interaction Network of CsGRAS Family
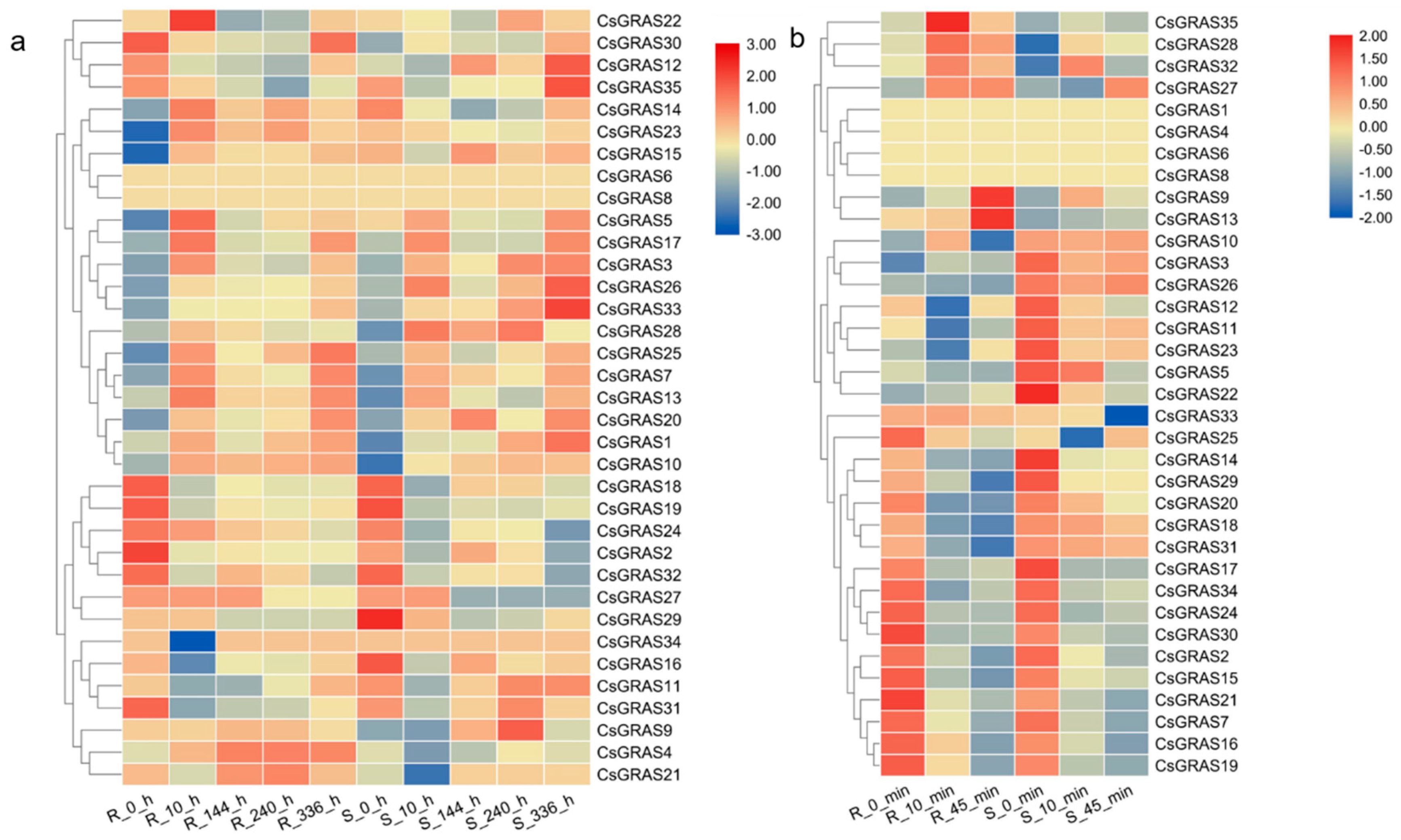
3.4. Expression Profiles of the Cucumber GRAS Genes under Various Abiotic Stresses
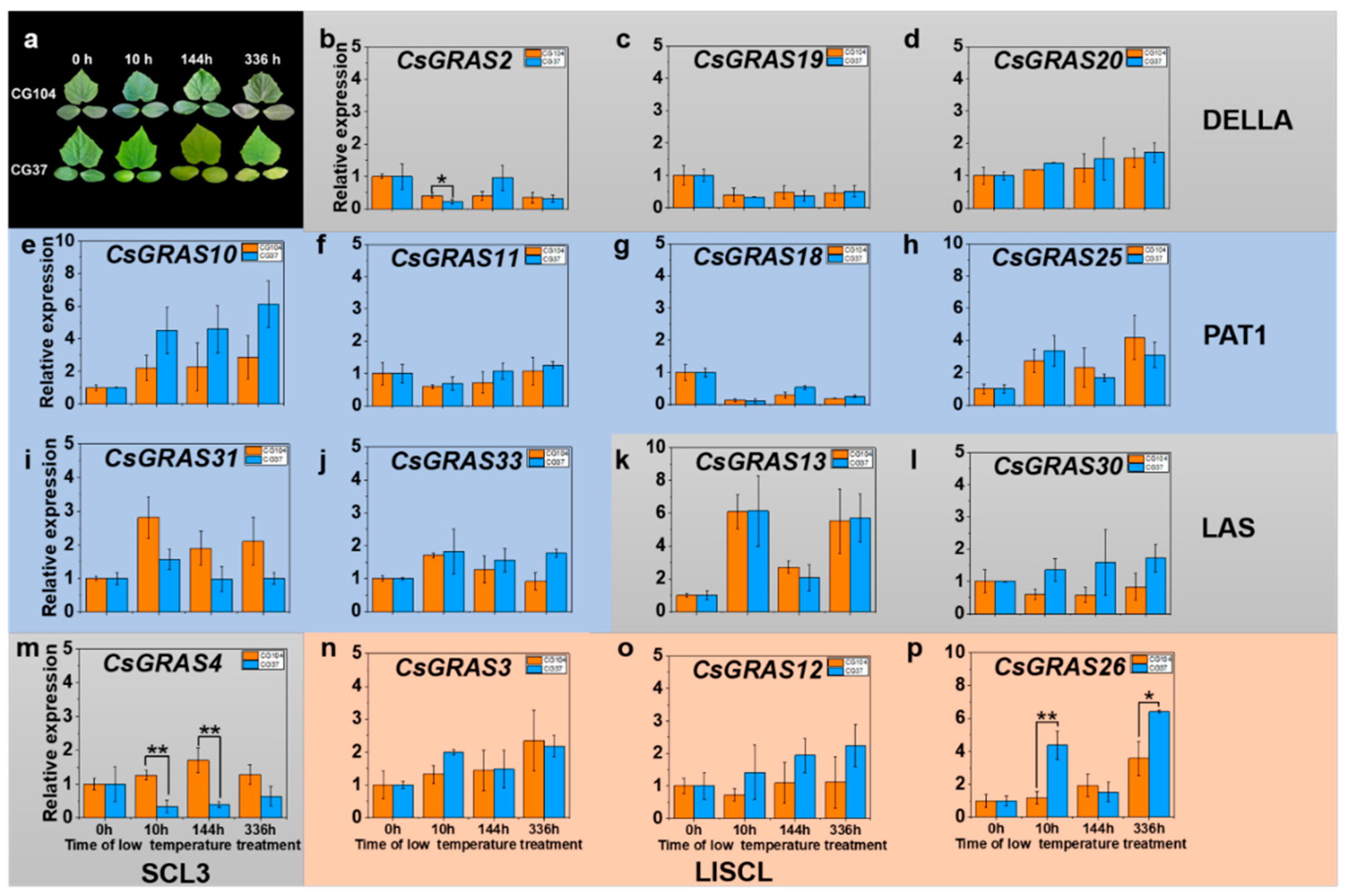
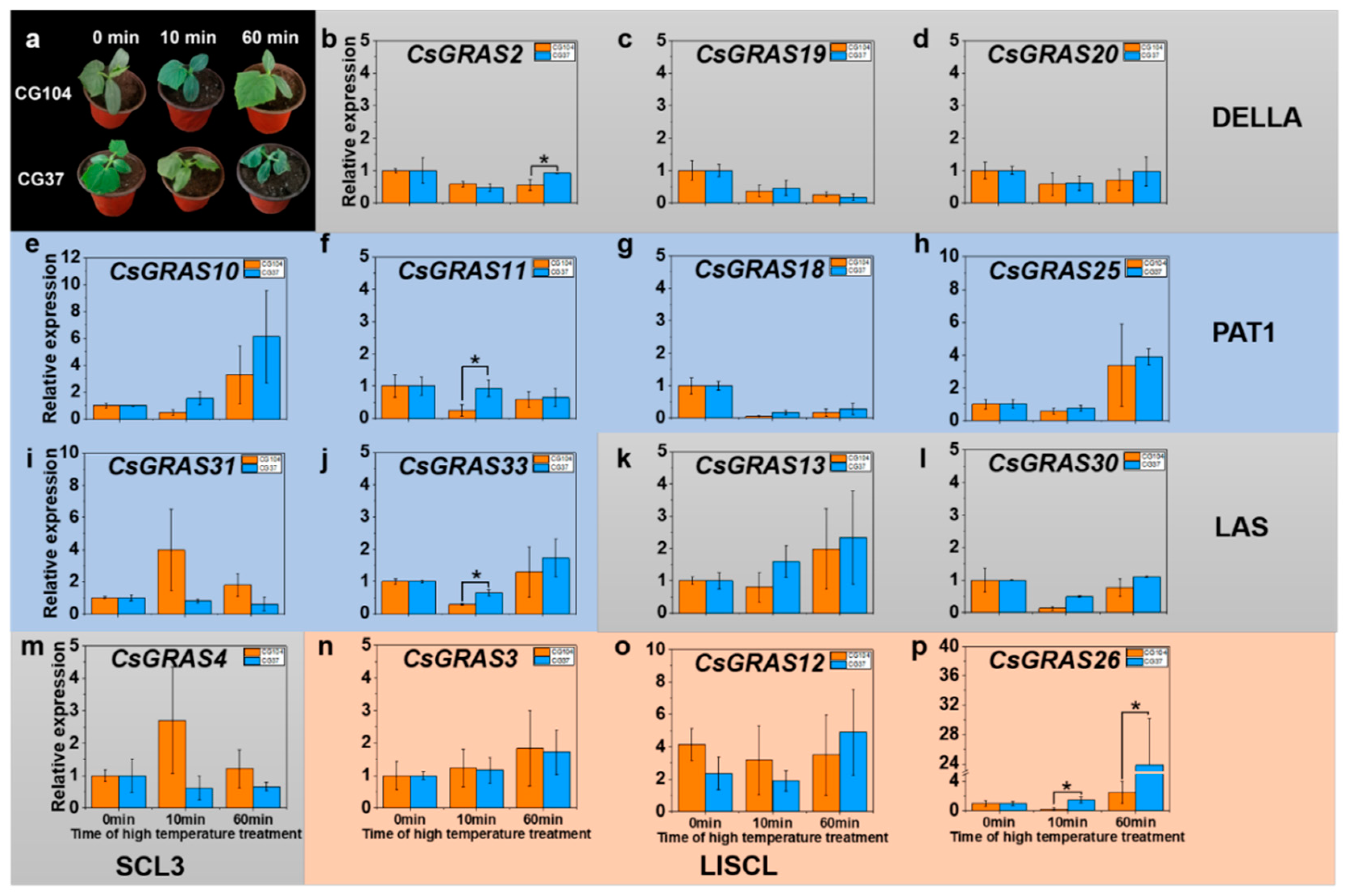
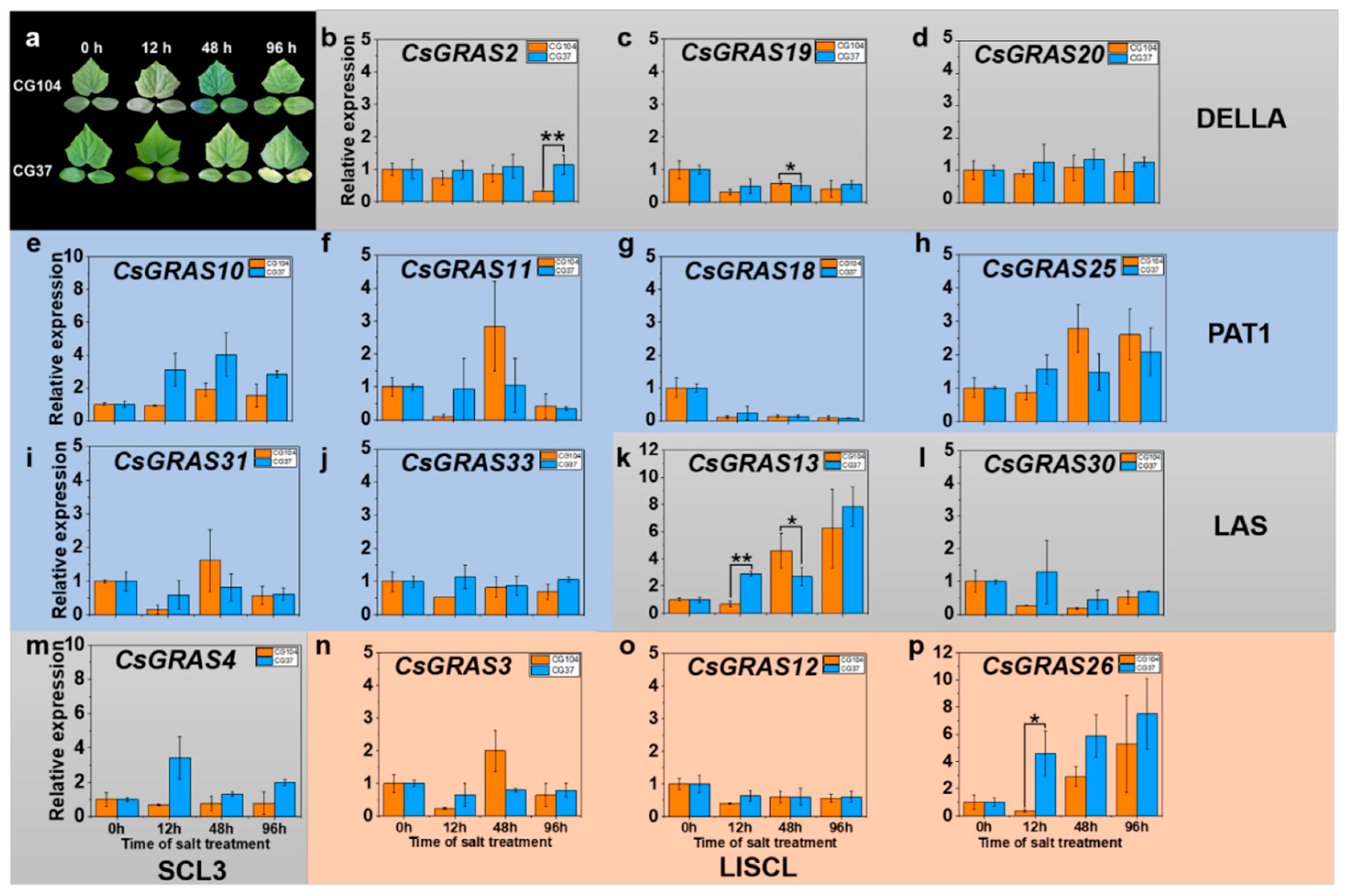
3.5. Expression Pattern of GRAS Genes under Various Abiotic Stresses in “CG104” and “CG37”
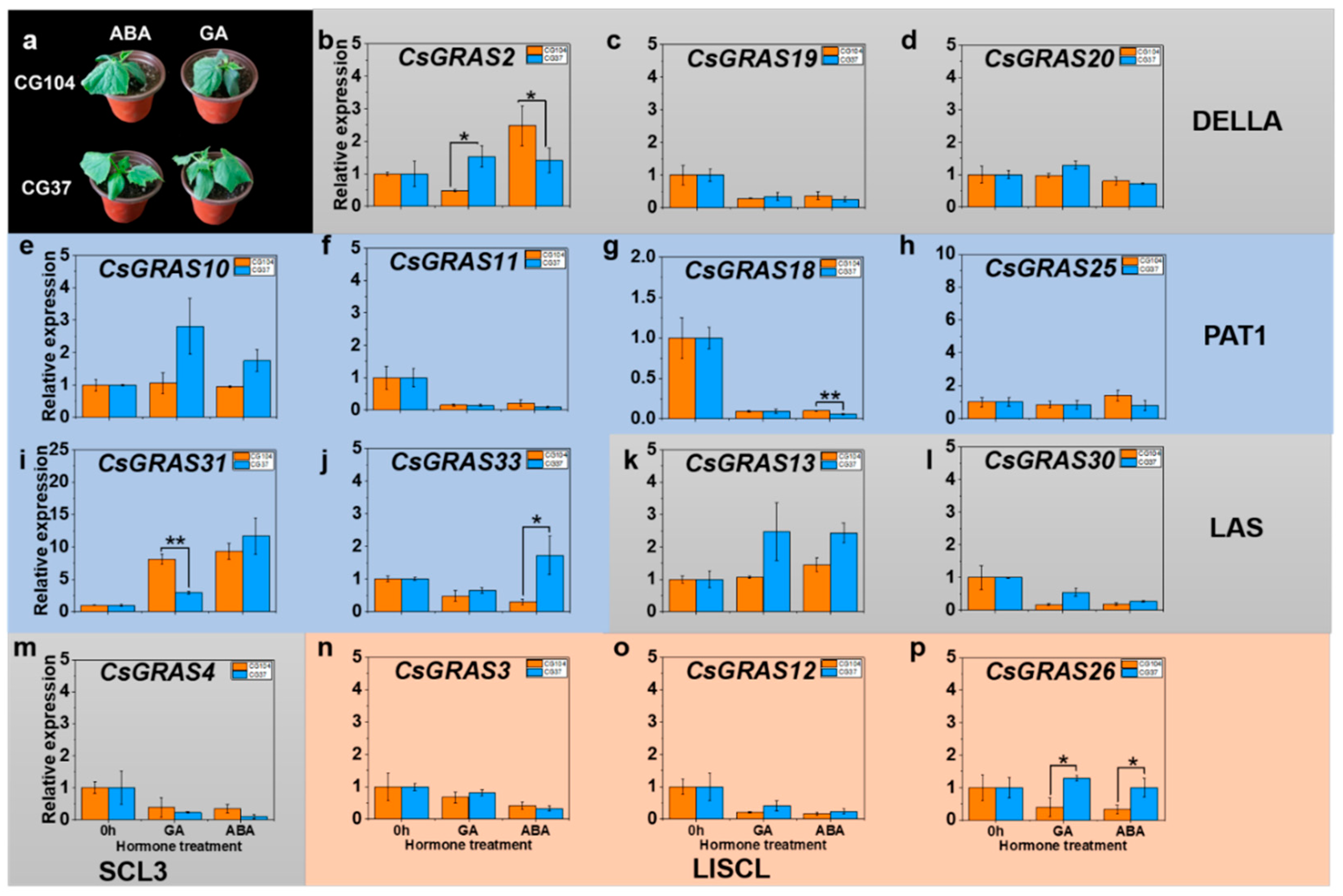
3.6. Expression Patterns of GRAS Genes after Exogenous Phytohormone Treatment in “CG104” and “CG37
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Casaretto, J.; Ho, T.-H.D. The transcription factors HvABI5 and HvVP1 are required for the abscisic acid induction of gene expression in barley aleurone cells. Plant Cell 2003, 15, 271–284. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.-K. Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol. 2002, 53, 247–273. [Google Scholar] [CrossRef] [PubMed]
- Ling, J.; Jiang, W.; Zhang, Y.; Yu, H.; Mao, Z.; Gu, X.; Huang, S.; Xie, B. Genome-wide analysis of WRKY gene family in Cucumis sativus. BMC Genom. 2011, 12, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Lei, N.; Yu, X.; Li, S.; Zeng, C.; Zou, L.; Liao, W.; Peng, M. Phylogeny and expression pattern analysis of TCP transcription factors in cassava seedlings exposed to cold and/or drought stress. Sci. Rep. 2017, 7, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Hu, W.; Yang, H.; Yan, Y.; Wei, Y.; Tie, W.; Ding, Z.; Zuo, J.; Peng, M.; Li, K. Genome-wide characterization and analysis of bZIP transcription factor gene family related to abiotic stress in cassava. Sci. Rep. 2016, 6, 22783. [Google Scholar] [CrossRef]
- Peng, X.; Hui, L.; Dan, W.; Shen, S. Genome-wide identification of the Jatropha curcas MYB family and functional analysis of the abiotic stress responsive gene JcMYB2. BMC Genom. 2016, 17, 251. [Google Scholar] [CrossRef]
- Zhuo, X.; Zheng, T.; Zhang, Z.; Zhang, Y.; Jiang, L.; Ahmad, S.; Sun, L.; Wang, J.; Cheng, T.; Zhang, Q. Genome-Wide Analysis of the NAC Transcription Factor Gene Family Reveals Differential Expression Patterns and Cold-Stress Responses in the Woody Plant Prunus mume. Genes 2018, 9, 494. [Google Scholar] [CrossRef]
- Liu, B.; Sun, Y.; Xue, J.; Jia, X.; Li, R. Genome-wide characterization and expression analysis of GRAS gene family in pepper (Capsicum annuum L.). PeerJ 2018, 6, e4796. [Google Scholar] [CrossRef]
- Peng, J.; Carol, P.; Richards, D.E.; King, K.E.; Cowling, R.J.; Murphy, G.P.; Harberd, N.P. The Arabidopsis GAI gene defines a signaling pathway that negatively regulates gibberellin responses. Genes Dev. 1997, 11, 3194–3205. [Google Scholar] [CrossRef]
- Silverstone, A.L.; Ciampaglio, C.N.; Sun, T.-P. The Arabidopsis RGA gene encodes a transcriptional regulator repressing the gibberellin signal transduction pathway. Plant Cell 1998, 10, 155–169. [Google Scholar] [CrossRef]
- Laurenzio, L.D.; Wysocka-Diller, J.; Malamy, J.E.; Pysh, L.; Helariutta, Y.; Freshour, G.; Hahn, M.G.; Feldmann, K.A.; Benfey, P.N. The SCARECROW Gene Regulates an Asymmetric Cell Division That Is Essential for Generating the Radial Organization of the Arabidopsis Root. Cell 1996, 86, 423–433. [Google Scholar] [CrossRef]
- Tian, C.; Wan, P.; Sun, S.; Li, J.; Chen, M. Genome-Wide Analysis of the GRAS Gene Family in Rice andArabidopsis. Plant Mol. Biol. 2004, 54, 519–532. [Google Scholar] [CrossRef] [PubMed]
- Guo, P.; Wen, J.; Yang, J.; Ke, Y.; Du, H. Genome-wide survey and expression analyses of the GRAS gene family in Brassica napus reveals their roles in root development and stress response. Planta 2019, 250, 1051–1072. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.; Xian, Z.; Kang, X.; Tang, N.; Li, Z. Genome-wide identification, phylogeny and expression analysis of GRAS gene family in tomato. BMC Plant Biol. 2015, 15, 1–18. [Google Scholar] [CrossRef]
- Guo, Y.; Hongyu, W.; Xiang, L.; Qi, L.; Xinyan, Z.; Xueqing, D.; Yanrong, A.; Wei, L.; Hailong, A.; Meng-xiang, S. Identification and expression of GRAS family genes in maize (Zea mays L.). PLoS ONE 2017, 12, e0185418. [Google Scholar] [CrossRef]
- Lu, J.; Wang, T.; Xu, Z.; Sun, L.; Zhang, Q. Genome-wide analysis of the GRAS gene family in Prunus mume. Mol. Genet. Genom. 2014, 290, 303–317. [Google Scholar] [CrossRef]
- Sun, X.; Xue, B.; Jones, W.T.; Rikkerink, E.; Dunker, A.K.; Uversky, V.N. A functionally required unfoldome from the plant kingdom: Intrinsically disordered N-terminal domains of GRAS proteins are involved in molecular recognition during plant development. Plant Mol. Biol. 2011, 77, 205–223. [Google Scholar] [CrossRef]
- Liu, X.; Widmer, A. Genome-wide Comparative Analysis of the GRAS Gene Family in Populus, Arabidopsisand Rice. Plant Mol. Biol. Report. 2014, 32, 1129–1145. [Google Scholar] [CrossRef]
- Song, L.; Tao, L.; Cui, H.; Ling, L.; Guo, C. Genome-wide identification and expression analysis of the GRAS family proteins in Medicago truncatula. Acta Physiol. Plant. 2017, 39, 93. [Google Scholar] [CrossRef]
- Alberto, C.; Mathieu, R. Evolutionary Analyses of GRAS Transcription Factors in Angiosperms. Front. Plant Sci. 2017, 8, 273. [Google Scholar] [CrossRef]
- Heo, J.O.; Chang, K.S.; Kim, I.A.; Lee, M.H.; Lee, S.A.; Song, S.K.; Lee, M.M.; Lim, J. Funneling of gibberellin signaling by the GRAS transcription regulator SCARECROW-LIKE 3 in the Arabidopsis root. Proc. Natl. Acad. Sci. USA 2011, 108, 2166–2171. [Google Scholar] [CrossRef] [PubMed]
- VanWallendael, A.; Soltani, A.; Emery, N.C.; Peixoto, M.M.; Olsen, J.; Lowry, D.B. A Molecular View of Plant Local Adaptation: Incorporating Stress-Response Networks. Annu. Rev. Plant Biol. 2019, 70, 559–583. [Google Scholar] [CrossRef] [PubMed]
- Sun, T.P.; Gubler, F. Molecular mechanism of gibberellin signaling in plants. Annu. Rev. Plant Biol. 2004, 55, 197–223. [Google Scholar] [CrossRef] [PubMed]
- Dill, A.; Sun, T.-P. Synergistic derepression of gibberellin signaling by removing RGA and GAI function in Arabidopsis thaliana. Genetics 2001, 159, 777–785. [Google Scholar] [PubMed]
- Zhang, Z.L.; Ogawa, M.; Fleet, C.M.; Zentella, R.; Hu, J.; Heo, J.O.; Lim, J.; Kamiya, Y.; Yamaguchi, S.; Sun, T. SCARECROW-LIKE 3 promotes gibberellin signaling by antagonizing master growth repressor DELLA in Arabidopsis. Proc. Natl. Acad. Sci. USA 2011, 108, 2160–2165. [Google Scholar] [CrossRef] [PubMed]
- Bolle, C.; Koncz, C.; Chua, N.H. PAT1, a new member of the GRAS family, is involved in phytochrome A signal transduction. Genome Res. 2000, 14, 1269–1278. [Google Scholar]
- Torres-Galea, P.; Hirtreiter, B.; Bolle, C. Two GRAS Proteins, SCARECROW-LIKE21 and PHYTOCHROME A SIGNAL TRANSDUCTION1, Function Cooperatively in Phytochrome A Signal Transduction. Plant Physiol. 2012, 161, 291–304. [Google Scholar] [CrossRef]
- Li, X.; Qian, Q.; Fu, Z.; Wang, Y.; Xiong, G.; Zeng, D.; Wang, X.; Liu, X.; Teng, S.; Hiroshi, F.; et al. Control of tillering in rice. Nat. Cell Biol. 2003, 422, 618–621. [Google Scholar] [CrossRef]
- Schumacher, K.; Schmitt, T.; Rossberg, M.; Schmitz, G.; Theres, K. The Lateral suppressor (Ls) gene of tomato encodes a new member of the VHIID protein family. Proc. Natl. Acad. Sci. USA 1999, 96, 290–295. [Google Scholar] [CrossRef]
- Greb, T.; Clarenz, O.; Schäfer, E.; Müller, D.; Herrero, R.; Schmitz, G.; Theres, K. Molecular analysis of the LATERAL SUPPRESSOR gene in Arabidopsis reveals a conserved control mechanism for axillary meristem formation. Genes Dev. 2003, 17, 1175–1187. [Google Scholar] [CrossRef]
- Ma, H.S.; Dan, L.; Peng, S.; Xia, X.L.; Yin, W.L. The salt− and drought−inducible poplar GRAS protein SCL7 confers salt and drought tolerance in Arabidopsis thaliana. J. Exp. Bot. 2010, 61, 4011–4019. [Google Scholar] [CrossRef] [PubMed]
- Park, H.; Jung, W.; Lee, S.; Song, J.; Kwon, S.Y.; Kim, H.R.; Kim, C.W.; Ahn, J.; Cho, H. Use of Heat Stress Responsive Gene Expression Levels for Early Selection of Heat Tolerant Cabbage (Brassica oleracea L.). Int. J. Mol. Sci. 2013, 14, 11871–11894. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.; Saltveit, M.E.; Beckles, D.M. Chilling-stress modifies DNA methylation level in cucumber (Cucumis sativus L.) seedling radicle to regulate elongation rate. Sci. Hortic. 2019, 252, 14–19. [Google Scholar] [CrossRef]
- Smeets, L.; Wehner, T.C. Environmental effects on genetic variation of chilling resistance in cucumber. Sci. Hortic. 2019, 252, 14–19. [Google Scholar] [CrossRef]
- Dong, S.; Wang, W.; Bo, K.; Miao, H.; Song, Z.; Wei, S.; Zhang, S.; Gu, X. Quantitative Trait Loci mapping and candidate gene analysis of low temperature tolerance in cucumber seedlings. Front. Plant Sci. 2019, 10, 1620. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.-C.; Wang, Y.-J. Inheritance of Heat Tolerance in Cucumber. Acta Agric. Boreali—Sin. 2003, 3. Available online: http://en.cnki.com.cn/Article_en/CJFDTotal-HBNB200303024.htm (accessed on 21 December 2020). (In Chinese).
- Yildirim, E.; Turan, M.; Guvenc, I. Effect of foliar salicylic acid applications on growth, chlorophyll, and mineral content of cucumber grown under salt stress. J. Plant Nutr. 2008, 31, 593–612. [Google Scholar] [CrossRef]
- Zhu, J.-K. Abiotic stress signaling and responses in plants. Cell 2016, 167, 313–324. [Google Scholar] [CrossRef]
- Magome, H.; Yamaguchi, S.; Hanada, A.; Kamiya, Y.; Oda, K. dwarf and delayed−flowering 1, a novel Arabidopsis mutant deficient in gibberellin biosynthesis because of overexpression of a putative AP2 transcription factor. Plant J. 2004, 37, 720–729. [Google Scholar] [CrossRef]
- Magome, H.; Yamaguchi, S.; Hanada, A.; Kamiya, Y.; Oda, K. The DDF1 transcriptional activator upregulates expression of a gibberellin−deactivating gene, GA2ox7, under high−salinity stress in Arabidopsis. Plant J. 2008, 56, 613–626. [Google Scholar] [CrossRef]
- Achard, P.; Gong, F.; Cheminant, S.; Alioua, M.; Hedden, P.; Genschik, P. The cold-inducible CBF1 factor–dependent signaling pathway modulates the accumulation of the growth-repressing DELLA proteins via its effect on gibberellin metabolism. Plant Cell 2008, 20, 2117–2129. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Chen, X.; Dong, S.; Jiang, X.; Wang, L.; Yu, J.; Zhou, Y. Crosstalk of PIF4 and DELLA modulates CBF transcript and hormone homeostasis in cold response in tomato. Plant Biotechnol. J. 2020, 18, 1041–1055. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Wang, T.; Ezra, B.; Kezia, B.; Dong, M.; Zhang, Y.; Yang, S.; Cai, Y.; Xue, S.; Weng, Y. Comprehensive analysis of NAC transcription factors and their expression during fruit spine development in cucumber (Cucumis sativus L.). Hortic. Res. 2018, 5, 31. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.Q.; Hu, L.F. Genome-wide analysis of the auxin response factor gene family in cucumber. Genet. Mol. Res. 2013, 12, 4317–4331. [Google Scholar] [CrossRef]
- Hu, L.; Liu, S. Genome-wide analysis of the MADS-box gene family in cucumber. Genome 2012, 55, 245–256. [Google Scholar] [CrossRef]
- Zhou, S.J.; Jing, Z.; Shi, J.L. Genome-wide identification, characterization, and expression analysis of the MLO gene family in Cucumis sativus. Genet. Mol. Res. 2013, 12, 6565–6578. [Google Scholar] [CrossRef]
- Schouten, H.J.; Krauskopf, J.; Visser, R.G.F.; Bai, Y. Identification of candidate genes required for susceptibility to powdery or downy mildew in cucumber. Euphytica 2014, 200, 475–486. [Google Scholar] [CrossRef]
- Huang, S.; Li, R.; Zhang, Z.; Li, L.; Gu, X.; Fan, W.; Lucas, W.J.; Wang, X.; Xie, B.; Ni, P.; et al. The genome of the cucumber, Cucumis sativus L. Nat. Genet. 2009, 41, 1275–1281. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Potter, S.C.; Aurélien, L.; Eddy, S.R.; Youngmi, P.; Rodrigo, L.; Finn, R.D. HMMER web server: 2018 update. Nucleic Acids Res. 2018, 46, W200–W204. [Google Scholar] [CrossRef]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef] [PubMed]
- Sudhir, K.; Glen, S.; Li, M.; Christina, K.; Koichiro, T. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Elisabeth, G.; Alexandre, G.; Christine, H.; Ivan, I.; Appel, R.D.; Amos, B. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003, 31, 3784–3788. [Google Scholar] [CrossRef]
- Bailey, T.L.; Nadya, W.; Chris, M.; Li, W.W. MEME: Discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 2006, 34, W369–W373. [Google Scholar] [CrossRef]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Franceschini, A.; Szklarczyk, D.; Frankild, S.; Kuhn, M.; Jensen, L.J. STRING V9.1: Protein-Protein Interaction Networks, with Increased Coverage and Integration. Nucleic Acids Res. 2012, 41, D808–D815. [Google Scholar] [CrossRef]
- Shannon, P. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; Van Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef]
- Woffelman, C. DNAMAN for Windows; Lynon Biosoft, Institute of Molecular Plant Sciences, Leiden University: Leiden, The Netherlands, 2004. [Google Scholar]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Rieu, I.; Powers, S.J. Real-time quantitative RT-PCR: Design, calculations, and statistics. Plant Cell 2009, 21, 1031–1033. [Google Scholar] [CrossRef]
- Koornneef, M.; Jorna, M.; Brinkhorst-Van der Swan, D.; Karssen, C. The isolation of abscisic acid (ABA) deficient mutants by selection of induced revertants in non-germinating gibberellin sensitive lines of Arabidopsis thaliana (L.) Heynh. Theor. Appl. Genet. 1982, 61, 385–393. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Jiang, Z.; Liu, S.; Lin, R. Interplay between REVEILLE1 and RGA−LIKE2 regulates seed dormancy and germination in Arabidopsis. New Phytol. 2019, 225, 1593–1605. [Google Scholar] [CrossRef] [PubMed]
- Achard, P.; Genschik, P. Releasing the brakes of plant growth: How GAs shutdown DELLA proteins. J. Exp. Bot. 2009, 60, 1085–1092. [Google Scholar] [CrossRef] [PubMed]
- Yao, T.; Bai, S.; Li, M.; Zhang, Y.; He, Y. DELLA contribute to tolerance to nitric oxide stress in Arabidopsis seedlings. Chin. Bull. Bot. 2011, 46, 481–488. [Google Scholar] [CrossRef]
- Xu, K.; Chen, S.; Li, T.; Ma, X.; Liang, X.; Ding, X.; Liu, H.; Luo, L. OsGRAS23, a rice GRAS transcription factor gene, is involved in drought stress response through regulating expression of stress-responsive genes. BMC Plant Biol. 2015, 15, 141. [Google Scholar] [CrossRef]
- Xu, W.; Chen, Z.; Ahmed, N.; Han, B.; Cui, Q.; Liu, A. Genome-wide identification, evolutionary analysis, and stress responses of the GRAS gene family in castor beans. Int. J. Mol. Sci. 2016, 17, 1004. [Google Scholar] [CrossRef]
- Yang, M.; Yang, Q.; Fu, T.; Zhou, Y. Overexpression of the Brassica napusBnLAS gene in Arabidopsis affects plant development and increases drought tolerance. Plant Cell Rep. 2011, 30, 373–388. [Google Scholar] [CrossRef]
- Torres-Galea, P.; Huang, L.-F.; Chua, N.-H.; Bolle, C. The GRAS protein SCL13 is a positive regulator of phytochrome-dependent red light signaling, but can also modulate phytochrome A responses. Mol. Genet. Genom. 2006, 276, 13–30. [Google Scholar] [CrossRef]
- Zhang, H.; Cao, Y.; Shang, C.; Li, J.; Wang, J.; Wu, Z.; Ma, L.; Qi, T.; Fu, C.; Bai, Z. Genome-wide characterization of GRAS family genes in Medicago truncatula reveals their evolutionary dynamics and functional diversification. PLoS ONE 2017, 12, e0185439. [Google Scholar] [CrossRef]
- Colebrook, E.H.; Thomas, S.G.; Phillips, A.L.; Hedden, P. The role of gibberellin signalling in plant responses to abiotic stress. J. Exp. Biol. 2013, 217, 67–75. [Google Scholar] [CrossRef]
- Achard, P.; Renou, J.-P.; Berthomé, R.; Harberd, N.P.; Genschik, P. Plant DELLAs restrain growth and promote survival of adversity by reducing the levels of reactive oxygen species. Curr. Biol. 2008, 18, 656–660. [Google Scholar] [CrossRef] [PubMed]
- Jung, J.-H.; Domijan, M.; Klose, C.; Biswas, S.; Ezer, D.; Gao, M.; Khattak, A.K.; Box, M.S.; Charoensawan, V.; Cortijo, S. Phytochromes function as thermosensors in Arabidopsis. Science 2016, 354, 886–889. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Guo, Z.; Li, H.; Wang, M.; Onac, E.; Zhou, J.; Xia, X.; Shi, K.; Yu, J.; Zhou, Y. Phytochrome A and B function antagonistically to regulate cold tolerance via abscisic acid-dependent jasmonate signaling. Plant Physiol. 2016, 170, 459–471. [Google Scholar] [CrossRef] [PubMed]
- Franklin, K.A.; Whitelam, G.C. Light-quality regulation of freezing tolerance in Arabidopsis thaliana. Nat. Genet. 2007, 39, 1410. [Google Scholar] [CrossRef] [PubMed]
- Jiang, B.; Shi, Y.; Peng, Y.; Jia, Y.; Yan, Y.; Dong, X.; Li, H.; Dong, J.; Li, J.; Gong, Z. Cold-Induced CBF-PIF3 Interaction Enhances Freezing Tolerance by Stabilizing the phyB Thermosensor in Arabidopsis. Mol. Plant 2020, 13, 894–906. [Google Scholar] [CrossRef]
- Bolle, C. The role of GRAS proteins in plant signal transduction and development. Planta 2004, 218, 683–692. [Google Scholar] [CrossRef]
- Tong, H.; Jin, Y.; Liu, W.; Li, F.; Fang, J.; Yin, Y.; Qian, Q.; Zhu, L.; Chu, C. DWARF AND LOW−TILLERING, a new member of the GRAS family, plays positive roles in brassinosteroid signaling in rice. Plant J. 2009, 58, 803–816. [Google Scholar] [CrossRef]
- Pysh, L.D.; Wysocka-Diller, J.W.; Camilleri, C.; Bouchez, D.; Benfey, P.N. The GRAS gene family in Arabidopsis: Sequence characterization and basic expression analysis of the scarecrow-like genes. Plant J. 1999, 18, 111–119. [Google Scholar] [CrossRef]
- Lu, X.; Liu, W.; Xiang, C.; Li, X.; Wang, Q.; Wang, T.; Liu, Z.; Zhang, J.; Gao, L.; Zhang, W. Genome-Wide Characterization of GRAS Family and Their Potential Roles in Cold Tolerance of Cucumber (Cucumis sativus L.). Int. J. Mol. Sci. 2020, 21, 3857. [Google Scholar] [CrossRef]
- Suzuki, N.; Bassil, E.; Hamilton, J.S.; Inupakutika, M.A.; Zandalinas, S.I.; Tripathy, D.; Luo, Y.; Dion, E.; Fukui, G.; Kumazaki, A. ABA is required for plant acclimation to a combination of salt and heat stress. PLoS ONE 2016, 11, e0147625. [Google Scholar] [CrossRef]
- Lim, S.; Park, J.; Lee, N.; Jeong, J.; Toh, S.; Watanabe, A.; Kim, J.; Kang, H.; Kim, D.H.; Kawakami, N.; et al. ABA-insensitive3, ABA-insensitive5, and DELLAs Interact to Activate the Expression of SOMNUS and Other High-Temperature-Inducible Genes in Imbibed Seeds in Arabidopsis. Plant Cell 2013, 25, 4863–4878. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.; Dong, X.; Yu, T.; Shi, X.; Li, Z.; Yang, W.; Widmer, A.; Karrenberg, S. A Single Nucleotide Deletion in Gibberellin20-oxidase1 Causes Alpine Dwarfism in Arabidopsis. Plant Physiol. 2015, 168, 930–937. [Google Scholar] [CrossRef] [PubMed]
- Lantzouni, O.; Alkofer, A.; Falter-Braun, P.; Schwechheimer, C. Growth-regulating factors interact with Dellas and Regulate Growth in Cold Stress. Plant Cell 2020, 32, 1018–1034. [Google Scholar] [CrossRef] [PubMed]
- Conti, L.; Nelis, S.; Zhang, C.; Woodcock, A.; Swarup, R.; Galbiati, M.; Tonelli, C.; Napier, R.; Hedden, P.; Bennett, M.; et al. Small Ubiquitin-like Modifier Protein SUMO Enables Plants to Control Growth Independently of the Phytohormone Gibberellin. Dev. Cell 2014, 28, 102–110. [Google Scholar] [CrossRef]
- Achard, P.; Baghour, M.; Chapple, A.; Hedden, P.; Van Der Straeten, D.; Genschik, P.; Moritz, T.; Harberd, N.P. The plant stress hormone ethylene controls floral transition via DELLA-dependent regulation of floral meristem-identity genes. Proc. Natl. Acad. Sci. USA 2007, 104, 6484–6489. [Google Scholar] [CrossRef]
- Fode, B.; Siemsen, T.; Thurow, C.; Weigel, R.; Gatz, C. The Arabidopsis GRAS Protein SCL14 Interacts with Class II TGA Transcription Factors and Is Essential for the Activation of Stress-Inducible Promoters. Plant Cell 2008, 20, 3122–3135. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, C.; Dong, S.; Liu, X.; Bo, K.; Miao, H.; Beckles, D.M.; Zhang, S.; Gu, X. Genome-Wide Characterization of Cucumber (Cucumis sativus L.) GRAS Genes and Their Response to Various Abiotic Stresses. Horticulturae 2020, 6, 110. https://doi.org/10.3390/horticulturae6040110
Li C, Dong S, Liu X, Bo K, Miao H, Beckles DM, Zhang S, Gu X. Genome-Wide Characterization of Cucumber (Cucumis sativus L.) GRAS Genes and Their Response to Various Abiotic Stresses. Horticulturae. 2020; 6(4):110. https://doi.org/10.3390/horticulturae6040110
Chicago/Turabian StyleLi, Caixia, Shaoyun Dong, Xiaoping Liu, Kailiang Bo, Han Miao, Diane M. Beckles, Shengping Zhang, and Xingfang Gu. 2020. "Genome-Wide Characterization of Cucumber (Cucumis sativus L.) GRAS Genes and Their Response to Various Abiotic Stresses" Horticulturae 6, no. 4: 110. https://doi.org/10.3390/horticulturae6040110
APA StyleLi, C., Dong, S., Liu, X., Bo, K., Miao, H., Beckles, D. M., Zhang, S., & Gu, X. (2020). Genome-Wide Characterization of Cucumber (Cucumis sativus L.) GRAS Genes and Their Response to Various Abiotic Stresses. Horticulturae, 6(4), 110. https://doi.org/10.3390/horticulturae6040110





