Developmental Variation in Fruit Polyphenol Content and Related Gene Expression of a Red-Fruited versus a White-Fruited Fragaria vesca Genotype
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. Fruit Extraction and Quantification of Target Phenolic Compounds by HPLC/MS
2.3. RNA Purification, cDNA Synthesis, and Cloning of Partial Sequence of Candidate Genes
2.4. Statistical Analyses
3. Results
3.1. Polyphenol Content—Development Stages
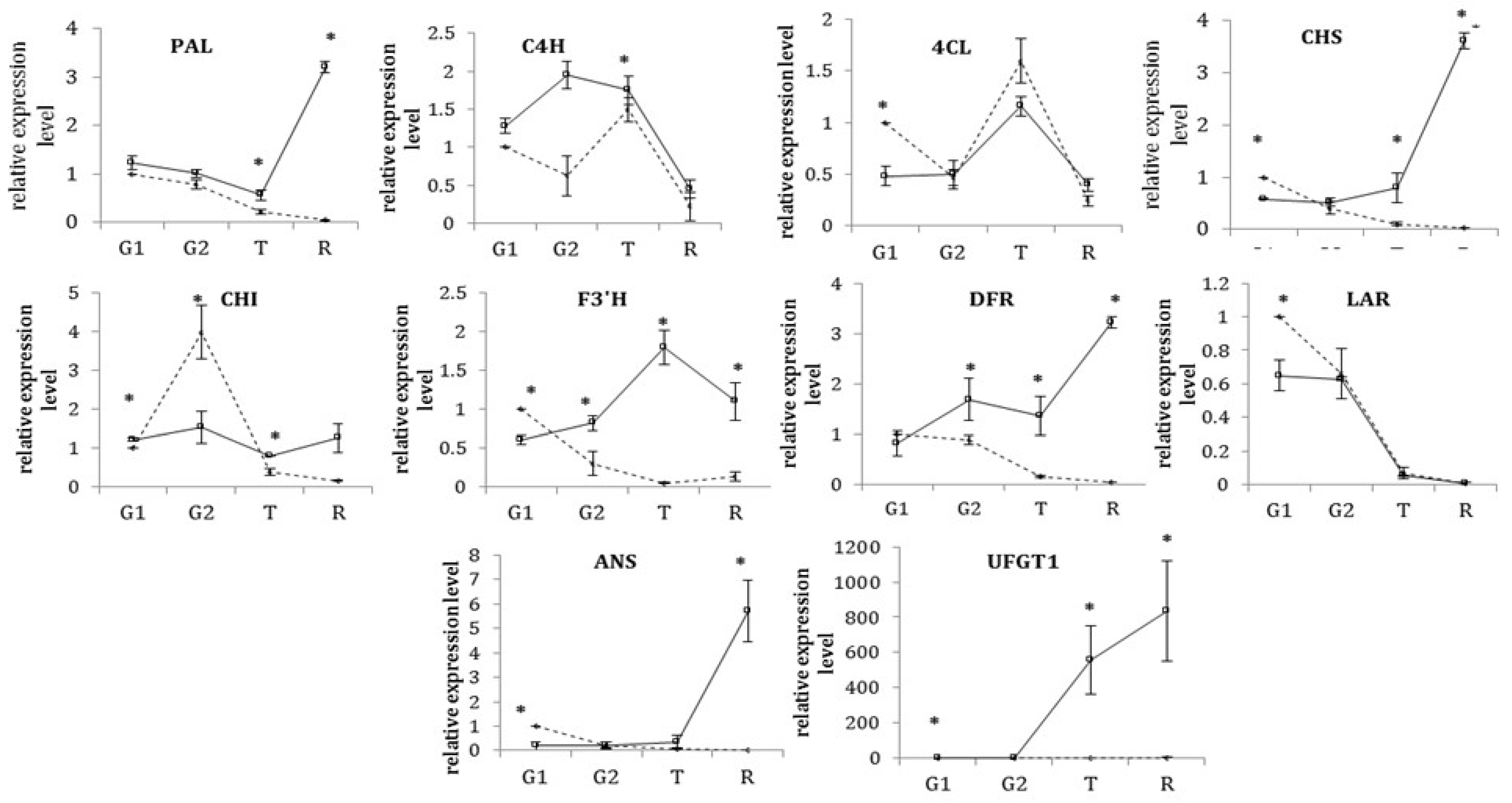
3.2. Transcriptional Profiles of the Structural Genes of the Phenylpropanoid/Flavonoid Biosynthetic Pathway during Development
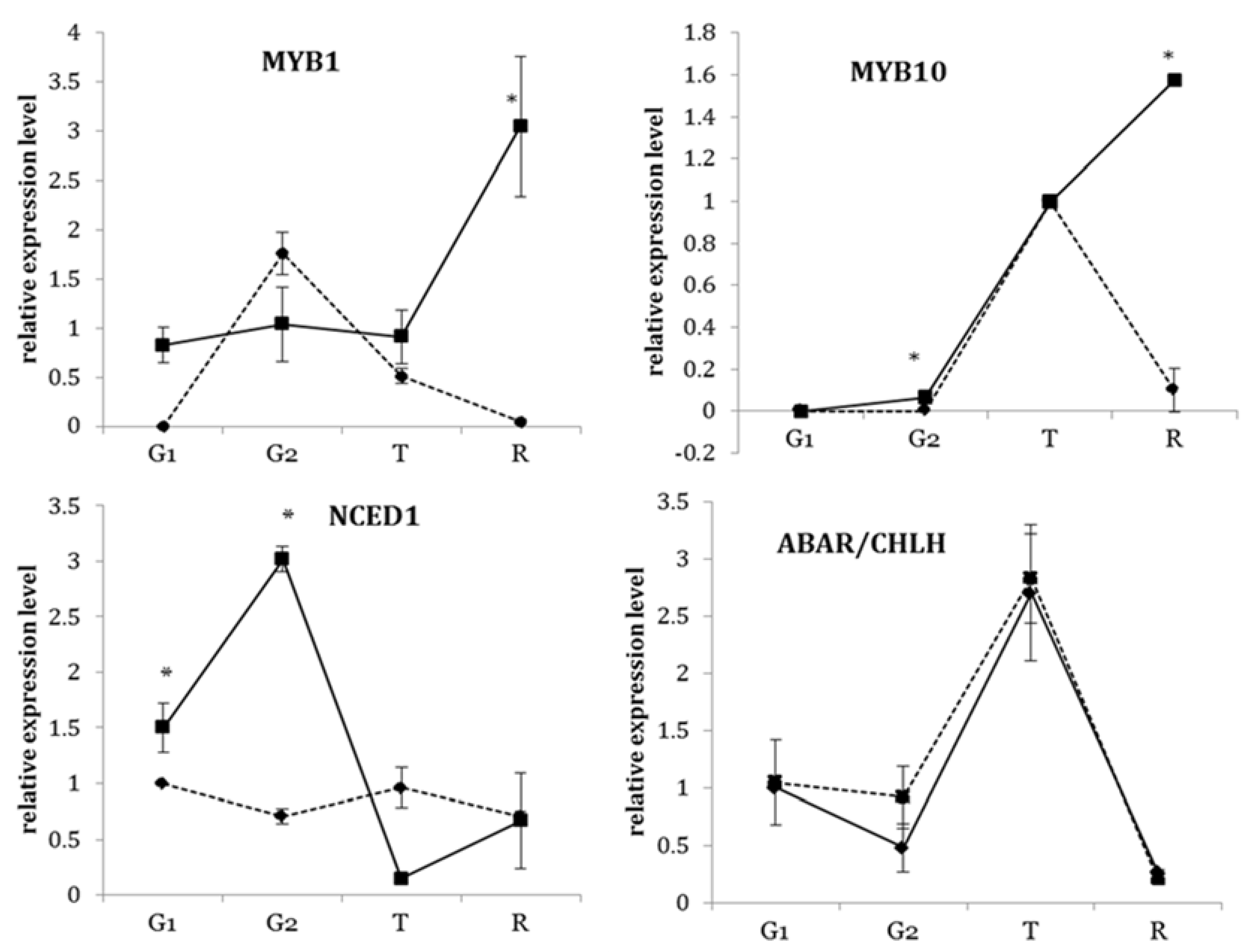
3.3. Transcriptional Profiles of Key Transcription Factors of the Flavonoid Biosynthetic Pathway during Development
3.4. Transcriptional Profiles of ABA-Related Genes in Strawberry Fruit at Different Developmental Stages
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Lattanzio, V.M.T.; Cardinali, A. Role of phenolics in the resistance mechanisms of plants against fungal pathogens and insects. Phytochem. Adv. Res. 2006, 661, 23–67. [Google Scholar]
- Buendía, B.; Gil, M.I.; Tudela, J.A.; Gady, A.; Medina, J.J.; Soria, C.; López, J.M.; Tomás-Barberán, F. HPLC-MS analysis of proanthocyanidin oligomers and other phenolics in 15 strawberry cultivars. J. Agric. Food Chem. 2010, 58, 3916–3926. [Google Scholar] [CrossRef] [PubMed]
- Aaby, K.; Mazur, S.; Nes, A.; Skrede, G. Phenolic compounds in strawberry (Fragaria x ananassa Duch.) fruits: Composition in 27 cultivars and changes during ripening. Food Chem. 2012, 132, 86–97. [Google Scholar] [CrossRef] [PubMed]
- Muňoz, C.; Sanchez-Sevilla, J.; Botella, M.A.; Hoffmann, T.; Schwab, W.; Valpuesta, V. Polyphenol composition in the ripe fruits of Fragaria species and transcriptional analyses of key genes in the pathway. J. Agric. Food Chem. 2011, 59, 12598–12604. [Google Scholar] [CrossRef] [PubMed]
- Simirgiotis, M.J.; Schmeda-Hirschmann, G. Determination of phenolic composition and antioxidant activity in fruits, rhizomes and leaves of the white strawberry (Fragaria chiloensis spp. chiloensis form chiloensis) using HPLC-DAD-ESI-MS and free radical quenching techniques. J. Food Comp. Anal. 2010, 23, 545–553. [Google Scholar] [CrossRef]
- Duan, W.; Sun, P.; Li, J. Expression of genes involved in the anthocyanin biosynthesis pathway in white and red fruits of Fragaria pentaphylla and genetic variation in the dihydroflavonol-4-reductase gene. Biochem. Syst. Ecol. 2017, 72, 40–46. [Google Scholar] [CrossRef]
- Del Bubba, M.; Checchini, L.; Chiuminatto, U.; Doumett, S.; Fibbi, D.; Giordani, E. Liquid chromatographic/electrospray ionization tandem mass spectrometric study of polyphenolic composition of four cultivars of Fragaria vesca L. berries and their comparative evaluation. J. Mass Spectrom. 2012, 47, 1207–1220. [Google Scholar] [CrossRef] [PubMed]
- Roy, S.; Wu, B.; Liu, W.; Archbold, D. Comparative analyses of polyphenolic composition of Fragaria spp. color mutants. Plant Physiol. Biochem. 2018, 125, 255–261. [Google Scholar] [CrossRef] [PubMed]
- Josuttis, M.; Verrall, S.; Stewart, D.; Krüger, E.; McDougall, G.J. Genetic and environmental effects on tannin composition in strawberry (Fragaria × ananassa) cultivars grown in different European locations. J. Agric. Food Chem. 2013, 61, 790–800. [Google Scholar] [CrossRef] [PubMed]
- Palmieri, L.; Masuero, D.; Martinatti, P.; Baratto, G.; Martens, S.; Vrhovsek, U. Genotype-by-environment effect on bioactive compounds in strawberry (Fragaria x ananassa Duch.). J. Sci. Food Agric. 2017, 97, 4180–4189. [Google Scholar] [CrossRef] [PubMed]
- Folta, K.M.; Davis, T.M. Strawberry Genes and Genomics. Crit. Rev. Plant Sci. 2006, 25, 399–415. [Google Scholar] [CrossRef]
- Slovin, J.P.; Schmitt, K.; Folta, K.M. An inbred line of the diploid strawberry Fragaria vesca f. semperflorens for genomic and molecular genetic studies in the Rosaceae. Plant Methods 2009, 5, 15. [Google Scholar] [CrossRef] [PubMed]
- Shulaev, V.; Sargent, D.J.; Crowhurst, R.N.; Mockler, T.C.; Folkerts, O.; Delcher, A.L.; Jaiswal, P.; Mockaitis, K.; Liston, A.; Mane, S.P.; et al. The genome of woodland strawberry (Fragaria vesca). Nat. Genet. 2011, 43, 109–116. [Google Scholar]
- Sun, J.; Liu, X.; Yang, T.; Slovin, J.; Chen, P. Profiling polyphenols of two diploid strawberry (Fragaria vesca) inbred lines using UHPLC-HRMSn. Food Chem. 2014, 146, 289–298. [Google Scholar] [CrossRef] [PubMed]
- Hartl, K.; Denton, A.; Franz-Oberdorf, K.; Hoffmann, T.; Spornraft, M.; Usadel, B.; Schwab, W. Early metabolic and transcriptional variations in fruit of natural white-fruited Fragaria vesca genotypes. Sci. Rep. 2017, 7, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Carbone, F.; Preuss, A.; De Vos, R.C.H.; D’Amico, E.; Perrotta, G.; Bovy, A. Developmental, genetic and environmental factors affect the expression of flavonoid genes, enzymes and metabolites in strawberry fruits. Plant Cell Environ. 2009, 32, 1117–1131. [Google Scholar] [CrossRef] [PubMed]
- Salvatierra, A.; Pimentel, P.; Moya-Leon, M.A.; Caligari, P.D.S.; Herrera, R. Comparison of transcriptional profiles of flavonoid genes and anthocyanin contents during fruit development of two botanical forms of Fragaria chiloensis ssp. chiloensis. Phytochemistry 2010, 71, 1839–1847. [Google Scholar] [CrossRef] [PubMed]
- Takos, A.M.; Jaffe, F.W.; Jacob, S.R.; Bogs, J.; Robinson, S.; Walker, A. Light–induced expression of a MYB gene regulates anthocyanin biosynthesis in red apples. Plant Physiol. 2006, 142, 1216–1232. [Google Scholar] [CrossRef] [PubMed]
- Ban, Y.; Honda, C.; Hatsuyama, Y.; Igarashi, M.; Bessho, H.; Moriguchi, T. Isolation and functional analysis of a MYB transcription factor gene that is a key regulator for the development of red coloration in apple skin. Plant Cell Physiol. 2007, 48, 958–970. [Google Scholar] [CrossRef] [PubMed]
- Changne, D.; Carlisle, C.M.; Blond, C.; Volz, R.K.; Whitworth, C.J.; Oraguzie, N.C.; Crowhurst, R.N.; Allan, A.C.; Espley, R.V.; Hellens, R.P.; et al. Mapping a candidate gene (MdMYB10) for red flesh and foliage colour in apple. BMC Genom. 2007, 8, 212. [Google Scholar] [CrossRef] [PubMed]
- Lin-Wang, K.; Bolitho, K.; Grafton, K.; Kortstee, A.; Karunairetnam, S.; McGhie, T.K.; Espley, R.V.; Hellens, R.P.; Allan, A.C. An R2R3 MYB transcription factor associated with regulation of the anthocyanin biosynthetic pathway in Rosaceae. BMC Plant Biol. 2010, 10, 549–553. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, S.; Goto-Yamamoto, N.; Hirochika, H. Retrotransposon induced mutations in grape skin color. Science 2004, 304, 982. [Google Scholar] [CrossRef] [PubMed]
- Walker, A.R.; Lee, E.; Bogs, J.; McDavid, D.A.J.; Thomas, M.R.; Robinson, S.P. White grapes arose through the mutation of two similar and adjacent regulatory genes. Plant J. 2007, 49, 772–785. [Google Scholar] [CrossRef] [PubMed]
- Rahim, M.A.; Busatto, N.; Trainotti, L. Regulation of anthocyanin biosynthesis in peach fruits. Planta 2014, 240, 913–929. [Google Scholar] [CrossRef] [PubMed]
- Ravaglia, D.; Espley, R.V.; Henry-Kirk, R.A.; Andreotti, C.; Ziosi, V.; Hellens, R.P.; Costa, G.; Allan, A.C. Transcriptional regulation of flavonoid biosynthesis in nectarine (Prunus persica) by a set of R2R3 MYB transcription factors. BMC Plant Biol. 2013, 13, 68. [Google Scholar] [CrossRef] [PubMed]
- Lin-Wang, K.; McGhie, T.K.; Wang, M.; Liu, Y.; Warren, B.; Storey, R.; Espley, R.V.; Allan, A.C. Engineering the anthocyanin regulatory complex of strawberry (Fragaria vesca). Front. Plant Sci. 2014, 5, 651. [Google Scholar] [CrossRef] [PubMed]
- Medina-Puche, L.; Cumplido-Laso, G.; Amil-Ruiz, F.; Hoffmann, T.; Ring, L.; Rodríguez-Franco, A.; Luis Caballero, J.; Schwab, W.; Muñoz-Blanco, J.; Blanco-Portales, R. MYB10 plays a major role in the regulation of flavonoid/phenylpropanoid metabolism during ripening of Fragaria x ananassa fruits. J. Exp. Bot. 2014, 65, 401–417. [Google Scholar] [CrossRef] [PubMed]
- Boss, P.K.; Davis, C.; Robinson, S.P. Analysis of the expression of anthocyanin pathway genes in developing Vitis vinifera. Plant Physiol. 1996, 111, 1059–1066. [Google Scholar] [CrossRef] [PubMed]
- Jia, H.F.; Chai, Y.M.; Li, C.L.; Lu, D.; Luo, J.J.; Qin, L.; Shen, Y.Y. Abscisic acid plays an important role in the regulation of strawberry fruit ripening. Plant Physiol. 2011, 157, 188–199. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Li, L.; Luo, Z.; Mou, W.; Mao, L.; Ying, T. Comparative transcriptome analysis reveals the influence of absicisic acid on the metabolism of pigments, ascorbic acid and folic acid during strawberry fruit ripening. PLoS ONE 2015, 10, e0130037. [Google Scholar] [CrossRef]
- Reid, K.; Olsson, N.; Schlosser, J.; Peng, F.; Lund, S. An optimized grapevine RNA isolation procedure and statistical determination of reference genes for real-time RT-PCR during berry development. BMC Plant Biol. 2006, 6, 27. [Google Scholar] [CrossRef] [PubMed]
- Pattanaik, S.; Kong, Q.; Zaitlin, D.; Werkman, J.R.; Xie, C.H.; Patr, B.; Yuan, L. Isolation and functional characterization of a floral tissue-specific R2R3 MYB regulator from tobacco. Planta 2010, 231, 1061–1076. [Google Scholar] [CrossRef] [PubMed]
- Almeida, J.R.M.; D’Amico, E.; Preuss, A.; Carbone, F.; de Vos, C.H.R.; Deiml, B.; Mourgues, F.; Perrotta, G.; Fischer, T.C.; Bovy, A.G.; et al. Characterization of major enzymes and genes involved in flavonoid and proanthocyanidin biosynthesis during fruit development in strawberry (Fragaria x ananassa). Arch. Biochem. Biophys. 2007, 465, 61–71. [Google Scholar] [CrossRef] [PubMed]
- Fait, A.; Hanhineva, K.; Beleggia, R.; Dai, N.; Rogachev, I.; Nikiforova, V.J.; Fernie, A.R.; Aharoni, A. Reconfiguration of the achene and receptacle metabolic networks during strawberry fruit development. Plant Physiol. 2008, 148, 730–750. [Google Scholar] [CrossRef] [PubMed]
- Gasperotti, M.; Masuero, D.; Guella, G.; Palmieri, L.; Martinatti, P.; Pojer, E.; Mattivi, F.; Vrhovsek, U. Evolution of ellagitannin content and profile during fruit ripening in Fragaria spp. J. Agric. Food Chem. 2013, 61, 8597–8607. [Google Scholar] [CrossRef] [PubMed]
- Kosar, M.; Kafkas, E.; Paydas, S.; Baser, K.H.C. Phenolic composition of strawberry genotypes at different maturation stages. J. Agric. Food Chem. 2004, 52, 1586–1589. [Google Scholar] [CrossRef] [PubMed]
- Bacchella, R.; Testoni, A.; ScalzoI, R. Influence of genetic and environmental factors on chemical profile and antioxidant potential of commercial strawberry (Fragaria × anannasa Duchesne). Electron. J. Environ. Agric. Food Chem. 2009, 8, 230–242. [Google Scholar]
- Doumett, S.; Fibbi, D.; Cincinelli, A.; Giordani, E.; Nin, S.; Del Bubba, M. Comparison of nutritional and nutraceutical properties in cultivated fruits of Fragaria vesca L. produced in Italy. Food Res. Int. 2011, 44, 1209–1216. [Google Scholar] [CrossRef]
- Urrutia, M.; Schwab, W.; Hoffmann, T.; Monfort, A. Genetic dissection of the (poly) phenol profile of diploid strawberry (Fragaria vesca) fruits using a NIL collection. Plant Sci. 2016, 242, 151–168. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Peng, H.; Yang, T.; Whitaker, B.; Huang, L.; Sun, J.; Chen, P. Effect of calcium on strawberry fruit flavonoid pathway gene expression and anthocyanin accumulation. Plant Physiol. Biochem. 2014, 82, 289–298. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Li, W.; Dou, Y.; Zhang, J.; Jiang, G.; Miao, L.; Han, G.; Liu, Y.; Li, H.; Zhang, Z. Transcript quantification by RNA-Seq reveals differentially expressed genes in the red and yellow fruits of Fragaria vesca. PLoS ONE 2015, 10, e0144356. [Google Scholar] [CrossRef] [PubMed]
- Saud, G.; Carbone, F.; Perrotta, G.; Figueroa, C.R.; Moya, M.; Herrera, R.; Retamales, J.B.; Carrasco, B.; Cheel, J.; Schmeda-Hirschmann, G.; et al. Transcript profiling suggests transcriptional repression of the flavonoid pathway in the white-fruited Chilean strawberry, Fragaria chiloensis (L.) Mill. Genet. Resour. Crop Evol. 2009, 56, 895–903. [Google Scholar] [CrossRef]
- Cheel, J.; Theoduloz, C.; Rodrıguez, J.A.; Caligari, P.D.S.; Schmeda-Hirschmann, G. Free radical scavenging activity and phenolic content in achenes and thalamu from Fragaria chiloensis ssp. chiloensis, F. vesca and F. x ananassa cv. Chandler. Food Chem. 2017, 102, 36–44. [Google Scholar] [CrossRef]
- Thill, J.; Silvija, M.; Gotame, T.P.; Mikulic-Petkovsek, M.; Gosch, C.; Verbric, R.; Preuss, A.; Schwab, W.; Stamper, F.; Stich, K.; et al. Differential expression of flavonoid 3’-hydroxylase during fruit development establishes the different B-ring hydroxylation patterns of flavonoids in Fragaria x ananassa and Fragaria vesca. Plant Physiol. Biochem. 2013, 72, 72–78. [Google Scholar] [CrossRef] [PubMed]
- Griesser, M.; Hoffmann, T.; Bellido, M.L.; Rosati, C.; Fink, B.; Kurtzer, R.; Aharoni, A.; Munoz-Blanco, J.; Schwab, W. Redirection of flavonoid biosynthesis through the down-regulation of an anthocyanidin glucosyltransferase in ripening strawberry fruit. Plant Physiol. 2008, 146, 1528–1539. [Google Scholar] [CrossRef] [PubMed]
- Halbwirth, H.; Puhl, I.; Haas, U.; Jezik, K.; Treutter, D.; Stich, K. Two-phase flavonoid formation in developing strawberry (Fragaria x ananassa) fruit. J. Agric. Food Chem. 2006, 541, 479–1485. [Google Scholar] [CrossRef] [PubMed]
- Hu, P.; Li, G.; Zhao, X.; Zhao, F.; Li, L.; Zhou, H. Transcriptome profiling by RNA-Seq reveals differentially expressed genes related to fruit development and ripening characteristics in strawberries (Fragaria x ananassa). Peer J. 2018, 6, e4976. [Google Scholar] [CrossRef] [PubMed]
- Starkevič, P.; Paukštytė, J.; Kazanavičiūtė, V.; Denkovskienė, E.; Stanys, V.; Bendokas, V. Expression and anthocyanin biosynthesis modulating potential of sweet cherry (Prunus avium L.) MYB10 and bHLH genes. PLoS ONE 2015, 10, e0126991. [Google Scholar] [CrossRef] [PubMed]
- Wei, H.; Chen, X.; Zong, X.; Shu, H.; Gao, D.; Liu, Q. Comparative transcriptome analysis of genes involved in anthocyanin biosynthesis in the red and yellow fruits of sweet cherry (Prunus avium L.). PLoS ONE 2015, 10, e0121164. [Google Scholar] [CrossRef] [PubMed]
- Schaart, J.G.; Dubos, C.; De La Fuente, I.R.; van Houwelingen, A.M.; de Vos, R.C.; Jonker, H.H.; Xu, W.; Routaboul, J.M.; Lepiniec, L.; Bovy, A.G. Identification and characterization of MYB-bHLH-WD40 regulatory complexes controlling proanthocyanidin biosynthesis in strawberry (Fragaria x ananassa) fruits. New Phytol. 2013, 197, 454–467. [Google Scholar] [CrossRef] [PubMed]
- Kadomura-Ishikawa, Y.; Miyawaki, K.; Takahashi, A.; Masuda, T.; Noji, S. Light and abscisic acid independently regulated FaMYB10 in Fragaria x ananassa fruit. Planta 2015, 241, 953–965. [Google Scholar] [CrossRef] [PubMed]
- Pillet, J.; Yu, H.W.; Chambers, A.H.; Vance, M.; Whitaker, M.; Folta, K.M. Identification of candidate flavonoid pathway genes using transcriptome correlation network analysis in ripe strawberry (Fragaria x ananassa) fruits. J. Exp. Bot. 2015, 66, 4455–4467. [Google Scholar] [CrossRef] [PubMed]

| Target Genes | Primers (F/R) | Amplicon Size (bp) | Accession No./Gene ID z |
|---|---|---|---|
| PAL1 | F5′-CACAACATTACTCCCTGCTTGC-3′ R5′-CCTTTTGGTCCAACCGACTTAG-3′ | 125 | XM_004304392.2/27263 |
| C4H | F5′-GACGGTTCCTTTCTTCACCAAC-3′ R5′-AAGTTTCACGAACAGAGGGTCC-3′ | 220 | DQ898278.1/28093 |
| 4CL | F5′-AGAGCTCAAAGTCATCGAACCC-3′ R 5′-GAGCTCCTTAACCCTGTCAACG-3′ | 217 | AF239685.1/15877 |
| CHS | F5′-TGTGTGAGTACATGGCACCTTC-3′ R5′-CCCATTCCTTAATGGCCTTG-3′ | 105 | AY017477.1/26826 |
| CHI | F5′-ACAATGATACTACCGCTGACGG-3′ R5′-CAATGGCTTTCGCTTCTGC-3′ | 112 | AB201755.1/23367 |
| F3′H | F5′-GAAGGACCTTTCGTGGTGAATC-3′ R5′-TGAGTTCTACATCCACGCACCT-3′ | 247 | AY017479.1/14611 |
| DFR | F-5′-CGGAGGGTGGTGTTTACATCTT-3′ R-5′-CCAGTCATCTTTACTTTCCGGC-3′ | 122 | AM691790.1/15174, 15176 |
| ANS | F5′-GCCTCAAACACCTTCCGACTAT-3′ R5′-TAACCCCCTCAGTTCCTTAGCA-3′ | 176 | AY017481.1/32347 |
| LAR | F5′-TTGAGAAGAGTGGGGTCCCTTA-3′ R5′-GATCTGGAACTGATCCAACGGT-3′ | 116 | DQ087253.1/03877 |
| UFGT1 | F5′-CTGCTTATCGTGGCTTGACA-3′ R5′-CCCGAAGTGACCACAAGAAT-3′ | 146 | AY695816.1/12591 |
| MYB1 | F5′-TTGCGTCGTTGTGGTAAGAG-3′ R5′-TCTGTCCTTCCAGGCAGTCT-3′ | 167 | AF401220.1 |
| MYB10 R2R3 | F5′-TTGCAGGCTTAAACAGATGC-3′ F5′-CGCATGCTTTACCTGAGAGA-3′ | 207 | EU155163.1 |
| NCED1 | F5′-CTACTTCAACGGCAGGCTTCTT-3′ R5′-GTCGTATCTCCCTTCGGTTTTG-3′ | 100 | HQ290318.1 |
| CHLH/ABAR | F5′-GCGATCACAGTGTTCAGTTTCC-3′ R5′-CAAAGCGTCTGAAGTCTCTGGA-3′ | 168 | GQ201451.1 |
| Actin | F5′-ACGAGCTGTTTTCCCTAGCA-3′ R5′-CTCTTTTGGATTGAGCCTCG-3′ | 107 | AB116565.1 |
| Compound y | Baron Solemacher | Pineapple Crush | ||||||
|---|---|---|---|---|---|---|---|---|
| Developmental Stage z | ||||||||
| G1 | G2 | T | R | G1 | G2 | T | R | |
| Hydroxycinnamic acid content (mg/100 g fresh weight) | ||||||||
| FA | 3.9 ± 0.2 f x | 14 ± 0.8 c | 4.9 ± 0.1 ef | 10.9 ± 0.04 d | 26.1 ± 0.3 b | 38.7 ± 1.8 a | 7.3 ± 0.3 e | 14.7 ± 0.9 c |
| ChA | 0.3 ± 0 e | 0.7 ± 0 a | 0.6 ± 0 b | 0.09 ± 0 g | 0.4 ± 0 d | 0.2 ± 0 f | 0.5 ± 0 c | 0.4 ± 0 d |
| p-Cou-hexose w | 0.4 ± 0 a | 0.2 ± 0 c | 0.2± 0 bc | 0.3 ± 0 b | 0.2 ± 0 c | 0.2 ± 0 c | 0.2 ± 0 c | 0.3 ± 0 b |
| Flavonol content (mg/100 g fresh weight) | ||||||||
| K-3-gluc | 5.5 ± 0.1 b y | 4.2 ± 0 c | 1.8 ± 0 e | 1.1 ± 0 f | 3.8 ± 0 cd | 4.6± 0 b | 6.2 ± 0 a | 1.6 ± 0 ef |
| K-3-act-glu | 1.4 ± 0 de | 1.4 ± 0 de | 1.2 ± 0 e | 1.9 ± 0 bc | 1.4 ± 0 de | 2.2 ± 0 b | 1.5 ± 0 cd | 2.8 ± 0 a |
| K-cou-hex v | 3.1 ± 0 ab | 3.3 ± 0 a | 1.8 ± 0 d | 1.1 ± 0 e | 2.4 ± 0 c | 3 ± 0 b | 2.4 ± 0 c | 1.1 ± 0 e |
| Quercetin | nd u | 0.3 ± 0 ab | nd | 0.2 ± 0 b | 0.3 ± 0ab | 0.7 ± 0 a | nd | 0.2 ± 0 a |
| Q-3-glu | 12.1 ± 0.2 a | 8.5 ± 0.2 b | 4.1 ± 0 e | 4.4 ± 0 e | 6.1 ± 0 d | 3.4 ± 0 f | 3.6 ± 0 f | 4.3± 0 e |
| Flavan-3-ol content (mg/100 g fresh weight) | ||||||||
| Catechin | 50 ± 1 e y | 115 ± 1 a | 93 ± 2 b | 86 ± 6 c | 61 ± 1 d | 114 ± 1 a | 96 ± 0 b | 40 ± 0 f |
| Epicatechin | 1.6 ± 0 f | 4.6 ± 0 b | 3.6 ± 0 d | 4 ± 0 c | 2.5 ± 0 e | 5.8 ± 0 a | 3.6 ± 0 d | 2.6± 0 e |
| PCD t | 210 ± 1 f | 484 ± 8 c | 460 ± 8 d | 326± 1 e | 213 ± 4 f | 545 ± 5 a | 505 ± 6 b | 167 ± 1 g |
| Ellagic acid (EA) and ellagitannin content (mg/100 g fresh weight) | ||||||||
| EA | 86 ± 1 b y | 104 ± 2 a | 30 ± 2 f | 33 ± 1 f | 108 ± 1 a | 56 ± 2 c | 37 ± 0 e | 44 ± 1 d |
| EADH s | 581 ± 4 a | 575 ± 3a | 245 ± 5 e | 254 ± 5 e | 571 ± 5 a | 424 ± 2 b | 359 ± 4 c | 308 ± 5 d |
| MEAP | 44 ± 1b | 33 ± 0 cd | 35 ± 0 c | 46 ± 0b | 25 ± 1 e | 29 ± 0 de | 44 ± 4 b | 53 ± 1 a |
| EAP | 107 ± 0 b | 117 ± 3 a | 53 ± 3 g | 77 ± 1 e | 110 ± 2 b | 67 ± 1 f | 87 ± 10 d | 93 ± 1 c |
| GHH | 217 ± 3 a | 151 ± 5 d | 83 ± 3 g | 43 ± 1 h | 161 ± 4 c | 178 ± 4 b | 125 ± 3 e | 114 ± 2 f |
| HGH | 536 ± 5 a y | 531 ± 8 a | 123 ± 1 e | 92 ± 1 f | 477 ± 3 b | 371 ± 5 c | 214 ± 2 d | 216 ± 4 d |
| Genotype | Stage z | Hydroxycinnamic Acids | Flavonols | Flavan-3-ols | Ellagic Acids | Ellagitannins |
|---|---|---|---|---|---|---|
| Baron Solemacher | G1 | 4.6 ± 0.2 f y | 22.1 ± 0.2 a | 260 ± 1 f | 816 ± 5 a | 757 ± 7a |
| G2 | 14.9 ± 0.8 c | 17.7 ± 0.2 b | 604 ± 8 b | 831 ± 8 a | 686 ± 8 b | |
| T | 5.7 ± 0 ef | 8.9 ± 0 e | 558 ± 9 c | 364 ± 8 f | 212 ± 3 f | |
| R | 11.2 ± 0 d | 8.7 ± 0.1 e | 417± 5 d | 409 ± 4 e | 140 ± 1 g | |
| Pineapple Crush | G1 | 26.7 ± 0.3 b | 14.0 ± 0.2 c | 277 ± 3 e | 814 ± 4 a | 643 ± 2 c |
| G2 | 39.1 ± 1.8 a | 13.9 ± 0.2 c | 665 ± 9a | 576 ± 2 b | 554 ± 8 d | |
| T | 8 ± 0.2 e | 13.7 ± 0.2 c | 605 ± 6 b | 529 ± 3 c | 344 ± 4 e | |
| R | 15.4 ± 0.9 c | 10 ± 0.2 d | 210 ± 1 g | 499 ± 4 d | 336 ± 6 e | |
| ANOVA (p) | ||||||
| Genotype | <0.001 | <0.001 | <0.001 | NS | <0.001 | |
| Stage | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | |
| Genotype X Stage | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Roy, S.; Singh, S.; Archbold, D.D. Developmental Variation in Fruit Polyphenol Content and Related Gene Expression of a Red-Fruited versus a White-Fruited Fragaria vesca Genotype. Horticulturae 2018, 4, 30. https://doi.org/10.3390/horticulturae4040030
Roy S, Singh S, Archbold DD. Developmental Variation in Fruit Polyphenol Content and Related Gene Expression of a Red-Fruited versus a White-Fruited Fragaria vesca Genotype. Horticulturae. 2018; 4(4):30. https://doi.org/10.3390/horticulturae4040030
Chicago/Turabian StyleRoy, Sutapa, Sanjay Singh, and Douglas D. Archbold. 2018. "Developmental Variation in Fruit Polyphenol Content and Related Gene Expression of a Red-Fruited versus a White-Fruited Fragaria vesca Genotype" Horticulturae 4, no. 4: 30. https://doi.org/10.3390/horticulturae4040030
APA StyleRoy, S., Singh, S., & Archbold, D. D. (2018). Developmental Variation in Fruit Polyphenol Content and Related Gene Expression of a Red-Fruited versus a White-Fruited Fragaria vesca Genotype. Horticulturae, 4(4), 30. https://doi.org/10.3390/horticulturae4040030






