Mealybug Wilt of Pineapple and Associated Viruses
Abstract
1. Mealybug Wilt of Pineapple
1.1. A History of Pineapple Production in Hawaii
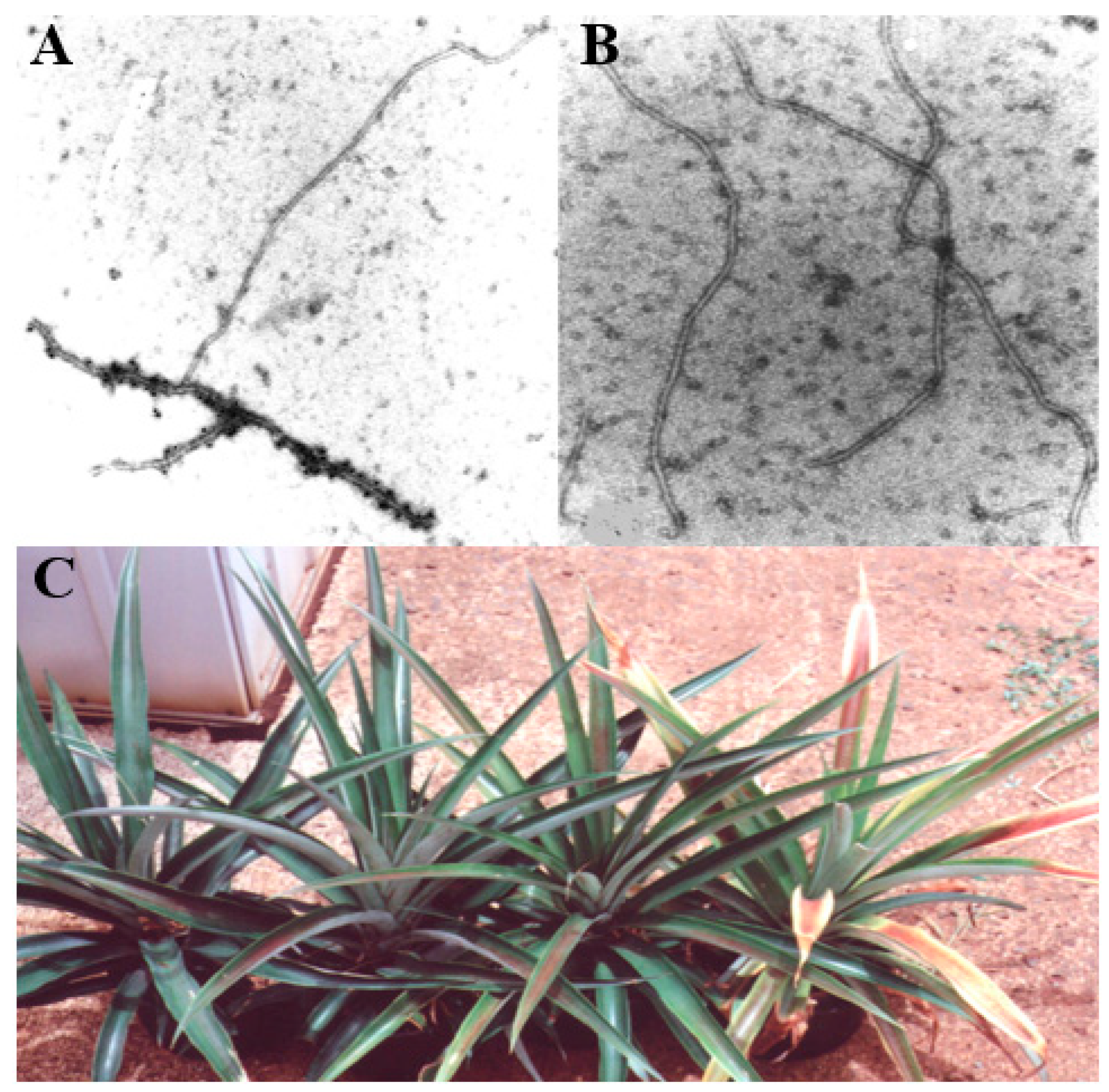
1.2. Disease and Symptoms of Mealybug Wilt of Pineapple
2. Etiology of MWP
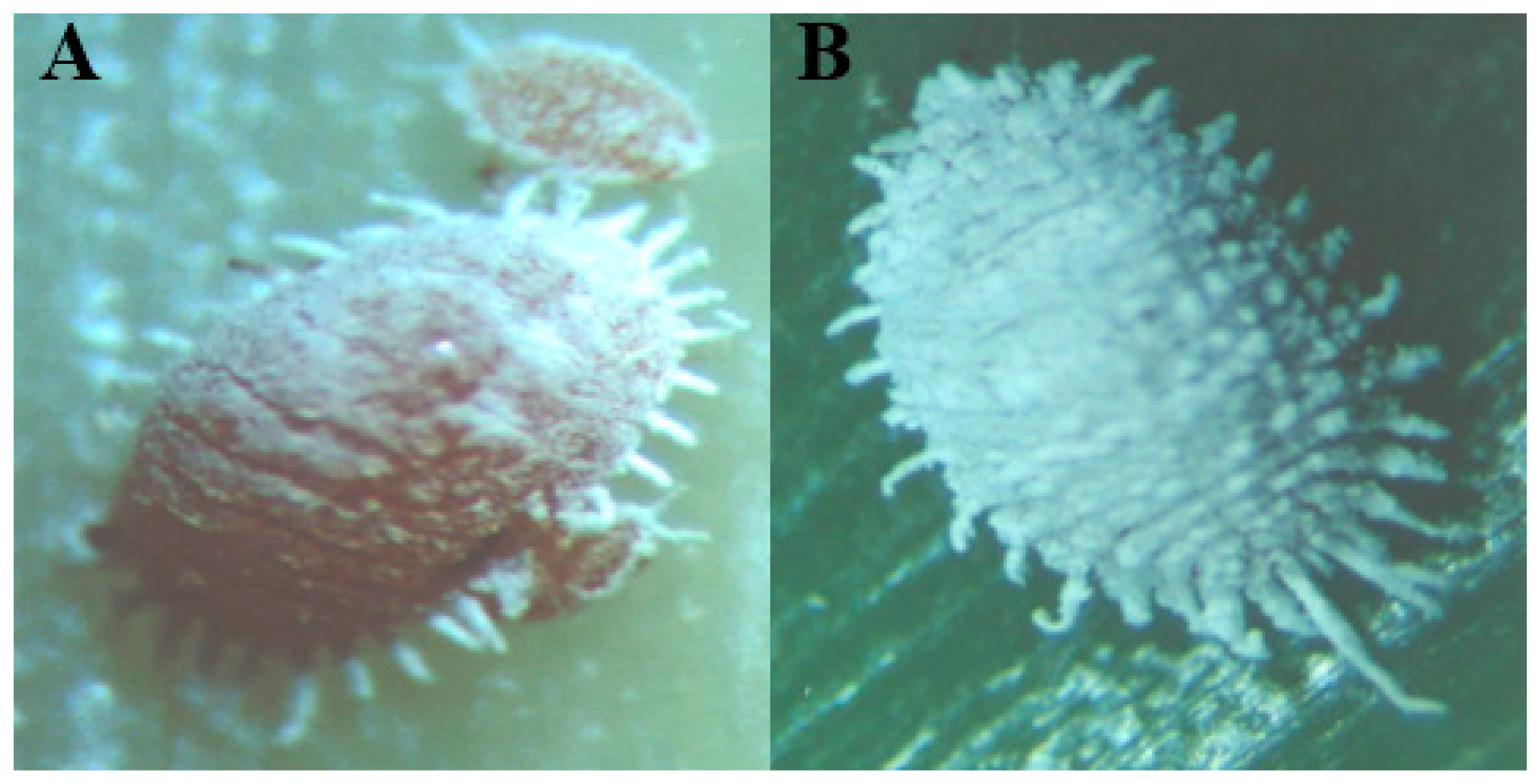
2.1. Association of Mealybug Vectors to MWP
2.2. Association of Ants to Mealybugs and MWP
2.3. Association of Viral Particles to MWP
2.4. Transmission and Interaction of PMWaVs
2.5. Detection of PMWaVs
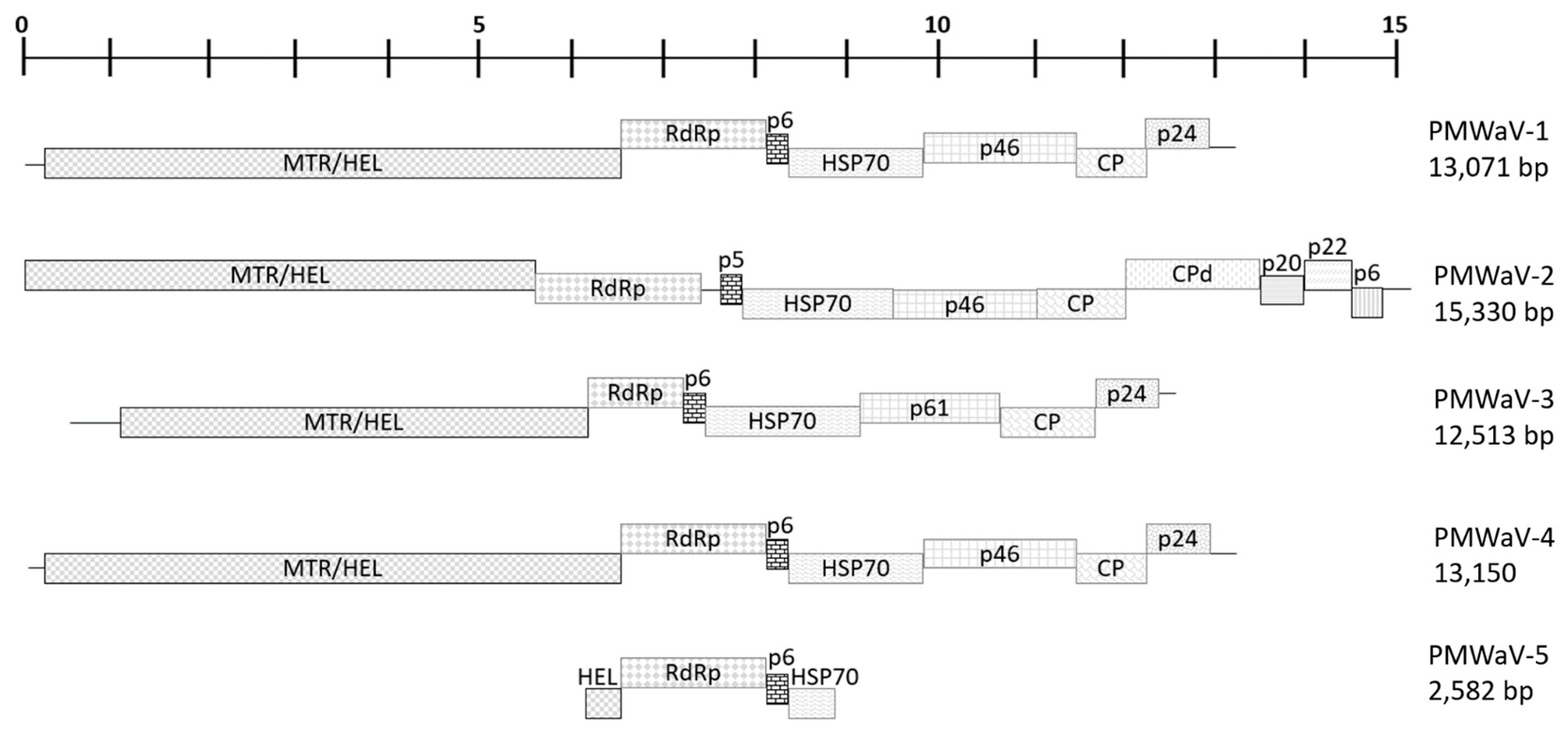
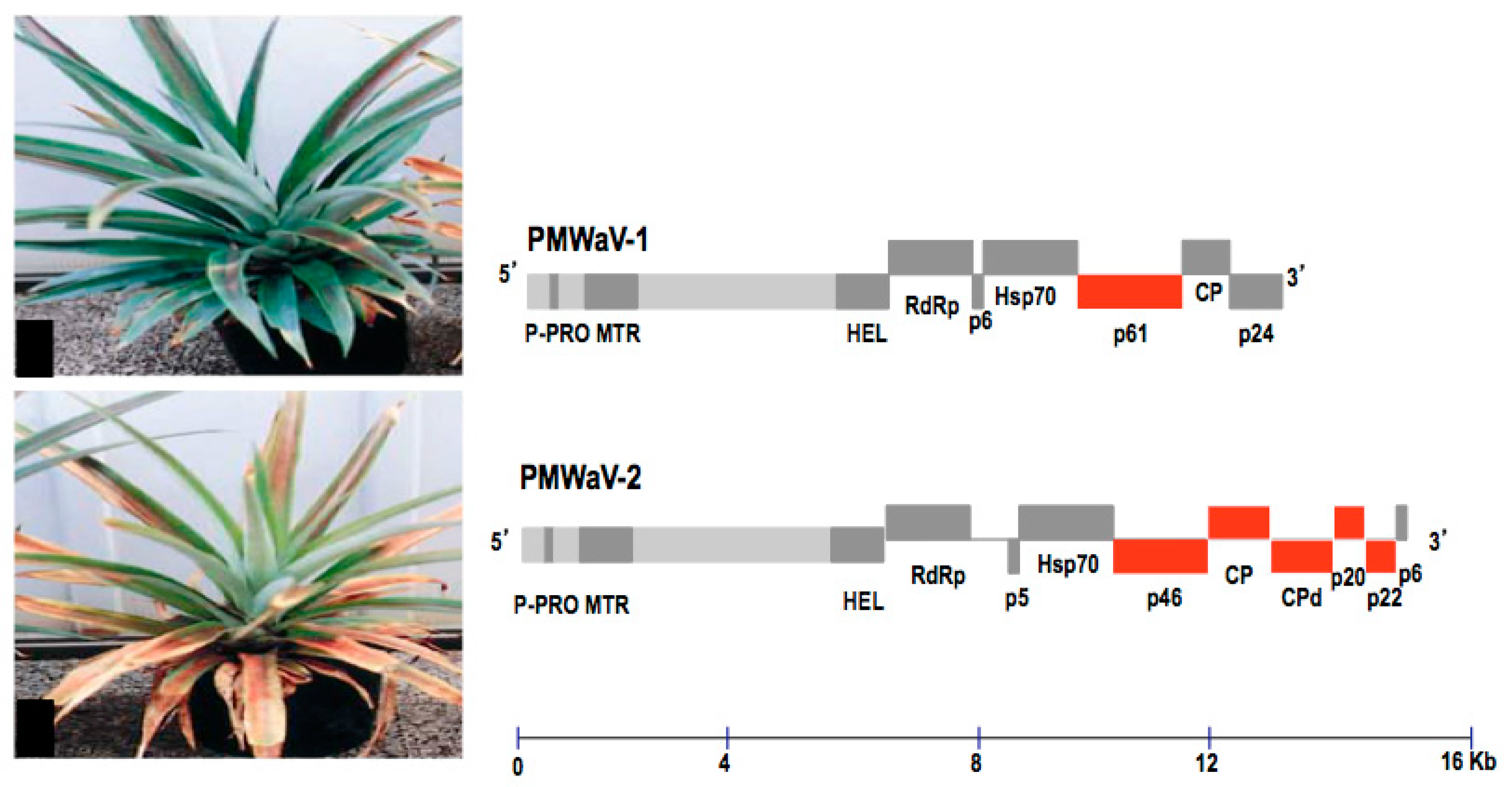
3. Genome Organization and Diversity of PMWaVs
3.1. PMWaV-2
3.2. PMWaV-1 and -3
3.3. Putative PMWaV-4
3.4. Putative PMWaV-5
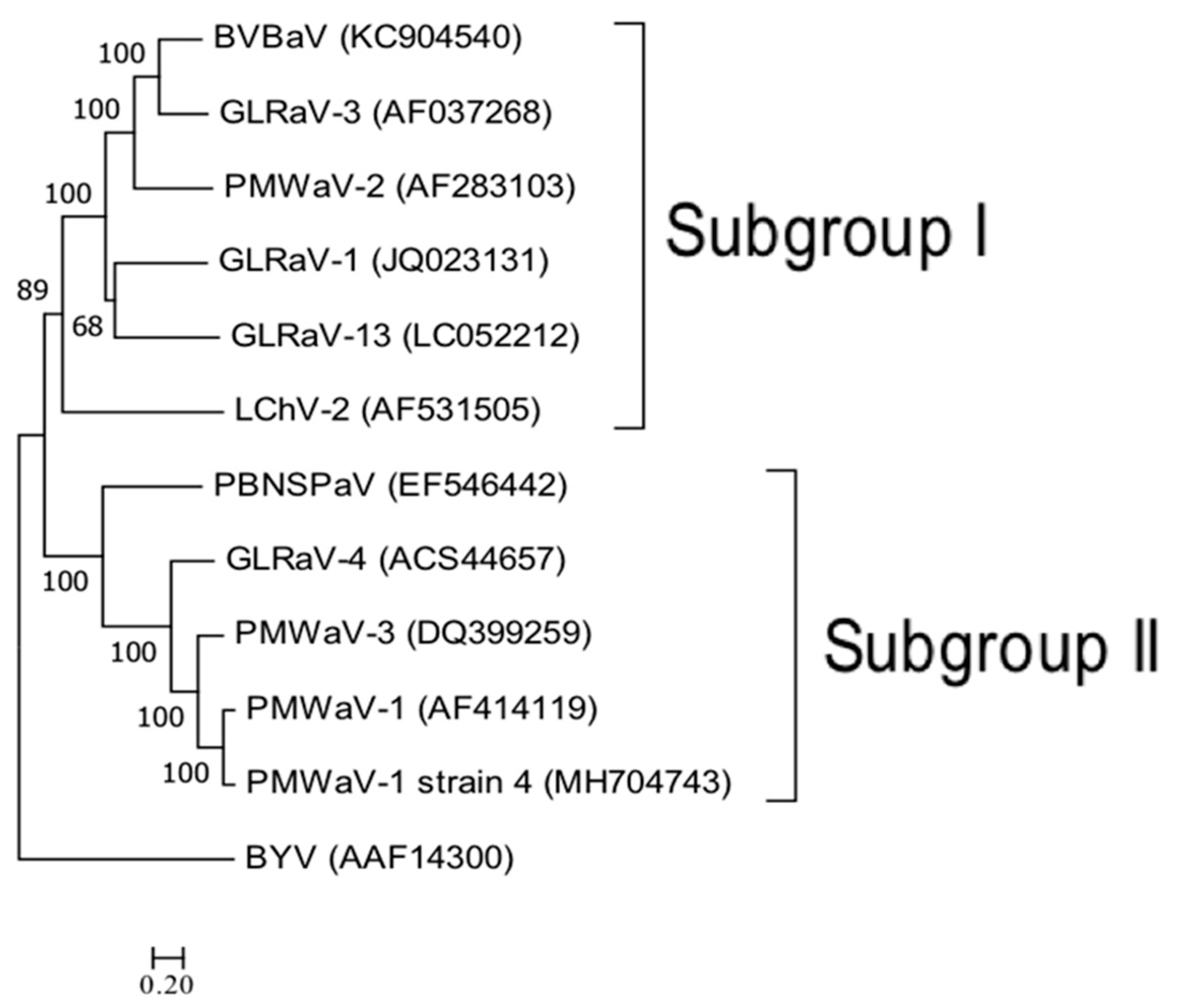
3.5. Phylogenetic Analysis of PMWaV Members
4. Role of RNA-Silencing Suppressors in the Etiology of MWP
5. Badnaviruses Infecting Pineapple
5.1. Badnaviruses
5.2. Synergism of Badnaviruses
5.3. Pineapple Bacilliform Viruses
6. Control of MWP
7. Conclusions and Future Perspectives
Author Contributions
Funding
Conflicts of Interest
References
- Krauss, B.H. Anatomy of the vegetative organs of the pineapple, Ananas comosus (L.) Merr Merr. I. Introduction, organography, the stem, and the lateral branch or axillary buds. Bot. Gazette 1948, 110, 159–217. [Google Scholar] [CrossRef]
- Samson, J.A. Pineapple. In Tropical Fruits. Gordon Wrigley; Logman Scientic and Technical; Longman Inc.: New York, NY, USA, 1986; pp. 190–215. [Google Scholar]
- Collins, J.L. The Pineapple; Leonard Hill Books Ltd.: London, UK, 1960. [Google Scholar]
- Larsen, J.L.; Marks, T.A. Hawaiian Pineapple Entrepreneurs 1894–2010; Creative Co.: McMinnville, OR, USA, 2010. [Google Scholar]
- Gortner, W.A. Field-Fresh Pineapple for Export; Pineapple Research Institute of Hawaii: Honolulu, HI, USA, 1963; PRI no.99. [Google Scholar]
- Bartholomew, D. MD-2’pineapple transforms the world’s pineapple fresh fruit export industry. Pineapple News 2009, 16, 2–5. [Google Scholar]
- Larsen, L.D. Diseases of pineapple. Hawaii Sugar Planters Association Pathol. Physiol. Ser. Exp. Stn. Bull. 1910, 10, 1–72. [Google Scholar]
- Rohrbach, K.G.; Beardsley, J.; German, T.; Reimer, N.; Sanford, W. Mealybug wilt, mealybugs, and ants of pineapple. Plant Dis. 1988, 72, 558–565. [Google Scholar] [CrossRef]
- German, T.L.; Ullman, D.E.; Gunashinghe, U. Advances in Disease Vector Research; Springer: Berlin, Germany, 1992; pp. 241–259. [Google Scholar]
- Carter, W. The pineapple mealy bug, Pseudococcus brevipes, and wilt of pineapples. Phytopathology 1933, 23, 207–242. [Google Scholar]
- Singh, S.; Sastry, K. Wilt of pineapple-a new virus disease in India. Indian Phytopathol. 1974, 27, 298–303. [Google Scholar]
- Carter, W. Some etiological aspects of mealybug wilt. Phytopathology 1944, 35, 305–315. [Google Scholar]
- Carter, W.; Collins, J. Resistance to mealybug wilt of pineapple with special reference to a Cayenne-Queen hybrid. Phytopathology 1947, 37, 332–348. [Google Scholar]
- Carter, W. Insects and Related Pests of Pineapple in Hawaii; A Manual for Fieldman; Pineapple Research Institute of Hawaii: Honolulu, HI, USA, 1967; pp. 36–53. [Google Scholar]
- Lyon, H. A survey of the pineapple problems. Hawaii Plant Rec. 1915, 13, 125–139. [Google Scholar]
- Williams, D.; Fleisch, H. Historical review of pineapple breeding in Hawaii. In Proceedings of the 1st International Pineapple Symposium, Honolulu, HI, USA, 2–6 November 1992; pp. 67–76. [Google Scholar]
- Ferris, G.F. Atlas of the Scale Insects of North America, V, The Pseudococcidae (Part I); Stanford University Press: Redwood City, CA, USA, 1950. [Google Scholar]
- Ito, K. Studies on the Life History of the Pineapple Mealybug, Pseudococcus brevipes (Ckll.). J. Econ. Entomol. 1983, 31, 291–298. [Google Scholar] [CrossRef]
- Beardsley, J.W. On the taxonomy of pineapple mealybugs in Hawaii, with a description of a previously unnamed species (Homoptera: Pseudococcidae). Proc. Hawaiian Entomol. Soc. 1959, 17, 29–37. [Google Scholar]
- Carter, W. Insects in Relation to Plant Disease; Interscience Publishers (John Wiley & Sons): New York, London, UK, 1962. [Google Scholar]
- Carter, W. Studies of populations of Pseudococcus brevipes (Ckl.) occurring on pineapple plants. Ecology 1932, 13, 296–304. [Google Scholar] [CrossRef]
- Collins, J.; Carter, W. Wilt resistant mutations in the Cayenne variety of pineapple. Phytopathology 1954, 44, 662–666. [Google Scholar]
- Beardsley, J.W. Notes on the pineapple mealybug complex, with descriptions of two new species (Homoptera: Pseudocoecidae). Proc. Hawaiian Entomol. Soc. 1965, 19, 55–68. [Google Scholar]
- Jahn, G.C. Gray Pineapple Mealybugs, Dysmicoccus neobrevipes Beardsley (Homoptera: Pseudococcidae), Inside Closed Pineapple Blossom Cups; University of Hawaii at Manoa: Honolulu, HI, USA, 1993. [Google Scholar]
- Carter, W. Studies on the biological control of Pseudococcus brevipes and wilt of pineapple. Phytopathology 1935, 28, 1037–1041. [Google Scholar]
- McKenzie, H.L. Mealybugs of California: With Taxonomy, Biology, and Control of North American Species (Homoptera, Coccoidea, Pseudococcidae); University of California Press: Berkeley, CA, USA, 1967. [Google Scholar]
- Gonzalez-Hernandez, H. The Status of Biological Control of Pineapple Mealybugs in Hawaii. Ph.D. Thesis, University of Hawaii at Manoa, Honolulu, HI, USA, 1995. [Google Scholar]
- Petty, G. The Pineapple Mealybug; Pineapple H.15; Farming in South Africa, Department of Agricultural Technical Services: South Africa, 1978.
- Carter, W. A Study of Mealybug Populations (Dysmicoccus brevipes (Ckl.)) in an Ant-Free Field1. J. Econ. Entomol. 1960, 53, 296–299. [Google Scholar] [CrossRef]
- Jahn, G.C. The Ecological Significance of the Big-Headed Ant in Mealybug Wilt Disease of Pineapple. Ph.D. Thesis, University of Hawaii at Manoa, Honolulu, HI, USA, 1992. [Google Scholar]
- Fluker, S.S.; Beardsley, J.W. Sympatric associations of three ants: Iridomyrmex humilis, Pheidole megacephala, and Anoplolepis longipes in Hawaii. Ann. Entomol. Soc. Am. 1970, 63, 1290–1296. [Google Scholar] [CrossRef]
- Glancey, B.; Reimer, N.; Banks, W. Studies on the effect of the IGR’s fenoxycarb and Sumitomo S-31183 on the queens of two species of myrmicine ants. In Applied Myrmecology: A World Perspective; Westview: Boulder, CO, USA, 1990; pp. 604–614. [Google Scholar]
- Reimer, N.J.; Beardsley, J. Effectiveness of hydramethylnon and fenoxycarb for control of bigheaded ant (Hymenoptera: Formicidae), an ant associated with mealybug wilt of pineapple in Hawaii. J. Econ. Entomol. 1990, 83, 74–80. [Google Scholar] [CrossRef]
- Reimer, N.J. Distribution and impact of alien ants in vulnerable Hawaiian ecosystems. In Exotic Ants: Biology, Impact and Control of Introduced Species; Westview Press: Boulder, CO, USA, 1994; pp. 11–22. [Google Scholar]
- Sether, D.; Hu, J. Closterovirus infection and mealybug exposure are necessary for the development of mealybug wilt of pineapple disease. Phytopathology 2002, 92, 928–935. [Google Scholar] [CrossRef] [PubMed]
- Sether, D.; Hu, J. Yield impact and spread of Pineapple mealybug wilt associated virus-2 and mealybug wilt of pineapple in Hawaii. Plant Dis. 2002, 86, 867–874. [Google Scholar] [CrossRef]
- Bruggencate, J.K.T. Hawai ‘i’s Pineapple Century: A History of the Crowned Fruit in the Hawaiian Islands; Mutual Publishing Co.: Honolulu, HI, USA, 2004. [Google Scholar]
- Rohrbach, K.; Schmitt, D.; Ploetz, R. Diseases of pineapple. In Diseases of Tropical Fruit Crops; University of Florida, IFAS: Homestead, FL, USA, 2003; pp. 443–464. [Google Scholar]
- Illingworth, J.F. Preliminary report on evidence that mealy bugs are an important factor in pineapple wilt. J. Econ. Entol. 1931, 24, 877–889. [Google Scholar] [CrossRef]
- Carter, W. The feeding sequence of pseudococcus brevipes- (ckl) in relation to mealybug wilt of pineapples in hawaii. Phytopathology 1951, 41, 769–780. [Google Scholar]
- Carter, W. Injuries to plants caused by insect toxins. II. The Bot. Rev. 1952, 18, 680–721. [Google Scholar] [CrossRef]
- Carter, W. Mealybug wilt of pineapple; a reappraisal. Ann. N. Y. Acad. Sci. 1963, 105, 741–764. [Google Scholar] [CrossRef]
- Ito, K. Additional immunological evidence supporting the virus nature of mealybug wilt. Pineapple Res. Inst. News 1962, 10, 158–162. [Google Scholar]
- Gunasinghe, U.; German, T. Association of virus-particles with mealybug-wilt of pineapple. Phytopathol. 1986, 76, 1073. [Google Scholar]
- Gunasinghe, U.B.; German, T.L. Further characterization of virus associated with mealybug-wilt of pineapple. Phytopathol. 1987, 77, 1776. [Google Scholar]
- Gunasinghe, U.; German, T. Purification and partial characterization of a virus from pineapple. Phytopathol. 1989, 79, 1337–1341. [Google Scholar] [CrossRef]
- Hu, J.; Gonsalves, A.; Sether, D.; Ullman, D. Detection of pineapple closterovirus, a possible cause of mealybug wilt of pineapple. Acta Hortic. 1992, 334, 411–416. [Google Scholar] [CrossRef]
- HU, J.S.; Wang, M.; Sether, D.; Xie, W.; Leonhardt, K.W. Use of polymerase chain reaction (PCR) to study transmission of banana bunchy top virus by the banana aphid (Pentalonia nigronervosa). Ann. Appl. Biol. 1996, 128, 55–64. [Google Scholar] [CrossRef]
- Hu, J.S.; Sether, D.M.; Liu, X.P.; Wang, M.; Zee, F.; Ullman, D.E. Use of a tissue blotting immunoassay to examine the distribution of pineapple closterovirus in Hawaii. Plant Dis. 1997, 81, 1150–1154. [Google Scholar] [CrossRef]
- Hu, J.; Sether, D.; Ullman, D. Detection of pineapple closterovirus in pineapple plants and mealybugs using monoclonal antibodies. Plant Pathol. 1996, 45, 829–836. [Google Scholar] [CrossRef]
- Agranovsky, A.A. Principles of molecular organization, expression, and evolution of closteroviruses: Over the barriers. Adv. Virus Res. 1996, 47, 119–158. [Google Scholar] [PubMed]
- Bar-Joseph, M.; Garnsey, S.M.; Gonsalves, D. The Closterovirus: A distinct Group of elongated plant viruses. Adv. Virus Res. 1979, 25, 93–168. [Google Scholar] [PubMed]
- Gambley, C.; Steele, V.; Geering, A.; Thomas, J. The genetic diversity of ampeloviruses in Australian pineapples and their association with mealybug wilt disease. Aust. Plant Pathol. 2008, 37, 95–105. [Google Scholar] [CrossRef]
- Sether, D.; Ullman, D.; Hu, J. Transmission of pineapple mealybug wilt-associated virus by two species of mealybug (Dysmicoccus spp.). Phytopathology 1998, 88, 1224–1230. [Google Scholar] [CrossRef] [PubMed]
- Sether, D.M.; Melzer, M.J.; Borth, W.B.; Hu, J.S. Genome organization and phylogenetic relationship of Pineapple mealybug wilt associated virus-3 with family Closteroviridae members. Virus Genes 2009, 38, 414–420. [Google Scholar] [CrossRef] [PubMed]
- Melzer, M.; Karasev, A.; Sether, D.; Hu, J. Nucleotide sequence, genome organization and phylogenetic analysis of pineapple mealybug wilt-associated virus-2. J. Gen. Virol. 2001, 82, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Melzer, M.; Sether, D.; Karasev, A.; Borth, W.; Hu, J. Complete nucleotide sequence and genome organization of pineapple mealybug wilt-associated virus-1. Arch. Virol. 2008, 153, 707–714. [Google Scholar] [CrossRef] [PubMed]
- Karasev, A.V. Genetic diversity and evolution of closteroviruses. Annu. Rev. Phytopathol. 2000, 38, 293–324. [Google Scholar] [CrossRef] [PubMed]
- Martelli, G.P.; Agranovsky, A.A.; Bar-Joseph, M.; Boscia, D.; Candresse, T.; Coutts, R.H.; Dolja, V.V.; Falk, B.W.; Gonsalves, D.; Jelkmann, W.; et al. The family Closteroviridae revised. Arch. Virol. 2002, 147, 2039–2044. [Google Scholar] [CrossRef] [PubMed]
- Wakman, W.; Teakle, D.; Thomas, J.; Dietzgen, R. Presence of a clostero-like virus and a bacilliform virus in pineapple plants in Australia. Crop Pasture Sci. 1995, 46, 947–958. [Google Scholar] [CrossRef]
- Hu, J.; Sether, D. Etiology of mealybug wilt of pineapple. In Proceedings of the Xth International Congress of Virology, Sydney, Australia, 9–13 August 1999; p. 321. [Google Scholar]
- Sether, D.M.; Karasev, A.V.; Okumura, C.; Arakawa, C.; Zee, F.; Kislan, M.M.; Busto, J.L.; Hu, J.S. Differentiation, distribution, and elimination of two different pineapple mealybug wilt-associated viruses found in pineapple. Plant Dis. 2001, 85, 856–864. [Google Scholar] [CrossRef]
- Hu, J.S.; Sether, D.M.; Metzer, M.J.; Perez, E.; Gonsalves, A.; Karasev, A.V.; Nagai, C. Pineapple mealybug wilt associated virus and mealbug wilt of pineapple. Acta Hortic. 2005. [Google Scholar] [CrossRef]
- Sether, D.M.; Perez, E.; Borth, W.; Melzer, M.J.; Subere, C.V.; Busto, J.L.; Hu, J. Pineapple mealybug wilt associated viruses1,3, and 4, and Grapevine leafroll associated viruses 4,5,6, and 9 are a distinct group in the genus Ampelovirus. In Abstracts of the XIII International Congress of Virology; American Society for Microbiology: Washington, DC, USA, 2005; Volume 89, pp. 121–122. [Google Scholar]
- Sether, D.M.; Melzer, M.J.; Busto, J.; Zee, F.; Hu, J.S. Diversity and Mealybug Transmissibility of Ampeloviruses in Pineapple. Plant Dis. 2005, 89, 450–456. [Google Scholar] [CrossRef]
- Fujiwara, K.; Ikeshiro, T. Detection of Pineapple mealybug wilt-associated virus 1, 2, 3 by LAMP Methods. Res. Bull. Pl. Prot. Jpn. 2017, 53, 33–38. [Google Scholar]
- Piyasak, C.; Peerasak, C. Complete lab on chip system for simple and rapid detection of pineapple mealybug wilt associated viruses. Pineapple News 2010, 17, 24. [Google Scholar]
- Dey, K.K.; Lin, H.; Borth, W.B.; Melzer, M.J.; Hu, J.S. A highly sensitive single-tube nested PCR assay for the detection of Pineapple mealybug wilt associated virus-2 (PMWaV-2). J. Virol. Methods 2012, 183, 215–218. [Google Scholar] [CrossRef] [PubMed]
- Adams, I.P.; Glover, R.H.; Monger, W.A.; Mumford, R.; Jackeviciene, E.; Navalinskiene, M.; Samuitiene, M.; Boonham, N. Next-generation sequencing and metagenomic analysis: A universal diagnostic tool in plant virology. Mol. Plant Pathol. 2009, 10, 537–545. [Google Scholar] [CrossRef] [PubMed]
- Al Rwahnih, M.; Daubert, S.; Golino, D.; Rowhani, A. Deep sequencing analysis of RNAs from a grapevine showing Syrah decline symptoms reveals a multiple virus infection that includes a novel virus. Virology 2009, 387, 395–401. [Google Scholar] [CrossRef] [PubMed]
- Coetzee, B.; Freeborough, M.J.; Maree, H.J.; Celton, J.M.; Rees, D.J.; Burger, J.T. Deep sequencing analysis of viruses infecting grapevines: Virome of a vineyard. Virology 2010, 400, 157–163. [Google Scholar] [CrossRef] [PubMed]
- Green, J.C.; Rwahnih, M.; Velarde, A.O.; Melzer, M.J.; Hamim, I.; Borth, W.B.; Brower, T.M.; Wall, M.; Hu, J.S. Further genomic characterization of pineapple mealybug wilt-associated viruses using high-throughput sequencing. 2018; submitted. [Google Scholar]
- Yu, N.; Luo, Z.; Fan, H.; Zhang, Z.; Li, X.; Wang, J.; Liu, Z.; He, F. Complete genomic sequence of a Pineapple mealybug wilt-associated virus-1 from Hainan Island, China. Eur. J. Plant Pathol. 2015, 141, 611–615. [Google Scholar] [CrossRef]
- Martelli, G.P.; Ghanem-Sabanadzovic, N.A.; Agranovsky, A.A.; Al Rwahnih, M.; Dolja, V.V.; Dovas, C.I.; Fuchs, M.; Gugerli, P.; Hu, J.S.; Jelkmann, W.; et al. Taxonomic revision of the family closteroviridae with special reference to the grapevine leafroll-associated members of the genus ampelovirus and the putative species unassigned to the family. J. Plant Pathol. 2012, 94, 7–19. [Google Scholar]
- Hamilton, A.J.; Baulcombe, D.C. A species of small antisense RNA in posttranscriptional gene silencing in plants. Science 1999, 286, 950–952. [Google Scholar] [CrossRef] [PubMed]
- Dolja, V.V.; Kreuze, J.F.; Valkonen, J. Comparative and functional genomics of closteroviruses. Virus Res. 2006, 117, 38–51. [Google Scholar] [CrossRef] [PubMed]
- Lu, R.; Folimonov, A.; Shintaku, M.; Li, W.X.; Falk, B.W.; Dawson, W.O.; Ding, S.W. Three distinct suppressors of RNA silencing encoded by a 20-kb viral RNA genome. Proc. Natl. Acad. Sci. USA 2004, 101, 15742–15747. [Google Scholar] [CrossRef] [PubMed]
- Cañizares, M.C.; Navas-Castillo, J.; Moriones, E. Multiple suppressors of RNA silencing encoded by both genomic RNAs of the crinivirus, Tomato chlorosis virus. Virology 2008, 379, 168–174. [Google Scholar] [CrossRef] [PubMed]
- Cuellar, W.J.; Tairo, F.; Kreuze, J.F.; Valkonen, J.P. Analysis of gene content in sweet potato chlorotic stunt virus RNA1 reveals the presence of the p22 RNA silencing suppressor in only a few isolates: Implications for viral evolution and synergism. J. Gen. Virol. 2008, 89, 573–582. [Google Scholar] [CrossRef] [PubMed]
- Dey, K.K.; Borth, W.B.; Melzer, M.J.; Wang, M.L.; Hu, J.S. Analysis of pineapple mealybug wilt associated virus-1 and-2 for potential RNA silencing suppressors and pathogenicity factor. Viruses 2015, 7, 969–995. [Google Scholar] [CrossRef] [PubMed]
- Brigneti, G.; Voinnet, O.; Li, W.X.; Ji, L.H.; Ding, S.H.; Baulcombe, D.C. Viral pathogenicity determinants are suppressors of transgenic silencing in Nicotiana benthamiana. EMBO J. 1998, 17, 6739–6746. [Google Scholar] [CrossRef] [PubMed]
- Anandalakshmi, R.; Pruss, G.J.; Ge, X.; Marathe, R.; Mallory, A.C.; Smith, T.H.; Vance, V.B. A viral suppressor of gene silencing in plants. Proc. Natl. Acad. Sci. USA 1998, 22, 13079–13084. [Google Scholar] [CrossRef]
- Dey, K.K. Further Characterization and Detection of Pineapple Mealybug Wilt Associated Viruses (PMWaVs). Ph.D. Desseration, University of Hawai’i at Manoa, Honolulu, HI, USA, 2014. [Google Scholar]
- Hernandez-Rodriguez, L.; Ramos-Gonzalez, P.; Garcia-Garcia, G.; Higginson, E.J.; Zamora-Rodriguez, V. First report of Pineapple bacilliform comosus virus (PBCoV) and endogenous Pineapple pararetrovirus-1 (ePPRV-1) in pineapple plants in Cuba. New Dis. Rep. 2013, 28, 2044-0588. [Google Scholar] [CrossRef]
- Hull, R.; Harper, G.; Lockhart, B. Viral sequences integrated into plant genomes. Trends Plant Sci. 2000, 5, 362–365. [Google Scholar] [CrossRef]
- Vanitharani, R.; Chellappan, P.; Fauquet, C.M. Geminiviruses and RNA silencing. Trends Plant Sci. 2005, 10, 144–151. [Google Scholar] [CrossRef] [PubMed]
- Bhat, A.; Hohn, T.; Selvarajan, R. Badnavirus: The current Global Scenario. Viruses 2016, 8, 177. [Google Scholar] [CrossRef] [PubMed]
- Hohn, T.; Richert-Pöggeler, K.R.; Staginnus, C.; Harper, G.; Schwarzacher, T.; Teo, C.H.; Teycheney, P.Y.; Iskra-Caruana, M.L.; Hull, R. Evolution of integrated plant viruses. In Plant Virus Evolution; Roosinck, M., Ed.; Academic Springer: Heidelberg, Germany, 2008; pp. 54–76. [Google Scholar]
- Staginnus, C.; Iskra-Caruana, M.; Lockhart, B.; Hohn, T.; Pooggeler, K.R. Suggestions for a nomenclature of endogenous pararetroviral sequence in plants. Arch. Virol. 2009, 154, 1189–1193. [Google Scholar] [CrossRef] [PubMed]
- Lheureux, F.; Carreel, F.; Jenny, C.; Lockhart, B.; Iskra-Caruana, M. Identification of genetic markers linked to banana streak disease expression in inter-specific Musa hybrids. Theor. Appl. Genet. 2003, 106, 594–598. [Google Scholar] [CrossRef] [PubMed]
- Cote, F.X.; Galzi, S.; Folliot, M.; Lamagnere, Y.; Teycheney, P.Y.; Iskra-Caruana, M.L. Micropropagation by tissue culture triggers differential expression of infectious endogenous Banana streak virus sequences (eBSV) present in the B genome of natural and synthetic interspecific banana plantains. Mol. Plant Pathol. 2010, 11, 137–144. [Google Scholar] [CrossRef] [PubMed]
- Geering, A.D.; Olszewski, N.E.; Harper, G.; Lockhart, B.E.; Hull, R.; Thomas, J.E. Banana contains a diverse array of endogenous badnaviruses. J. Gen. Virol. 2005, 86, 511–520. [Google Scholar] [CrossRef] [PubMed]
- Gregor, W.; Mette, M.; Staginnus, C.; Matzke, M.; Matzke, A. A distinct endogenous pararetrovirus family in Nicotiana tomentosiformus, a diploid progenitor of polyploidy tobacco. Plant Physiol. 2004, 134, 1191–1199. [Google Scholar] [CrossRef] [PubMed]
- Ndowora, T.; Dahal, G.; LaFleur, D.; Harper, G.; Hull, R.; Olszewski, N.E.; Lockhart, B. Evidence That Badnavirus Infection in Musa Can Originate from Integrated Pararetroviral Sequences. Virology 1999, 255, 214–220. [Google Scholar] [CrossRef] [PubMed]
- Lockhart, B.; Olszewski, N. Serological and genomic heterogeneity of banana streak badnavirus: Implications for virus detection in Musa germplasm. In Breeding Banana and Plantain for Resistance to Diseases and Pests; CIRAD/INIBAP: Montpellier, France, 2013; pp. 105–113. [Google Scholar]
- Daniells, J.; Thomas, J.; Smith, M. Seed transmission of banana streak virus confirmed. Infomusa 1995, 4, 7. [Google Scholar]
- Lockhart, B. Purification and serology of a bacilliform virus associated with banana streak disease. Phytopathology 1986, 76, 995–999. [Google Scholar] [CrossRef]
- Harper, G.; Hull, R. Cloning and sequence analysis of banana streak virus DNA. Virus Genes 1998, 17, 271–278. [Google Scholar] [CrossRef] [PubMed]
- Brunt, A. Cacao swollen shoot virus. CMI/AAB Descr. Plant Viruses 1970, 10, 4. [Google Scholar]
- Lockhart, B.; Autrey, L. Occurrence in sugarcane of a bacilliform virus related serologically to banana streak virus. Plant Dis. 1988, 72, 230–233. [Google Scholar] [CrossRef]
- Harrison, B.; Roberts, I. Association of virus-like particles with internal brown spot of yam (Dioscorea alata). Trop. Agric. Trinidad 1973, 50, 335–340. [Google Scholar]
- Phillips, S.; Briddon, R.; Brunt, A.; Hull, R. The partial characterization of a badnavirus infecting the greater asiatic or water yam (Dioscorea alata). J. Phytopathol. 1999, 147, 265–269. [Google Scholar] [CrossRef]
- Hearon, S.; Locke, J. Graft, pollen, and seed transmission of an agent associated with top spotting in Kalanchoë blossfeldiana. Plant Dis. 1984, 68, 346–350. [Google Scholar] [CrossRef]
- Lockhart, B.E.; Kiratiya-Angul, K.; Jones, P.; Eng, L.; De Silva, P.; Olszewski, N.E.; Lockhart, N.; Deema, N.; Sangalang, J. Identification of Piper yellow mottle virus, a mealybug-transmitted badnavirus infecting Piper spp. in Southeast Asia. Eur. J. Plant Pathol. 1997, 103, 303–311. [Google Scholar] [CrossRef]
- Lockhart, B.E.L.; Olszewski, N.E. A widely distributed mealy-bug-transmitted badnavirus occurring in Scheflera and Aralia. Acta Hortic. 1996, 432, 196–203. [Google Scholar] [CrossRef]
- Huang, Q.; Hartung, J.S. Cloning and sequence analysis of an infectious clone of Citrus yellow mosaic virus that can infect sweet orange via Agrobacterium-mediated inoculation. J. General Virol. 2001, 82, 2549–2558. [Google Scholar] [CrossRef] [PubMed]
- Druka, A.; Hull, R. Variation of rice tungro viruses: Further evidence of two rice tungro bacilliform virus strains and possibly several rice tungro spherical virus variants. J. Phytopathol. 1998, 146, 175–178. [Google Scholar] [CrossRef]
- Gambley, C.F.; Geering, A.D.; Steele, V.; Thomas, J.E. Identification of viral and non-viral reverse transcribing elements in pineapple (Ananas comosus), including members of two new badnavirus species. Arch. Virol. 2008, 153, 1599–1604. [Google Scholar] [CrossRef] [PubMed]
- Thomson, K.; Dietzgen, R.; Thomas, J.; Teakle, D. Detection of pineapple bacilliform virus using the polymerase chain reaction. Ann. Appl. Biol. 1996, 129, 57–69. [Google Scholar] [CrossRef]
- Sether, D.; Melzer, M.; Borth, W.; Hu, J. Pineapple bacilliform CO virus: Diversity, Detection, Distribution, and Transmission. Plant Dis. 2012, 96, 1798–1804. [Google Scholar] [CrossRef]
- Li, Y.; Mo, Y.; Xi, J.; Zhang, L.; Sun, G. Detection of Pineapple mealybug wilt-associated viruses with RT-PCR. Chin. J. Trop. Crops 2010, 31, 1003–1008. (In Chinese) [Google Scholar]
- Subere, C.V.Q. Characterization of Pineapple Mealybug Wilt Associated Viruses in Hawaii; University of Hawaii at Manoa: Honolulu, HI, USA, 2009. [Google Scholar]
- Hughes, W.O.; Howse, P.E.; Vilela, E.F.; Knapp, J.J.; Goulson, D. Field evaluation of potential of alarm pheromone compounds to enhance baits for control of grass-cutting ants (Hymenoptera: Formicidae). J. Econ. Entomol. 2002, 95, 537–543. [Google Scholar] [CrossRef] [PubMed]
- Jouvenaz, D.; Lofgren, C.; Banks, W. Biological control of imported fire ants: A review of current knowledge. Bull. ESA 1981, 27, 203–209. [Google Scholar] [CrossRef]
- D’Eeckenbrugge, G.C.; Sanewski, G.M.; Smith, M.K.; Duval, M.; Leal, F. Wild Crop Relatives: Genomic and Breeding Resources; Springer: Berlin, Germany, 2011; pp. 21–41. [Google Scholar]
- Ko, L.; Eccleston, K.; O’Hare, T.; Wong, L.; Giles, J.; Smith, M. Field evaluation of transgenic pineapple (Ananas comosus (L.) Merr.) cv.‘Smooth Cayenne’for resistance to blackheart under subtropical conditions. Sci. Hortic. 2013, 159, 103–108. [Google Scholar] [CrossRef]
- Meng, B.; Li, C.; Wang, W.; Goszczynski, D.; Gonsalves, D. Complete genome sequences of two new variants of Grapevine rupestris stem pitting-associated virus and comparative analyses. J. Gen. Virol. 2006, 86, 1555–1560. [Google Scholar] [CrossRef] [PubMed]
- Waterhouse, P.M.; Wang, M.B.; Lough, T. Gene silencing as an adaptive defence against viruses. Nature 2001, 411, 834–842. [Google Scholar] [CrossRef] [PubMed]
- Ng, T.F.F.; Duffy, S.; Polston, J.E.; Bixby, E.; Vallad, G.E.; Breitbart, M. Exploring the diversity of plant DNA viruses and their satellites using vector-enabled metagenomics on whiteflies. PLoS ONE 2011, 6, e19050. [Google Scholar] [CrossRef] [PubMed]


© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dey, K.K.; Green, J.C.; Melzer, M.; Borth, W.; Hu, J.S. Mealybug Wilt of Pineapple and Associated Viruses. Horticulturae 2018, 4, 52. https://doi.org/10.3390/horticulturae4040052
Dey KK, Green JC, Melzer M, Borth W, Hu JS. Mealybug Wilt of Pineapple and Associated Viruses. Horticulturae. 2018; 4(4):52. https://doi.org/10.3390/horticulturae4040052
Chicago/Turabian StyleDey, Kishore K., James C. Green, Michael Melzer, Wayne Borth, and John S. Hu. 2018. "Mealybug Wilt of Pineapple and Associated Viruses" Horticulturae 4, no. 4: 52. https://doi.org/10.3390/horticulturae4040052
APA StyleDey, K. K., Green, J. C., Melzer, M., Borth, W., & Hu, J. S. (2018). Mealybug Wilt of Pineapple and Associated Viruses. Horticulturae, 4(4), 52. https://doi.org/10.3390/horticulturae4040052




