Modulation of the Genetic Response in Vitis vinifera L. Against the Oomycete Plasmopara viticola, Causing Grapevine Downy Mildew, Through the Action of Different Basic Substances
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material and Pathogen Inoculum
2.2. Commercial Products
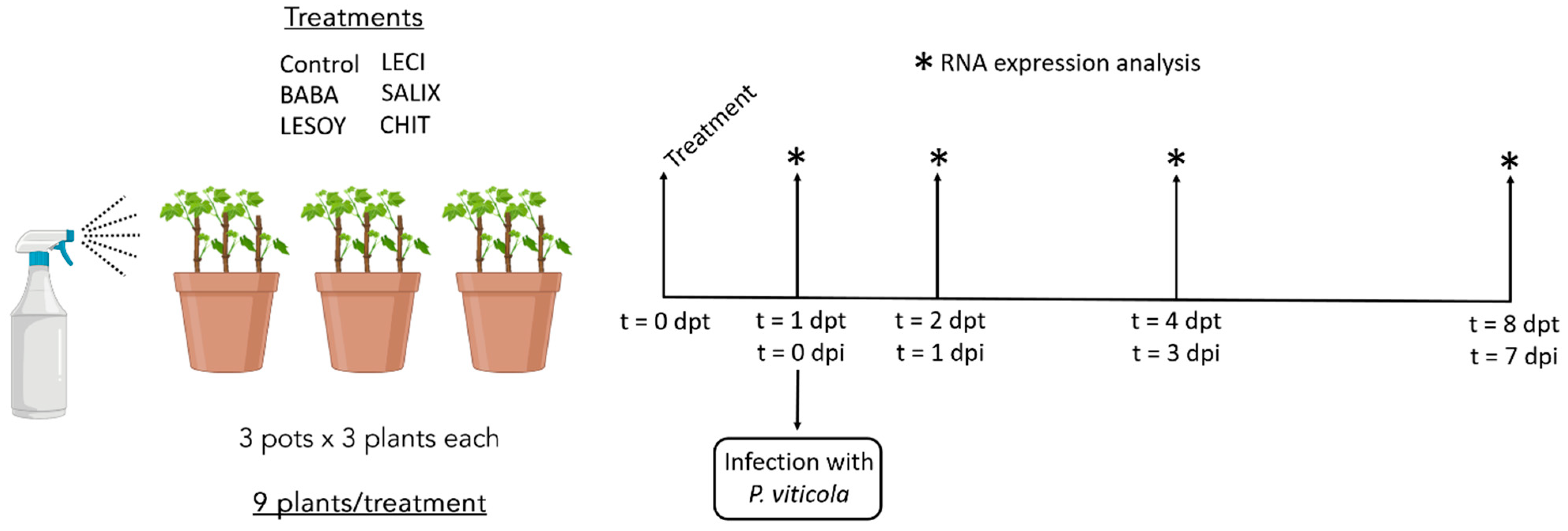
2.3. Experimental Assay
2.4. RNA Extraction and Reverse Transcription
2.5. Gene Expression
2.6. Data Analysis
3. Results
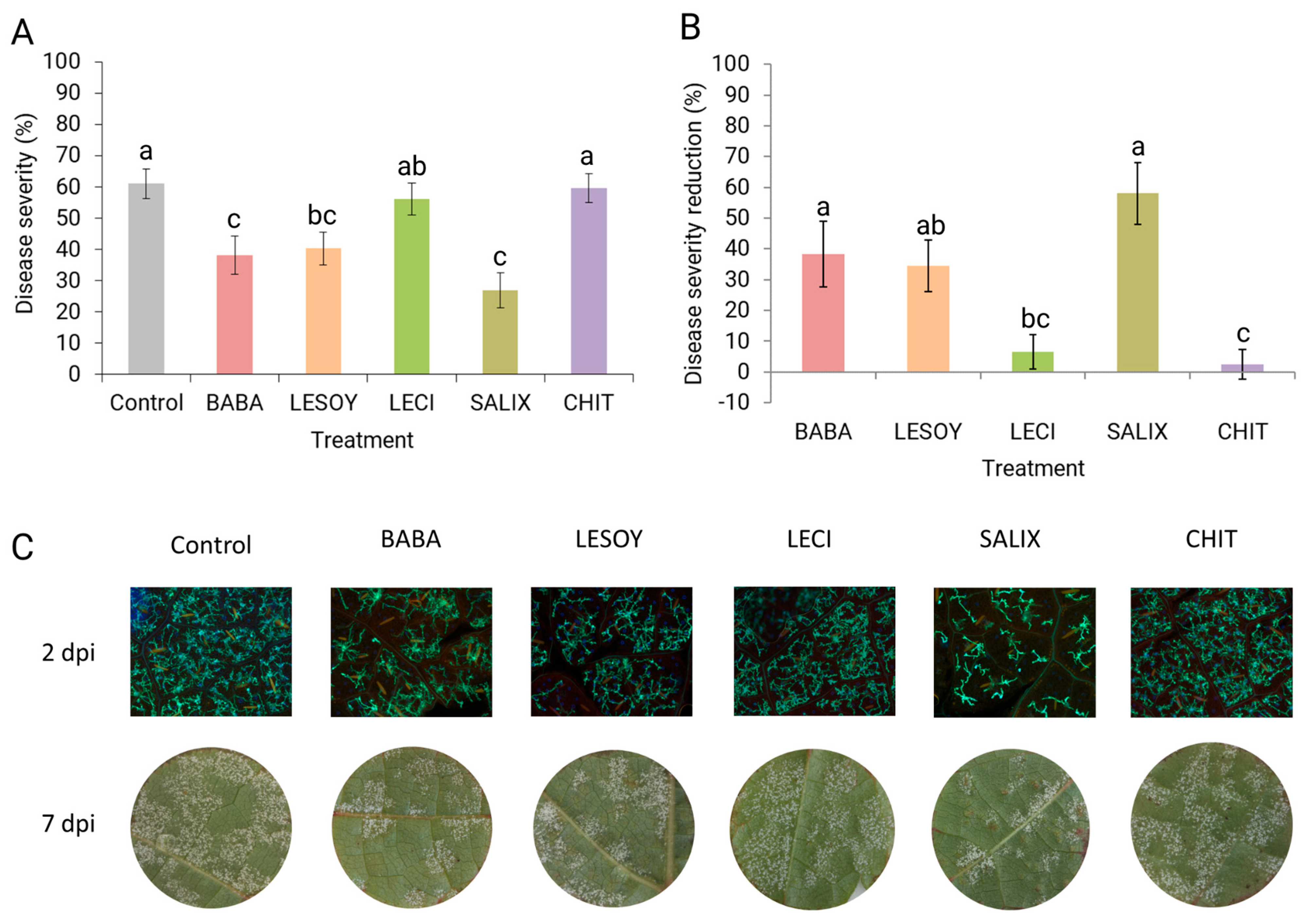
3.1. Disease Severity After Product Application
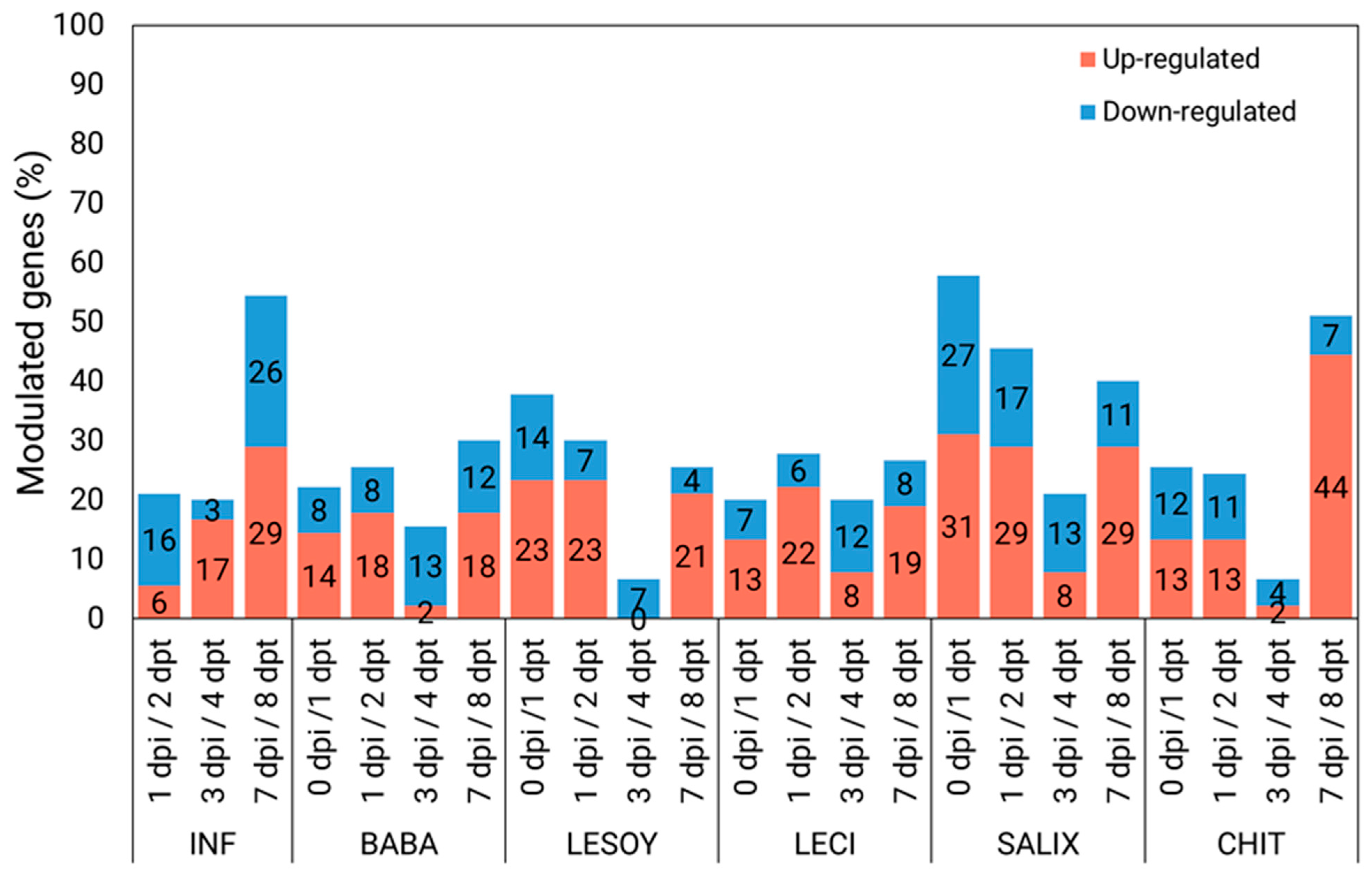
3.2. General Gene Modulation
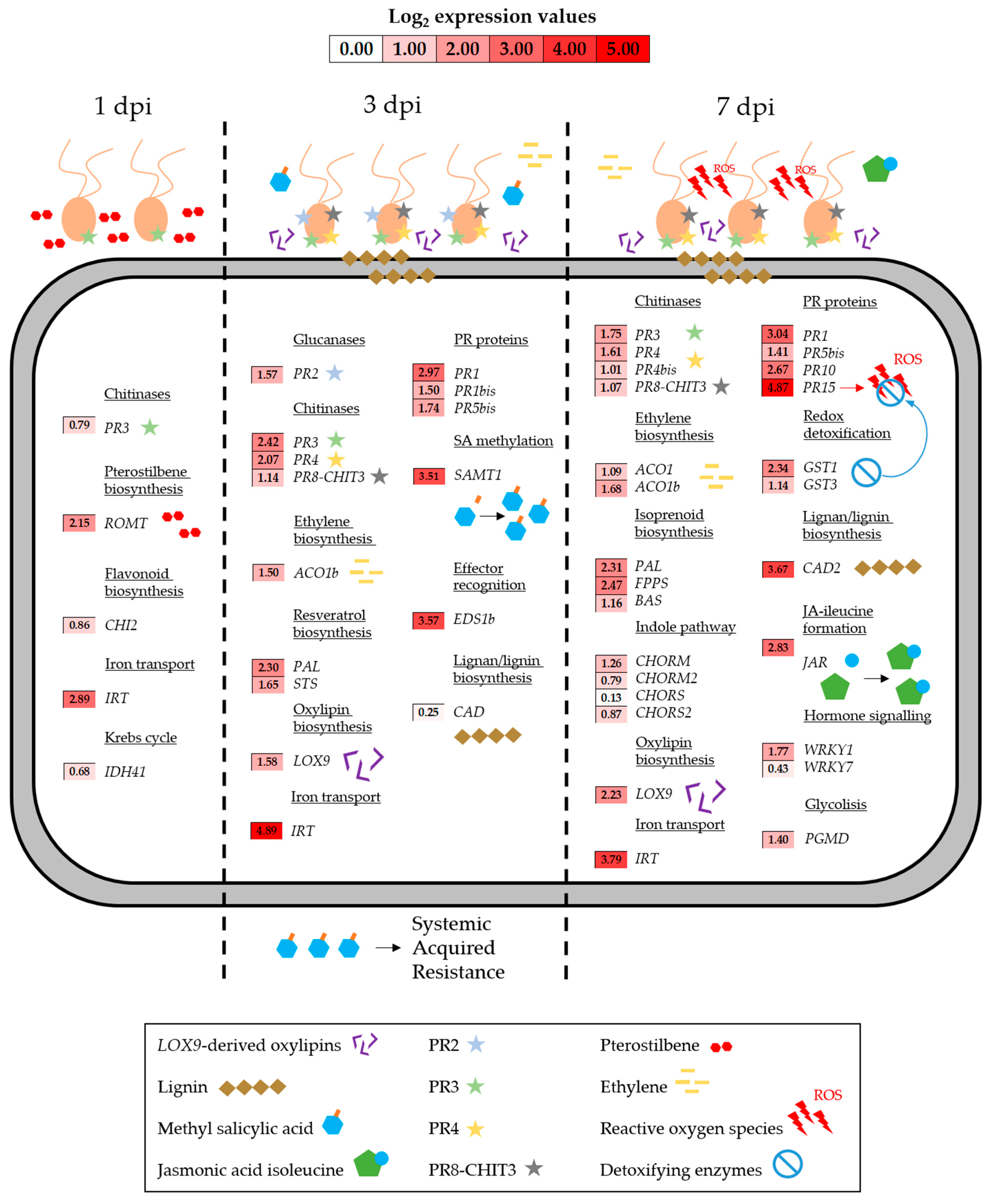
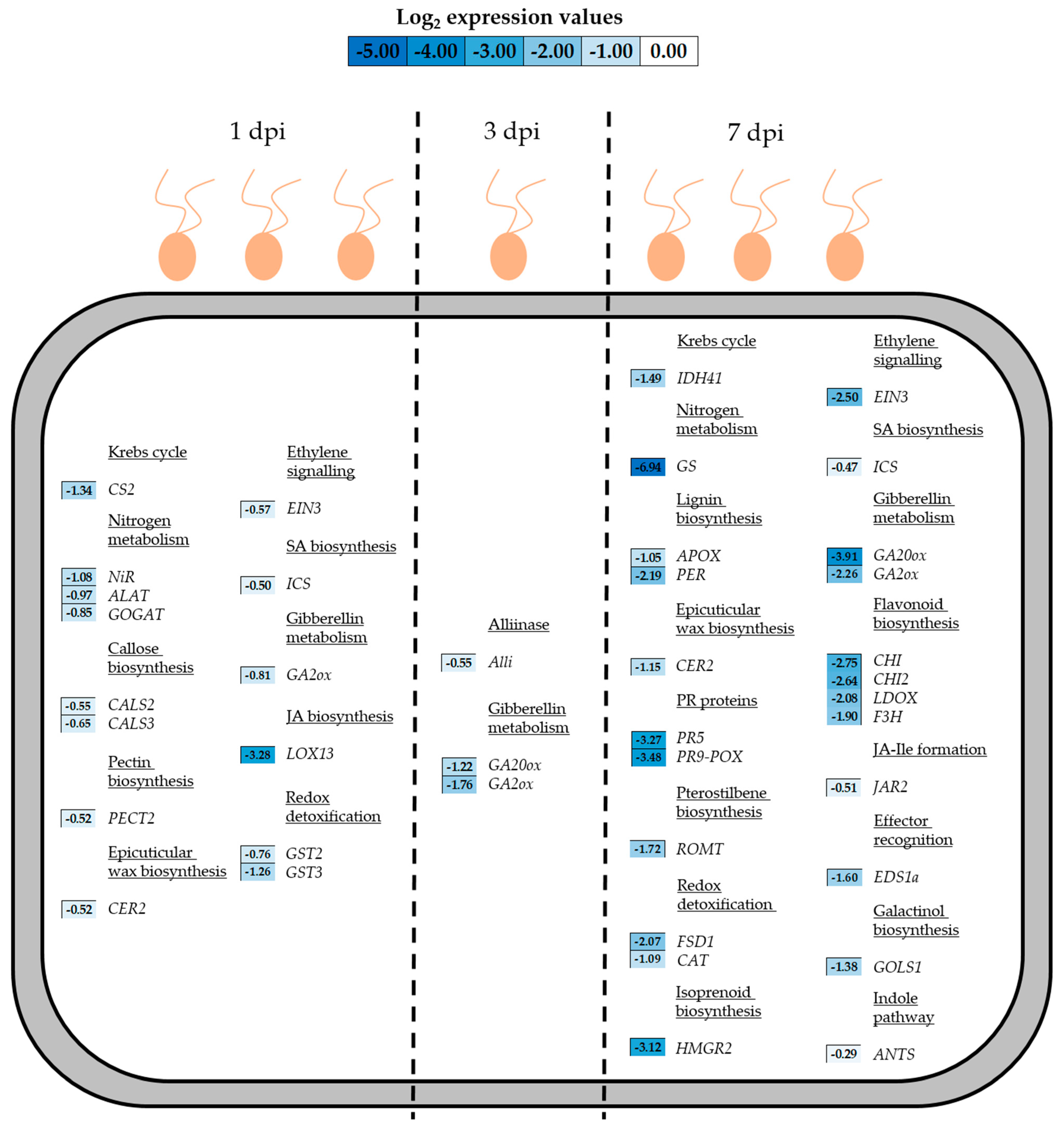
3.3. Gene Expression During P. viticola Infection
3.3.1. Up-Regulated Genes in Untreated Plants During Infection
3.3.2. Down-Regulated Genes in Untreated Plants During Infection
3.4. Gene Expression During P. viticola Infection in Grapevine Plants Previously Treated with the Products
3.4.1. Up-Regulated Genes in Treated Plants During Infection
3.4.2. Down-Regulated Genes in Treated Plants During Infection
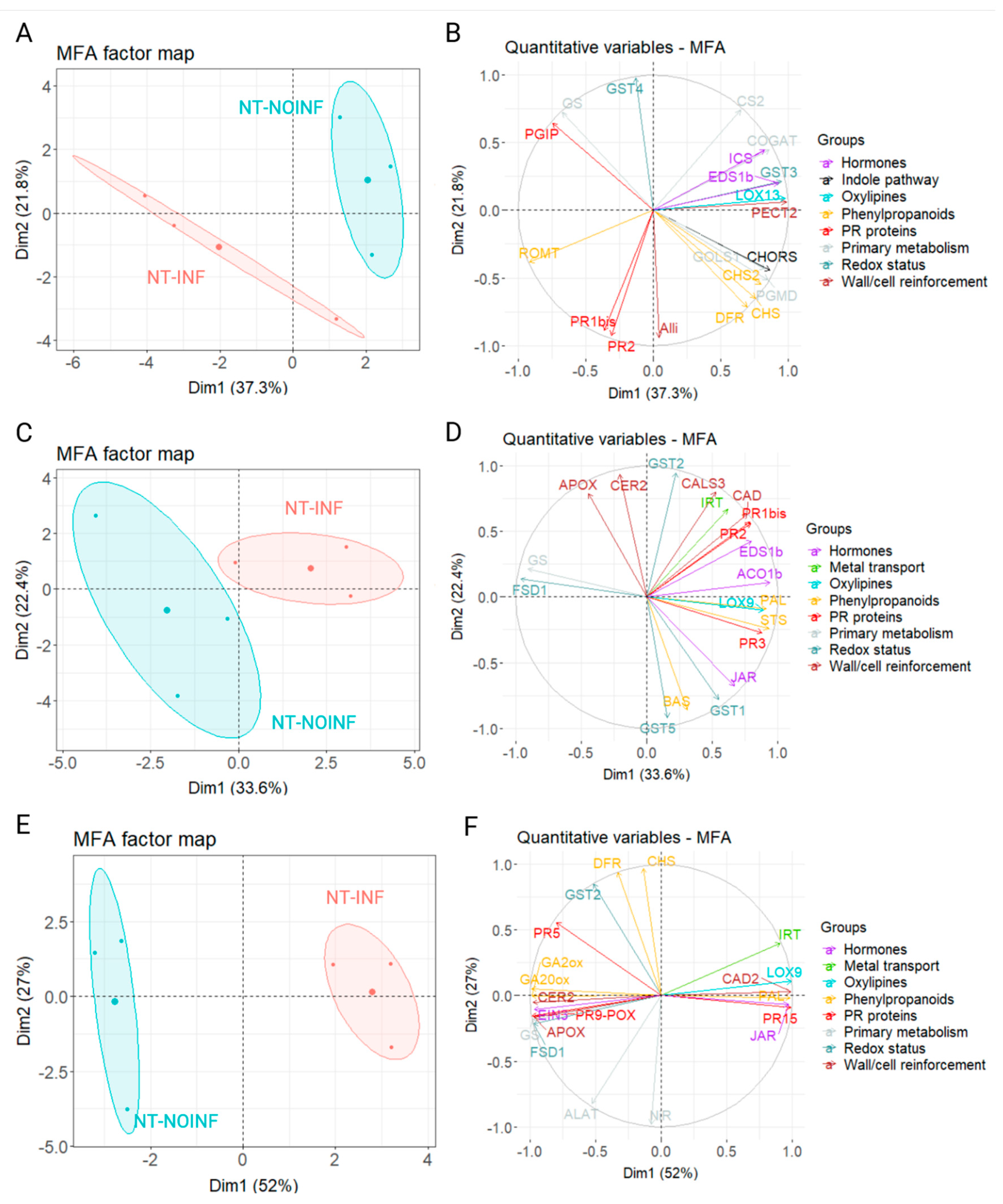
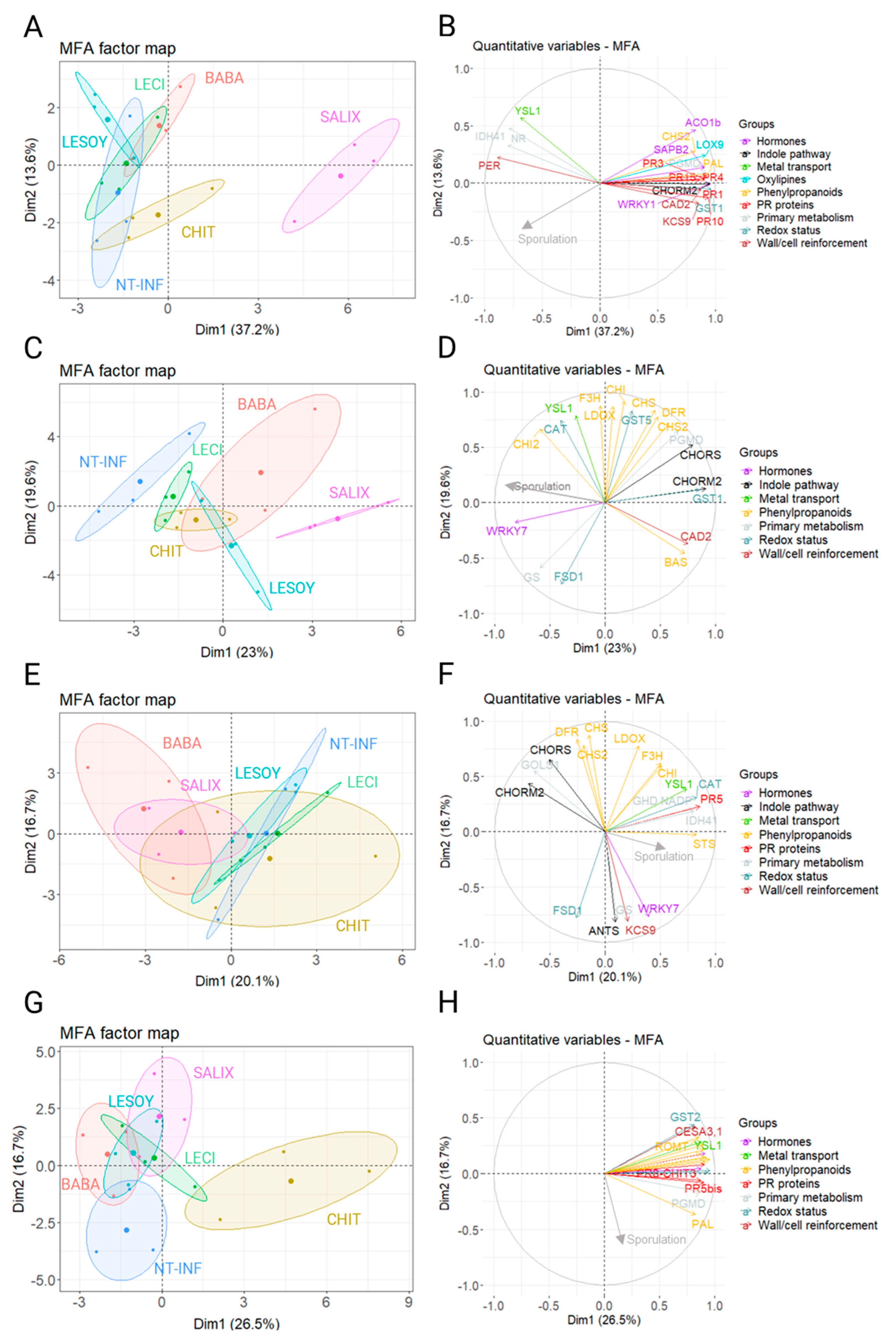
3.5. Multiple Factor Analysis
4. Discussion
4.1. Up-Regulated Genes
4.1.1. Pathogenesis-Related Proteins (PR Proteins) Are Induced Early and Strongly by Infection and Treatments
4.1.2. Stilbene Biosynthesis Is Anticipatively Activated in LESOY- and SALIX-Treated Plants
4.1.3. BABA and SALIX Enhance Oxylipin-Related Gene Expression Prior to Infection
4.1.4. LECI and SALIX Accelerate Terpene Biosynthesis During Early Infection
4.1.5. Salicylic Acid (SA)-Mediated Defence Is Promoted Earlier by LE SOY and SALIX
4.1.6. Ethylene (ET) Pathway Shows Limited Activation but Is Partially Modulated by Treatments
4.1.7. Jasmonic Acid (JA)-Related Genes Show Late Induction, Minimally Affected by Products
4.1.8. LECI and SALIX Induce Lignin Biosynthesis, with a Potential Enhancement on Physical Defence
4.2. Down-Regulated Genes During P. viticola Infection in Tempranillo Genotype
4.2.1. Reduced Cellulose Biosynthesis by SALIX and Other Treatments Could Enhance Lignin Accumulation to Restrict P. viticola
4.2.2. BABA, LESOY, LECI and SALIX Could Moderately Increase Cuticular Wax, Potentially Reinforcing the Physical Barrier
4.2.3. BABA and SALIX Might Slightly Promote Callose Deposition, Although This Effect Could Be Limited in Tempranillo
4.2.4. Transient Reduction in Gibberellin Activity by LESOY and SALIX May Redirect Metabolism Toward Defence
4.3. Comparison Between LESOY, LECI, SALIX and CHIT and Other PDSs
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| BABA | β-aminobutyric acid |
| BS | substances |
| BTH | benzothiadiazole or acibenzolar-S-methyl |
| CHIT | chitosan hydrochloride 1% |
| dpi | days post-inoculation |
| dpt | days post-treatment |
| JA | jasmonic acid |
| LECI | soy lecithin 25% + E. arvense extract 15% |
| LESOY | soy lecithin 20% |
| MeJA | methyl jasmonate |
| MFA | multiple factor analysis |
| n/a | not applicable |
| NT-INF | non-treated infected |
| NT-NOINF | non-treated non-infected |
| PDS | plant defence stimulator |
| PR | pathogenesis-related |
| SA | salicylic acid |
| SALIX | Salix cortex extract 42% + chitosan hydrochloride 0.5% |
| sp | sporangia |
| T-INF | treated infected |
References
- Gessler, C.; Pertot, I.; Perazzolli, M. Plasmopara viticola: A Review of Knowledge on Downy Mildew of Grapevine and Effective Disease Management. Phytopathol. Mediterr. 2011, 50, 3–44. [Google Scholar]
- Boso, S.; Santiago, J.L.; Martínez, M.C. Resistance of Eight Different Clones of the Grape Cultivar Albariño to Plasmopara viticola. Plant Dis. 2004, 88, 741–744. [Google Scholar] [CrossRef] [PubMed]
- Unger, S.; Büche, C.; Boso, S.; Kassemeyer, H.-H. The Course of Colonization of Two Different Vitis Genotypes by Plasmopara viticola Indicates Compatible and Incompatible Host-Pathogen Interactions. Phytopathology 2007, 97, 780–786. [Google Scholar] [CrossRef] [PubMed]
- Buonassisi, D.; Colombo, M.; Migliaro, D.; Dolzani, C.; Peressotti, E.; Mizzotti, C.; Velasco, R.; Masiero, S.; Perazzolli, M.; Vezzulli, S. Breeding for Grapevine Downy Mildew Resistance: A Review of “Omics” Approaches. Euphytica 2017, 213, 103. [Google Scholar] [CrossRef]
- Peng, J.; Wang, X.; Wang, H.; Li, X.; Zhang, Q.; Wang, M.; Yan, J. Advances in Understanding Grapevine Downy Mildew: From Pathogen Infection to Disease Management. Mol. Plant Pathol. 2024, 25, e13401. [Google Scholar] [CrossRef]
- Koledenkova, K.; Esmaeel, Q.; Jacquard, C.; Nowak, J.; Clément, C.; Ait Barka, E. Plasmopara viticola the Causal Agent of Downy Mildew of Grapevine: From Its Taxonomy to Disease Management. Front. Microbiol. 2022, 13, 889472. [Google Scholar] [CrossRef]
- European Parliament and the Council of the European Union Regulation (EC) No 1107/2009 of the European Parliament and of the Council of 21 October 2009 Concerning the Placing of Plant Protection Products on the Market and Repealing Council Directives 79/117/EEC and 91/414/EEC. Off. J. Eur. Union 2009, 309, 1–50.
- Kookana, R.S.; Baskaran, S.; Naidu, R. Pesticide Fate and Behaviour in Australian Soils in Relation to Contamination and Management of Soil and Water: A Review. Soil Res. 1998, 36, 715–764. [Google Scholar] [CrossRef]
- Komárek, M.; Čadková, E.; Chrastný, V.; Bordas, F.; Bollinger, J.-C. Contamination of Vineyard Soils with Fungicides: A Review of Environmental and Toxicological Aspects. Environ. Int. 2010, 36, 138–151. [Google Scholar] [CrossRef]
- Bora, F.-D.; Bunea, C.-I.; Rusu, T.; Pop, N. Vertical Distribution and Analysis of Micro-, Macroelements and Heavy Metals in the System Soil-Grapevine-Wine in Vineyard from North-West Romania. Chem. Cent. J. 2015, 9, 19. [Google Scholar] [CrossRef]
- Karimi, B.; Masson, V.; Guilland, C.; Leroy, E.; Pellegrinelli, S.; Giboulot, E.; Maron, P.-A.; Ranjard, L. Ecotoxicity of Copper Input and Accumulation for Soil Biodiversity in Vineyards. Environ. Chem. Lett. 2021, 19, 2013–2030. [Google Scholar] [CrossRef]
- Corio-Costet, M.F. Fungicide Resistance in Plasmopara viticola in France and Anti-Resistance Measures. In Fungicide Resistance in Crop Protection: Risk and Management; Thind, T.S., Ed.; CABI: Wallingford, UK, 2012; pp. 157–171. [Google Scholar]
- Sánchez-Zelaia, H.; Nanni, I.M.; Oggiano, I.; Hernández, M.; Díez-Navajas, A.M.; Collina, M. Droplet Digital PCR: A New Molecular Method to Detect G1105S/V Mutations in Plasmopara viticola CesA3 Gene. Biology 2024, 13, 919. [Google Scholar] [CrossRef] [PubMed]
- Wightwick, A.M.; Mollah, M.R.; Partington, D.L.; Allinson, G. Copper Fungicide Residues in Australian Vineyard Soils. J. Agric. Food Chem. 2008, 56, 2457–2464. [Google Scholar] [CrossRef] [PubMed]
- Garde-Cerdán, T.; Mancini, V.; Carrasco-Quiroz, M.; Servili, A.; Gutiérrez-Gamboa, G.; Foglia, R.; Pérez-Álvarez, E.P.; Romanazzi, G. Chitosan and Laminarin as Alternatives to Copper for Plasmopara viticola Control: Effect on Grape Amino Acid. J. Agric. Food Chem. 2017, 65, 7379–7386. [Google Scholar] [CrossRef] [PubMed]
- European Commission a European Green Deal. Striving to Be the First Climate-Neutral Continent. Available online: https://commission.europa.eu/strategy-and-policy/priorities-2019-2024/european-green-deal_en (accessed on 26 August 2022).
- European Parliament and the Council of the European Union Directive 2009/128/EC of the European Parliament and of the Council of 21 October 2009 Establishing a Framework for Community Action to Achieve the Sustainable Use of Pesticides. Off. J. Eur. Union 2009, 309, 71–86.
- Halleux, V. Sustainable Use of Plant Protection Products; European Parliamentary Research Service: Brussels, Belgium, 2023. [Google Scholar]
- European Commission EU Pesticides Database. Available online: https://ec.europa.eu/food/plant/pesticides/eu-pesticides-database/start/screen/active-substances (accessed on 5 October 2022).
- Aziz, A.; Trotel-Aziz, P.; Dhuicq, L.; Jeandet, P.; Couderchet, M.; Vernet, G. Chitosan Oligomers and Copper Sulfate Induce Grapevine Defense Reactions and Resistance to Gray Mold and Downy Mildew. Phytopathology 2006, 96, 1188–1194. [Google Scholar] [CrossRef]
- Aziz, A.; Gauthier, A.; Bezier, A.; Poinssot, B.; Joubert, J.-M.; Pugin, A.; Heyraud, A.; Baillieul, F. Elicitor and Resistance-Inducing Activities of β-1,4 Cellodextrins in Grapevine, Comparison with β-1,3 Glucans and α-1,4 Oligogalacturonides. J. Exp. Bot. 2007, 58, 1463–1472. [Google Scholar] [CrossRef]
- De Bona, G.S.; Vincenzi, S.; De Marchi, F.; Angelini, E.; Bertazzon, N. Chitosan Induces Delayed Grapevine Defense Mechanisms and Protects Grapevine against Botrytis cinerea. J. Plant Dis. Prot. 2021, 128, 715–724. [Google Scholar] [CrossRef]
- Scholfield, C.R. Composition of Soybean Lecithin. J. Am. Oil Chem. Soc. 1981, 58, 889–892. [Google Scholar] [CrossRef]
- Laxalt, A.M.; Munnik, T. Phospholipid Signalling in Plant Defence. Curr. Opin. Plant Biol. 2002, 5, 332–338. [Google Scholar] [CrossRef]
- Canonne, J.; Froidure-Nicolas, S.; Rivas, S. Phospholipases in Action during Plant Defense Signaling. Plant Signal. Behav. 2011, 6, 13–18. [Google Scholar] [CrossRef] [PubMed]
- Cavaco, A.R.; Laureano, G.; Duarte, B.; Marques da Silva, J.; Gameiro, C.; Cunha, J.; Eiras Dias, J.; Matos, A.R.; Figueiredo, A. First Assessment of Leaf Lipids and Fatty Acids as Biomarkers of Grapevine Tolerance/Susceptibility to Plasmopara viticola. Physiol. Mol. Plant Pathol. 2023, 124, 101948. [Google Scholar] [CrossRef]
- Wasternack, C.; Feussner, I. The Oxylipin Pathways: Biochemistry and Function. Annu. Rev. Plant Biol. 2018, 69, 363–386. [Google Scholar] [CrossRef] [PubMed]
- Ali, K.; Maltese, F.; Figueiredo, A.; Rex, M.; Fortes, A.M.; Zyprian, E.; Pais, M.S.; Verpoorte, R.; Choi, Y.H. Alterations in Grapevine Leaf Metabolism upon Inoculation with Plasmopara viticola in Different Time-Points. Plant Sci. 2012, 191–192, 100–107. [Google Scholar] [CrossRef]
- Figueiredo, A.; Monteiro, F.; Sebastiana, M. First Clues on a Jasmonic Acid Role in Grapevine Resistance against the Biotrophic Fungus Plasmopara viticola. Eur. J. Plant Pathol. 2015, 142, 645–652. [Google Scholar] [CrossRef]
- Guerreiro, A.; Figueiredo, J.; Sousa Silva, M.; Figueiredo, A. Linking Jasmonic Acid to Grapevine Resistance against the Biotrophic Oomycete Plasmopara viticola. Front. Plant Sci. 2016, 7, 565. [Google Scholar] [CrossRef]
- Kammerer, B.; Kahlich, R.; Biegert, C.; Gleiter, C.H.; Heide, L. HPLC-MS/MS Analysis of Willow Bark Extracts Contained in Pharmaceutical Preparations. Phytochem. Anal. 2005, 16, 470–478. [Google Scholar] [CrossRef]
- Ramos, P.A.B.; Moreirinha, C.; Santos, S.A.O.; Almeida, A.; Freire, C.S.R.; Silva, A.M.S.; Silvestre, A.J.D. Valorisation of Bark Lipophilic Fractions from Three Portuguese Salix Species: A Systematic Study of the Chemical Composition and Inhibitory Activity on Escherichia coli. Ind. Crops Prod. 2019, 132, 245–252. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, X. Salicylic Acid: Biosynthesis, Perception, and Contributions to Plant Immunity. Curr. Opin. Plant Biol. 2019, 50, 29–36. [Google Scholar] [CrossRef]
- Derckel, J.-P.; Legendre, L.; Audran, J.-C.; Haye, B.; Lambert, B. Chitinases of the Grapevine (Vitis vinifera L.): Five Isoforms Induced in Leaves by Salicylic Acid Are Constitutively Expressed in Other Tissues. Plant Sci. 1996, 119, 31–37. [Google Scholar] [CrossRef]
- Derckel, J.-P.; Audran, J.-C.; Haye, B.; Lambert, B.; Legendre, L. Characterization, Induction by Wounding and Salicylic Acid, and Activity against Botrytis cinerea of Chitinases and β-1,3-Glucanases of Ripening Grape Berries. Physiol. Plant. 1998, 104, 56–64. [Google Scholar] [CrossRef]
- Renault, A.-S.; Deloire, A.; Bierne, J. Pathogenesis-Related Proteins in Grapevines Induced by Salicylic Acid and Botrytis cinerea. Vitis 1996, 35, 49–52. [Google Scholar] [CrossRef]
- Dufour, M.C.; Lambert, C.; Bouscaut, J.; Mérillon, J.M.; Corio-Costet, M.F. Benzothiadiazole-Primed Defence Responses and Enhanced Differential Expression of Defence Genes in Vitis vinifera Infected with Biotrophic Pathogens Erysiphe necator and Plasmopara viticola: Elicitation and Grapevine Responses to Mildews. Plant Pathol. 2013, 62, 370–382. [Google Scholar] [CrossRef]
- Bodin, E.; Bellée, A.; Dufour, M.-C.; André, O.; Corio-Costet, M.-F. Grapevine Stimulation: A Multidisciplinary Approach to Investigate the Effects of Biostimulants and a Plant Defense Stimulator. J. Agric. Food Chem. 2020, 68, 15085–15096. [Google Scholar] [CrossRef] [PubMed]
- Burdziej, A.; Bellée, A.; Bodin, E.; Valls Fonayet, J.; Magnin, N.; Szakiel, A.; Richard, T.; Cluzet, S.; Corio-Costet, M.-F. Three Types of Elicitors Induce Grapevine Resistance against Downy Mildew via Common and Specific Immune Responses. J. Agric. Food Chem. 2021, 69, 1781–1795. [Google Scholar] [CrossRef]
- Mimica-Dukic, N.; Simin, N.; Cvejic, J.; Jovin, E.; Orcic, D.; Bozin, B. Phenolic Compounds in Field Horsetail (Equisetum arvense L.) as Natural Antioxidants. Molecules 2008, 13, 1455–1464. [Google Scholar] [CrossRef] [PubMed]
- Kolomiets, N.E.; Yusubov, M.S.; Kalinkina, G.I. Flavonoid Composition of Equisetum Arvense and E. x litorale Studied by High-Performance Liquid Chromatography–Mass Spectrometry. Chem. Nat. Compd. 2012, 48, 135–136. [Google Scholar] [CrossRef]
- Garcia, D.; Ramos, A.J.; Sanchis, V.; Marín, S. Equisetum arvense Hydro-Alcoholic Extract: Phenolic Composition and Antifungal and Antimycotoxigenic Effect against Aspergillus flavus and Fusarium verticillioides in Stored Maize: Equisetum arvense Hydro-Alcoholic Extract. J. Sci. Food Agric. 2013, 93, 2248–2253. [Google Scholar] [CrossRef]
- Langa-Lomba, N.; Buzón-Durán, L.; Martín-Ramos, P.; Casanova-Gascón, J.; Martín-Gil, J.; Sánchez-Hernández, E.; González-García, V. Assessment of Conjugate Complexes of Chitosan and Urtica dioica or Equisetum arvense Extracts for the Control of Grapevine Trunk Pathogens. Agronomy 2021, 11, 976. [Google Scholar] [CrossRef]
- Llamazares De Miguel, D.; Mena-Petite, A.; Díez-Navajas, A.M. Toxicity and Preventive Activity of Chitosan, Equisetum arvense, Lecithin and Salix Cortex against Plasmopara viticola, the Causal Agent of Downy Mildew in Grapevine. Agronomy 2022, 12, 3139. [Google Scholar] [CrossRef]
- De Miccolis Angelini, R.M.; Rotolo, C.; Gerin, D.; Abate, D.; Pollastro, S.; Faretra, F. Global Transcriptome Analysis and Differentially Expressed Genes in Grapevine after Application of the Yeast-derived Defense Inducer Cerevisane. Pest. Manag. Sci. 2019, 75, 2020–2033. [Google Scholar] [CrossRef] [PubMed]
- Pagliarani, C.; Moine, A.; Chitarra, W.; Meloni, G.R.; Abbà, S.; Nerva, L.; Pugliese, M.; Gullino, M.L.; Gambino, G. The Molecular Priming of Defense Responses Is Differently Regulated in Grapevine Genotypes Following Elicitor Application against Powdery Mildew. Int. J. Mol. Sci. 2020, 21, 6776. [Google Scholar] [CrossRef] [PubMed]
- Adrian, M.; Lucio, M.; Roullier-Gall, C.; Héloir, M.-C.; Trouvelot, S.; Daire, X.; Kanawati, B.; Lemaître-Guillier, C.; Poinssot, B.; Gougeon, R.; et al. Metabolic Fingerprint of PS3-Induced Resistance of Grapevine Leaves against Plasmopara viticola Revealed Differences in Elicitor-Triggered Defenses. Front. Plant Sci. 2017, 8, 101. [Google Scholar] [CrossRef] [PubMed]
- Lemaître-Guillier, C.; Hovasse, A.; Schaeffer-Reiss, C.; Recorbet, G.; Poinssot, B.; Trouvelot, S.; Daire, X.; Adrian, M.; Héloir, M.-C. Proteomics towards the Understanding of Elicitor Induced Resistance of Grapevine against Downy Mildew. J. Proteom. 2017, 156, 113–125. [Google Scholar] [CrossRef]
- Krzyzaniak, Y.; Trouvelot, S.; Negrel, J.; Cluzet, S.; Valls, J.; Richard, T.; Bougaud, A.; Jacquens, L.; Klinguer, A.; Chiltz, A.; et al. A Plant Extract Acts Both as a Resistance Inducer and an Oomycide Against Grapevine Downy Mildew. Front. Plant Sci. 2018, 9, 1085. [Google Scholar] [CrossRef]
- Reuveni, M.; Zahavi, T.; Cohen, Y. Controlling Downy Mildew (Plasmopara viticola) in Field-Grown Grapevine with β-Aminobutyric Acid (BABA). Phytopamsitica 2001, 29, 125–133. [Google Scholar] [CrossRef]
- Cohen, Y.; Reuveni, M.; Baider, A. Local and Systemic Activity of Baba (DL-3-Aminobutyric Acid) Against Plasmopara viticola in Grapevines. In Advances in Downy Mildew Research; Spencer-Phillips, P.T.N., Gisi, U., Lebeda, A., Eds.; Kluwer Academic Publishers: Dordrecht, The Netherlands, 2002; pp. 207–224. [Google Scholar]
- Aziz, A.; Poinssot, B.; Daire, X.; Adrian, M.; Bézier, A.; Lambert, B.; Joubert, J.-M.; Pugin, A. Laminarin Elicits Defense Responses in Grapevine and Induces Protection Against Botrytis cinerea and Plasmopara viticola. Mol. Plant-Microbe Interact. 2003, 16, 1118–1128. [Google Scholar] [CrossRef]
- Hamiduzzaman, M.M.; Jakab, G.; Barnavon, L.; Neuhaus, J.-M.; Mauch-Mani, B. β-Aminobutyric Acid-Induced Resistance Against Downy Mildew in Grapevine Acts Through the Potentiation of Callose Formation and Jasmonic Acid Signaling. Mol. Plant-Microbe Interact. 2005, 18, 819–829. [Google Scholar] [CrossRef]
- Trotel-Aziz, P.; Couderchet, M.; Vernet, G.; Aziz, A. Chitosan Stimulates Defense Reactions in Grapevine Leaves and Inhibits Development of Botrytis cinerea. Eur. J. Plant Pathol. 2006, 114, 405–413. [Google Scholar] [CrossRef]
- Delaunois, B.; Farace, G.; Jeandet, P.; Clément, C.; Baillieul, F.; Dorey, S.; Cordelier, S. Elicitors as Alternative Strategy to Pesticides in Grapevine? Current Knowledge on Their Mode of Action from Controlled Conditions to Vineyard. Environ. Sci. Pollut. Res. 2014, 21, 4837–4846. [Google Scholar] [CrossRef]
- van Aubel, G.; Buonatesta, R.; Van Cutsem, P. COS-OGA: A Novel Oligosaccharidic Elicitor That Protects Grapes and Cucumbers against Powdery Mildew. Crop Prot. 2014, 65, 129–137. [Google Scholar] [CrossRef]
- Baccelli, I.; Mauch-Mani, B. Beta-Aminobutyric Acid Priming of Plant Defense: The Role of ABA and Other Hormones. Plant Mol. Biol. 2016, 91, 703–711. [Google Scholar] [CrossRef] [PubMed]
- Dufour, M.-C.; Magnin, N.; Dumas, B.; Vergnes, S.; Corio-Costet, M.-F. High-Throughput Gene-Expression Quantification of Grapevine Defense Responses in the Field Using Microfluidic Dynamic Arrays. BMC Genom. 2016, 17, 957. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Pfaffl, M.W. A New Mathematical Model for Relative Quantification in Real-Time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef]
- Díez-Navajas, A.M.; Greif, C.; Poutaraud, A.; Merdinoglu, D. Two Simplified Fluorescent Staining Techniques to Observe Infection Structures of the Oomycete Plasmopara viticola in Grapevine Leaf Tissues. Micron 2007, 38, 680–683. [Google Scholar] [CrossRef]
- Chen, L.; Wang, W.-S.; Wang, T.; Meng, X.-F.; Chen, T.; Huang, X.-X.; Li, Y.; Hou, B.-K. Methyl Salicylate Glucosylation Regulates Plant Defense Signaling and Systemic Acquired Resistance. Plant Physiol. 2019, 180, 2167–2181. [Google Scholar] [CrossRef]
- Sun, X.; Lapin, D.; Feehan, J.M.; Stolze, S.C.; Kramer, K.; Dongus, J.A.; Rzemieniewski, J.; Blanvillain-Baufumé, S.; Harzen, A.; Bautor, J.; et al. Pathogen Effector Recognition-Dependent Association of NRG1 with EDS1 and SAG101 in TNL Receptor Immunity. Nat. Commun. 2021, 12, 3335. [Google Scholar] [CrossRef]
- Tian, H.; Wu, Z.; Chen, S.; Ao, K.; Huang, W.; Yaghmaiean, H.; Sun, T.; Xu, F.; Zhang, Y.; Wang, S.; et al. Activation of TIR Signalling Boosts Pattern-Triggered Immunity. Nature 2021, 598, 500–503. [Google Scholar] [CrossRef]
- Gullner, G.; Komives, T.; Király, L.; Schröder, P. Glutathione S-Transferase Enzymes in Plant-Pathogen Interactions. Front. Plant Sci. 2018, 9, 1836. [Google Scholar] [CrossRef]
- Yao, T.; Feng, K.; Xie, M.; Barros, J.; Tschaplinski, T.J.; Tuskan, G.A.; Muchero, W.; Chen, J.-G. Phylogenetic Occurrence of the Phenylpropanoid Pathway and Lignin Biosynthesis in Plants. Front. Plant Sci. 2021, 12, 704697. [Google Scholar] [CrossRef]
- Barros, J.; Dixon, R.A. Plant Phenylalanine/Tyrosine Ammonia-Lyases. Trends Plant Sci. 2020, 25, 66–79. [Google Scholar] [CrossRef] [PubMed]
- Bernard, S.M.; Habash, D.Z. The Importance of Cytosolic Glutamine Synthetase in Nitrogen Assimilation and Recycling. New Phytol. 2009, 182, 608–620. [Google Scholar] [CrossRef] [PubMed]
- Joglekar, S.; Suliman, M.; Bartsch, M.; Halder, V.; Maintz, J.; Bautor, J.; Zeier, J.; Parker, J.E.; Kombrink, E. Chemical Activation of EDS1/PAD4 Signaling Leading to Pathogen Resistance in Arabidopsis. Plant Cell Physiol. 2018, 59, 1592–1607. [Google Scholar] [CrossRef] [PubMed]
- Yu, F.; Utsumi, R. Diversity, Regulation, and Genetic Manipulation of Plant Mono- and Sesquiterpenoid Biosynthesis. Cell. Mol. Life Sci. 2009, 66, 3043–3052. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Wang, Z.; Cui, Z.; Qi, Q.; Hou, J. Progress and Perspectives for Microbial Production of Farnesene. Bioresour. Technol. 2022, 347, 126682. [Google Scholar] [CrossRef]
- Falcone Ferreyra, M.L.; Rius, S.P.; Casati, P. Flavonoids: Biosynthesis, Biological Functions, and Biotechnological Applications. Front. Plant Sci. 2012, 3, 222. [Google Scholar] [CrossRef]
- Aranega-Bou, P.; De La O Leyva, M.; Finiti, I.; García-Agustín, P.; González-Bosch, C. Priming of Plant Resistance by Natural Compounds. Hexanoic Acid as a Model. Front. Plant Sci. 2014, 5, 488. [Google Scholar] [CrossRef]
- Conrath, U. Chapter 9 Priming of Induced Plant Defense Responses. In Advances in Botanical Research; Elsevier: Amsterdam, The Netherlands, 2009; Volume 51, pp. 361–395. [Google Scholar]
- Legay, G.; Marouf, E.; Berger, D.; Neuhaus, J.-M.; Mauch-Mani, B.; Slaughter, A. Identification of Genes Expressed during the Compatible Interaction of Grapevine with Plasmopara viticola through Suppression Subtractive Hybridization (SSH). Eur. J. Plant Pathol. 2011, 129, 281–301. [Google Scholar] [CrossRef]
- Kortekamp, A. Expression Analysis of Defence-Related Genes in Grapevine Leaves after Inoculation with a Host and a Non-Host Pathogen. Plant Physiol. Biochem. 2006, 44, 58–67. [Google Scholar] [CrossRef]
- Milli, A.; Cecconi, D.; Bortesi, L.; Persi, A.; Rinalducci, S.; Zamboni, A.; Zoccatelli, G.; Lovato, A.; Zolla, L.; Polverari, A. Proteomic Analysis of the Compatible Interaction between Vitis Vinifera and Plasmopara viticola. J. Proteom. 2012, 75, 1284–1302. [Google Scholar] [CrossRef]
- Liu, S.; Wu, J.; Zhang, P.; Hasi, G.; Huang, Y.; Lu, J.; Zhang, Y. Response of Phytohormones and Correlation of SAR Signal Pathway Genes to the Different Resistance Levels of Grapevine against Plasmopara viticola Infection. Plant Physiol. Biochem. 2016, 107, 56–66. [Google Scholar] [CrossRef] [PubMed]
- Mestre, P.; Arista, G.; Piron, M.-C.; Rustenholz, C.; Ritzenthaler, C.; Merdinoglu, D.; Chich, J.-F. Identification of a Vitis vinifera Endo-β-1,3-Glucanase with Antimicrobial Activity against Plasmopara viticola: Antimicrobial Activity of a Vitis Endoglucanase. Mol. Plant Pathol. 2017, 18, 708–719. [Google Scholar] [CrossRef] [PubMed]
- Li, M.-Y.; Jiao, Y.-T.; Wang, Y.-T.; Zhang, N.; Wang, B.-B.; Liu, R.-Q.; Yin, X.; Xu, Y.; Liu, G.-T. CRISPR/Cas9-Mediated VvPR4b Editing Decreases Downy Mildew Resistance in Grapevine (Vitis vinifera L.). Hortic. Res. 2020, 7, 149. [Google Scholar] [CrossRef] [PubMed]
- Werner, S.; Steiner, U.; Becher, R.; Kortekamp, A.; Zyprian, E.; Deising, H.B. Chitin Synthesis during in Planta Growth and Asexual Propagation of the Cellulosic Oomycete and Obligate Biotrophic Grapevine Pathogen Plasmopara viticola. FEMS Microbiol. Lett. 2002, 208, 169–173. [Google Scholar] [CrossRef]
- Ali, S.; Ganai, B.A.; Kamili, A.N.; Bhat, A.A.; Mir, Z.A.; Bhat, J.A.; Tyagi, A.; Islam, S.T.; Mushtaq, M.; Yadav, P.; et al. Pathogenesis-Related Proteins and Peptides as Promising Tools for Engineering Plants with Multiple Stress Tolerance. Microbiol. Res. 2018, 212–213, 29–37. [Google Scholar] [CrossRef]
- Manghwar, H.; Hussain, A. Mechanism of Tobacco Osmotin Gene in Plant Responses to Biotic and Abiotic Stress Tolerance: A Brief History. Biocell 2022, 46, 623–632. [Google Scholar] [CrossRef]
- He, R.; Wu, J.; Zhang, Y.; Agüero, C.B.; Li, X.; Liu, S.; Wang, C.; Walker, M.A.; Lu, J. Overexpression of a Thaumatin-like Protein Gene from Vitis amurensis Improves Downy Mildew Resistance in Vitis vinifera Grapevine. Protoplasma 2017, 254, 1579–1589. [Google Scholar] [CrossRef]
- He, M.; Xu, Y.; Cao, J.; Zhu, Z.; Jiao, Y.; Wang, Y.; Guan, X.; Yang, Y.; Xu, W.; Fu, Z. Subcellular Localization and Functional Analyses of a PR10 Protein Gene from Vitis pseudoreticulata in Response to Plasmopara viticola Infection. Protoplasma 2013, 250, 129–140. [Google Scholar] [CrossRef]
- Shimizu, T.; Suzaki, K. Multiple Gene Clusters Responsive to Plasmopara viticola Infection in Grapevines. Eur. J. Plant Pathol. 2020, 158, 681–691. [Google Scholar] [CrossRef]
- Corio-Costet, M.F.; Bellée, A.; Dufour, M.C.; Merdinoglu, D.; Coppin, C. Use of the NeoViGen96 Chip to Understand the Defense Status of Cultivars and Resistant Genotypes of Vitis vinifera. Acta Hortic. 2019, 1248, 555–560. [Google Scholar] [CrossRef]
- Bernier, F.; Berna, A. Germins and Germin-like Proteins: Plant Do-All Proteins. But What Do They Do Exactly? Plant Physiol. Biochem. 2001, 39, 545–554. [Google Scholar] [CrossRef]
- Hu, X.; Bidney, D.L.; Yalpani, N.; Duvick, J.P.; Crasta, O.; Folkerts, O.; Lu, G. Overexpression of a Gene Encoding Hydrogen Peroxide-Generating Oxalate Oxidase Evokes Defense Responses in Sunflower. Plant Physiol. 2003, 133, 170–181. [Google Scholar] [CrossRef] [PubMed]
- Verma, R.; Kaur, J. Expression of Barley Oxalate Oxidase Confers Resistance against Sclerotinia sclerotiorum in Transgenic Brassica juncea cv Varuna. Transgenic Res. 2021, 30, 143–154. [Google Scholar] [CrossRef]
- Vannozzi, A.; Dry, I.B.; Fasoli, M.; Zenoni, S.; Lucchin, M. Genome-Wide Analysis of the Grapevine Stilbene Synthase Multigenic Family: Genomic Organization and Expression Profiles upon Biotic and Abiotic Stresses. BMC Plant Biol. 2012, 12, 130. [Google Scholar] [CrossRef] [PubMed]
- Valletta, A.; Iozia, L.M.; Leonelli, F. Impact of Environmental Factors on Stilbene Biosynthesis. Plants 2021, 10, 90. [Google Scholar] [CrossRef]
- Dubrovina, A.S.; Kiselev, K.V. Regulation of Stilbene Biosynthesis in Plants. Planta 2017, 246, 597–623. [Google Scholar] [CrossRef]
- Boso, S.; Alonso-Villaverde, V.; Martínez, M.-C.; Kassemeyer, H.-H. Quantification of Stilbenes in Vitis Genotypes with Different Levels of Resistance to Plasmopara viticola Infection. Am. J. Enol. Vitic. 2012, 63, 419–423. [Google Scholar] [CrossRef]
- Goufo, P.; Singh, R.K.; Cortez, I. A Reference List of Phenolic Compounds (Including Stilbenes) in Grapevine (Vitis vinifera L.) Roots, Woods, Canes, Stems, and Leaves. Antioxidants 2020, 9, 398. [Google Scholar] [CrossRef]
- Pezet, R.; Gindro, K.; Viret, O.; Richter, H. Effects of Resveratrol, Viniferins and Pterostilbene on Plasmopara viticola Zoospore Mobility and Disease Development. Vitis 2004, 43, 145–148. [Google Scholar]
- Gindro, K.; Spring, J.L.; Pezet, R.; Richter, H.; Viret, O. Histological and Biochemical Criteria for Objective and Early Selection of Grapevine Cultivars Resistant to Plasmopara viticola. Vitis 2006, 45, 191–196. [Google Scholar]
- Malacarne, G.; Vrhovsek, U.; Zulini, L.; Cestaro, A.; Stefanini, M.; Mattivi, F.; Delledonne, M.; Velasco, R.; Moser, C. Resistance to Plasmopara viticola in a Grapevine Segregating Population Is Associated with Stilbenoid Accumulation and with Specific Host Transcriptional Responses. BMC Plant Biol. 2011, 11, 114. [Google Scholar] [CrossRef] [PubMed]
- Pezet, R.; Gindro, K.; Viret, O.; Spring, J.-L. Glycosylation and Oxidative Dimerization of Resveratrol Are Respectively Associated to Sensitivity and Resistance of Grapevine Cultivars to Downy Mildew. Physiol. Mol. Plant Pathol. 2004, 65, 297–303. [Google Scholar] [CrossRef]
- Ciaffi, M.; Paolacci, A.R.; Paolocci, M.; Alicandri, E.; Bigini, V.; Badiani, M.; Muganu, M. Transcriptional Regulation of Stilbene Synthases in Grapevine Germplasm Differentially Susceptible to Downy Mildew. BMC Plant Biol. 2019, 19, 404. [Google Scholar] [CrossRef]
- Morales, M.; Ros Barceló, A. A Basic Peroxidase Isoenzyme from Vacuoles and Cell Walls of Vitis vinifera. Phytochemistry 1997, 45, 229–232. [Google Scholar] [CrossRef]
- Wilkens, A.; Paulsen, J.; Wray, V.; Winterhalter, P. Structures of Two Novel Trimeric Stilbenes Obtained by Horseradish Peroxidase Catalyzed Biotransformation of Trans-Resveratrol and (−)-ε-Viniferin. J. Agric. Food Chem. 2010, 58, 6754–6761. [Google Scholar] [CrossRef]
- Park, S.H.; Jeong, Y.J.; Park, S.-C.; Kim, S.; Kim, Y.-G.; Shin, G.; Jeong, H.J.; Ryu, Y.B.; Lee, J.; Lee, O.R.; et al. Highly Efficient Bioconversion of trans-Resveratrol to δ-Viniferin Using Conditioned Medium of Grapevine Callus Suspension Cultures. Int. J. Mol. Sci. 2022, 23, 4403. [Google Scholar] [CrossRef]
- Christensen, S.A.; Huffaker, A.; Kaplan, F.; Sims, J.; Ziemann, S.; Doehlemann, G.; Ji, L.; Schmitz, R.J.; Kolomiets, M.V.; Alborn, H.T.; et al. Maize Death Acids, 9-Lipoxygenase–Derived Cyclopente(a)Nones, Display Activity as Cytotoxic Phytoalexins and Transcriptional Mediators. Proc. Natl. Acad. Sci. USA 2015, 112, 11407–11412. [Google Scholar] [CrossRef]
- Thakur, M.; Udayashankar, A.C. Lipoxygenases and Their Function in Plant Innate Mechanism. In Bioactive Molecules in Plant Defense: Signaling in Growth and Stress; Jogaiah, S., Abdelrahman, M., Eds.; Springer International Publishing: Cham, Switzerland, 2019. [Google Scholar]
- Vellosillo, T.; Martínez, M.; López, M.A.; Vicente, J.; Cascón, T.; Dolan, L.; Hamberg, M.; Castresana, C. Oxylipins Produced by the 9-Lipoxygenase Pathway in Arabidopsis Regulate Lateral Root Development and Defense Responses through a Specific Signaling Cascade. Plant Cell 2007, 19, 831–846. [Google Scholar] [CrossRef]
- Göbel, C.; Feussner, I.; Schmidt, A.; Scheel, D.; Sanchez-Serrano, J.; Hamberg, M.; Rosahl, S. Oxylipin Profiling Reveals the Preferential Stimulation of the 9-Lipoxygenase Pathway in Elicitor-Treated Potato Cells. J. Biol. Chem. 2001, 276, 6267–6273. [Google Scholar] [CrossRef]
- Saubeau, G.; Goulitquer, S.; Barloy, D.; Potin, P.; Andrivon, D.; Val, F. Differential Induction of Oxylipin Pathway in Potato and Tobacco Cells by Bacterial and Oomycete Elicitors. Plant Cell Rep. 2013, 32, 579–589. [Google Scholar] [CrossRef] [PubMed]
- Ninkuu, V.; Zhang, L.; Yan, J.; Fu, Z.; Yang, T.; Zeng, H. Biochemistry of Terpenes and Recent Advances in Plant Protection. Int. J. Mol. Sci. 2021, 22, 5710. [Google Scholar] [CrossRef] [PubMed]
- Nagegowda, D.A.; Gupta, P. Advances in Biosynthesis, Regulation, and Metabolic Engineering of Plant Specialized Terpenoids. Plant Sci. 2020, 294, 110457. [Google Scholar] [CrossRef] [PubMed]
- Kwun, M.S.; Lee, H.J.; Lee, D.G. β-Amyrin-Induced Apoptosis in Candida albicans Triggered by Calcium. Fungal Biol. 2021, 125, 630–636. [Google Scholar] [CrossRef]
- Algarra Alarcon, A.; Lazazzara, V.; Cappellin, L.; Bianchedi, P.L.; Schuhmacher, R.; Wohlfahrt, G.; Pertot, I.; Biasioli, F.; Perazzolli, M. Emission of Volatile Sesquiterpenes and Monoterpenes in Grapevine Genotypes Following Plasmopara viticola Inoculation in vitro: VOCs of Grapevines with Plasmopara viticola. J. Mass Spectrom. 2015, 50, 1013–1022. [Google Scholar] [CrossRef]
- Steimetz, E.; Trouvelot, S.; Gindro, K.; Bordier, A.; Poinssot, B.; Adrian, M.; Daire, X. Influence of Leaf Age on Induced Resistance in Grapevine against Plasmopara viticola. Physiol. Mol. Plant Pathol. 2012, 79, 89–96. [Google Scholar] [CrossRef]
- Lefevere, H.; Bauters, L.; Gheysen, G. Salicylic Acid Biosynthesis in Plants. Front. Plant Sci. 2020, 11, 338. [Google Scholar] [CrossRef]
- Zhang, Y.; Fan, W.; Kinkema, M.; Li, X.; Dong, X. Interaction of NPR1 with Basic Leucine Zipper Protein Transcription Factors That Bind Sequences Required for Salicylic Acid Induction of the PR-1 Gene. Proc. Natl. Acad. Sci. USA 1999, 96, 6523–6528. [Google Scholar] [CrossRef]
- Binder, B.M. Ethylene Signaling in Plants. J. Biol. Chem. 2020, 295, 7710–7725. [Google Scholar] [CrossRef]
- Toffolatti, S.L.; De Lorenzis, G.; Costa, A.; Maddalena, G.; Passera, A.; Bonza, M.C.; Pindo, M.; Stefani, E.; Cestaro, A.; Casati, P.; et al. Unique Resistance Traits against Downy Mildew from the Center of Origin of Grapevine (Vitis vinifera). Sci. Rep. 2018, 8, 12523. [Google Scholar] [CrossRef]
- Marchive, C.; Léon, C.; Kappel, C.; Coutos-Thévenot, P.; Corio-Costet, M.-F.; Delrot, S.; Lauvergeat, V. Over-Expression of VvWRKY1 in Grapevines Induces Expression of Jasmonic Acid Pathway-Related Genes and Confers Higher Tolerance to the Downy Mildew. PLoS ONE 2013, 8, e54185. [Google Scholar] [CrossRef] [PubMed]
- Encinas-Villarejo, S.; Maldonado, A.M.; Amil-Ruiz, F.; de los Santos, B.; Romero, F.; Pliego-Alfaro, F.; Muñoz-Blanco, J.; Caballero, J.L. Evidence for a Positive Regulatory Role of Strawberry (Fragaria × Ananassa) Fa WRKY1 and Arabidopsis At WRKY75 Proteins in Resistance. J. Exp. Bot. 2009, 60, 3043–3065. [Google Scholar] [CrossRef] [PubMed]
- Miedes, E.; Vanholme, R.; Boerjan, W.; Molina, A. The Role of the Secondary Cell Wall in Plant Resistance to Pathogens. Front. Plant Sci. 2014, 5, 358. [Google Scholar] [CrossRef] [PubMed]
- Wróbel-Kwiatkowska, M.; Starzycki, M.; Zebrowski, J.; Oszmiański, J.; Szopa, J. Lignin Deficiency in Transgenic Flax Resulted in Plants with Improved Mechanical Properties. J. Biotechnol. 2007, 128, 919–934. [Google Scholar] [CrossRef]
- Bhuiyan, N.H.; Selvaraj, G.; Wei, Y.; King, J. Gene Expression Profiling and Silencing Reveal That Monolignol Biosynthesis Plays a Critical Role in Penetration Defence in Wheat against Powdery Mildew Invasion. J. Exp. Bot. 2009, 60, 509–521. [Google Scholar] [CrossRef]
- Tronchet, M.; Balagué, C.; Kroj, T.; Jouanin, L.; Roby, D. Cinnamyl Alcohol Dehydrogenases-C and D, Key Enzymes in Lignin Biosynthesis, Play an Essential Role in Disease Resistance in Arabidopsis. Mol. Plant Pathol. 2010, 11, 83–92. [Google Scholar] [CrossRef]
- Liu, R.; Weng, K.; Dou, M.; Chen, T.; Yin, X.; Li, Z.; Li, T.; Zhang, C.; Xiang, G.; Liu, G.; et al. Transcriptomic Analysis of Chinese Wild Vitis pseudoreticulata in Response to Plasmopara viticola. Protoplasma 2019, 256, 1409–1424. [Google Scholar] [CrossRef]
- Xie, D.; Yang, X.; He, R.; Huo, H.; Ye, Z.; Ren, X.; Yuan, H.; Dai, Z.; Sun, J.; Su, J. Comprehensive Analysis of the UDP-glycosyltransferase Gene Family in Flax [Linum usitatissimum L.] and Functional Verification of the Role of LuUGT175 in the Regulation of Lignin Biosynthesis. Ind. Crops Prod. 2022, 188, 115720. [Google Scholar] [CrossRef]
- Cano-Delgado, A.; Penfield, S.; Smith, C.; Catley, M.; Bevan, M. Reduced Cellulose Synthesis Invokes Lignification and Defense Responses in Arabidopsis Thaliana. Plant J. 2003, 34, 351–362. [Google Scholar] [CrossRef]
- Hernández-Blanco, C.; Feng, D.X.; Hu, J.; Sánchez-Vallet, A.; Deslandes, L.; Llorente, F.; Berrocal-Lobo, M.; Keller, H.; Barlet, X.; Sánchez-Rodríguez, C.; et al. Impairment of Cellulose Synthases Required for Arabidopsis Secondary Cell Wall Formation Enhances Disease Resistance. Plant Cell 2007, 19, 890–903. [Google Scholar] [CrossRef]
- Menna, A.; Dora, S.; Sancho-Andrés, G.; Kashyap, A.; Meena, M.K.; Sklodowski, K.; Gasperini, D.; Coll, N.S.; Sánchez-Rodríguez, C. A Primary Cell Wall Cellulose-Dependent Defense Mechanism against Vascular Pathogens Revealed by Time-Resolved Dual Transcriptomics. BMC Biol. 2021, 19, 161. [Google Scholar] [CrossRef] [PubMed]
- Grenville-Briggs, L.J.; Anderson, V.L.; Fugelstad, J.; Avrova, A.O.; Bouzenzana, J.; Williams, A.; Wawra, S.; Whisson, S.C.; Birch, P.R.J.; Bulone, V.; et al. Cellulose Synthesis in Phytophthora infestans Is Required for Normal Appressorium Formation and Successful Infection of Potato. Plant Cell 2008, 20, 720–738. [Google Scholar] [CrossRef] [PubMed]
- Blum, M.; Waldner, M.; Gisi, U. A Single Point Mutation in the Novel PvCesA3 Gene Confers Resistance to the Carboxylic Acid Amide Fungicide Mandipropamid in Plasmopara viticola. Fungal Genet. Biol. 2010, 47, 499–510. [Google Scholar] [CrossRef] [PubMed]
- Haslam, T.M.; Mañas-Fernández, A.; Zhao, L.; Kunst, L. Arabidopsis ECERIFERUM2 Is a Component of the Fatty Acid Elongation Machinery Required for Fatty Acid Extension to Exceptional Lengths. Plant Physiol. 2012, 160, 1164–1174. [Google Scholar] [CrossRef]
- Keller, M. The Science of Grapevines, 3rd ed.; Academic Press: London, UK, 2020. [Google Scholar]
- Marois, J.J.; Nelson, J.K.; Morrison, J.C.; Lile, L.S.; Bledsoe, A.M. The Influence of Berry Contact within Grape Clusters on the Development of Botrytis cinerea and Epicuticular Wax. Am. J. Enol. Vitic. 1986, 37, 293–296. [Google Scholar] [CrossRef]
- Yu, Y.; Zhang, Y.; Yin, L.; Lu, J. The Mode of Host Resistance to Plasmopara viticola Infection of Grapevines. Phytopathology 2012, 102, 1094–1101. [Google Scholar] [CrossRef]
- Regnault, T.; Davière, J.-M.; Wild, M.; Sakvarelidze-Achard, L.; Heintz, D.; Carrera Bergua, E.; Lopez Diaz, I.; Gong, F.; Hedden, P.; Achard, P. The Gibberellin Precursor GA12 Acts as a Long-Distance Growth Signal in Arabidopsis. Nat. Plants 2015, 1, 15073. [Google Scholar] [CrossRef]
- De Bruyne, L.; Höfte, M.; De Vleesschauwer, D. Connecting Growth and Defense: The Emerging Roles of Brassinosteroids and Gibberellins in Plant Innate Immunity. Mol. Plant 2014, 7, 943–959. [Google Scholar] [CrossRef]
- Corio-Costet, M.-F.; Burdziej, A.; Laurens, M.; Bodin, E.; Bellée, A.; Da Costa, G.; Le Mao, I.; Cluzet, S. Effects of Three Elicitors on Primary Metabolism Six Days after Treatment in Healthy Vitis vinifera Leaves and Eight Days after Treatment in Healthy and Downy Mildew-Inoculated Leaves. OENO One 2024, 58. [Google Scholar] [CrossRef]
- Trouvelot, S.; Varnier, A.-L.; Allègre, M.; Mercier, L.; Baillieul, F.; Arnould, C.; Gianinazzi-Pearson, V.; Klarzynski, O.; Joubert, J.-M.; Pugin, A.; et al. A β-1,3 Glucan Sulfate Induces Resistance in Grapevine against Plasmopara viticola Through Priming of Defense Responses, Including HR-like Cell Death. Mol. Plant-Microbe Interact. 2008, 21, 232–243. [Google Scholar] [CrossRef]
- Boubakri, H.; Chong, J.; Poutaraud, A.; Schmitt, C.; Bertsch, C.; Mliki, A.; Masson, J.E.; Soustre-Gacougnolle, I. Riboflavin (Vitamin B2) Induces Defence Responses and Resistance to Plasmopara viticola in Grapevine. Eur. J. Plant Pathol. 2013, 136, 837–855. [Google Scholar] [CrossRef]
- Boubakri, H.; Wahab, M.A.; Chong, J.; Bertsch, C.; Mliki, A.; Soustre-Gacougnolle, I. Thiamine Induced Resistance to Plasmopara viticola in Grapevine and Elicited Host–Defense Responses, Including HR like-Cell Death. Plant Physiol. Biochem. 2012, 57, 120–133. [Google Scholar] [CrossRef]
- Dubreuil-Maurizi, C.; Trouvelot, S.; Frettinger, P.; Pugin, A.; Wendehenne, D.; Poinssot, B. β-Aminobutyric Acid Primes an NADPH Oxidase–Dependent Reactive Oxygen Species Production During Grapevine-Triggered Immunity. Mol. Plant-Microbe Interact. 2010, 23, 1012–1021. [Google Scholar] [CrossRef] [PubMed]
- Harm, A.; Kassemeyer, H.-H.; Seibicke, T.; Regner, F. Evaluation of Chemical and Natural Resistance Inducers against Downy Mildew (Plasmopara viticola) in Grapevine. Am. J. Enol. Vitic. 2011, 62, 184–192. [Google Scholar] [CrossRef]
- Slaughter, A.R.; Hamiduzzaman, M.M.; Gindro, K.; Neuhaus, J.-M.; Mauch-Mani, B. Beta-Aminobutyric Acid-Induced Resistance in Grapevine against Downy Mildew: Involvement of Pterostilbene. Eur. J. Plant Pathol. 2008, 122, 185–195. [Google Scholar] [CrossRef]
- Wendehenne, D.; Durner, J.; Chen, Z.; Klessig, D.F. Benzothiadiazole, an Inducer of Plant Defenses, Inhibits Catalase and Ascorbate Peroxidase. Phytochemistry 1998, 47, 651–657. [Google Scholar] [CrossRef]
- Scheel, D.; Parker, J.E. Elicitor Recognition and Signal Transduction in Plant Defense Gene Activation. Z. Naturforschung C 1990, 45, 569–575. [Google Scholar] [CrossRef]
- Jones, J.D.G.; Dangl, J.L. The Plant Immune System. Nature 2006, 444, 323–329. [Google Scholar] [CrossRef]
- Testerink, C.; Munnik, T. Molecular, Cellular, and Physiological Responses to Phosphatidic Acid Formation in Plants. J. Exp. Bot. 2011, 62, 2349–2361. [Google Scholar] [CrossRef]
- Zhang, Q.; Xiao, S. Lipids in Salicylic Acid-Mediated Defense in Plants: Focusing on the Roles of Phosphatidic Acid and Phosphatidylinositol 4-Phosphate. Front. Plant Sci. 2015, 6, 387. [Google Scholar] [CrossRef]
- Paasela, T.; Lim, K.-J.; Pavicic, M.; Harju, A.; Venäläinen, M.; Paulin, L.; Auvinen, P.; Kärkkäinen, K.; Teeri, T.H. Transcriptomic Analysis Reveals Novel Regulators of the Scots Pine Stilbene Pathway. Plant Cell Physiol. 2023, 64, 1204–1219. [Google Scholar] [CrossRef]
- Corio-Costet, M.-F.; Bodin, E.; Laurens, M.; Dubois, L.; Malembic-Maher, S.; Eveillard, S. Year and Flavescence Dorée Phytoplasma Infection: Impact on Gene Expression in Two Grapevine Varieties. Sci. Hortic. 2025, 347, 114190. [Google Scholar] [CrossRef]
- Wiesel, L.; Newton, A.C.; Elliott, I.; Booty, D.; Gilroy, E.M.; Birch, P.R.J.; Hein, I. Molecular Effects of Resistance Elicitors from Biological Origin and Their Potential for Crop Protection. Front. Plant Sci. 2014, 5, 655. [Google Scholar] [CrossRef]
- Sozoniuk, M.; Petrova, M.; Mishev, K.; Miladinova-Georgieva, K.; Geneva, M. Identification and Validation of Reference Genes with Stable Expression under Elicitor Treatments of the Medicinal Plant Arnica montana L. BMC Plant Biol. 2025, 25, 546. [Google Scholar] [CrossRef]
- Tresas, T.; Isaioglou, I.; Roussis, A.; Haralampidis, K. A Brief Overview of the Epigenetic Regulatory Mechanisms in Plants. Int. J. Mol. Sci. 2025, 26, 4700. [Google Scholar] [CrossRef]

| Abbr. | Company | Content | Type of Extraction | Chemical Composition | Dose (in 1 L) | Category |
|---|---|---|---|---|---|---|
| BABA | Sigma-Aldrich (Steinheim, Germany) | β-aminobutyric acid | n/a | n/a | 0.20624 g | Positive control |
| LESOY | Idai Nature SL (La pobla de Vallbona, Spain) | Soy lecithin 20% | Aqueous | Phosphatidylcholine and phosphatidylethanolamine | 3.75 mL | BS |
| LECI | Econatur (La Carlota, Spain) | Soy lecithin 25% + E. arvense extract 15% | Aqueous | Phosphatidylcholine, phosphatidylethanolamine and polyphenols such as flavonoids (quercetin, kaempferol, apigenin), phenolic acids (ferulic and caffeic acids). Presence of saponins and free amino acids (proline and glutamine) | 2.5 mL | Mixture of BS |
| SALIX | Econatur (La Carlota, Spain) | Salix cortex extract 42% + chitosan hydrochloride 0.5% | Aqueous | Phenolic extract including flavonoids (isorhamnetin and luteolin), hydrolysable tannins and SA and related salicylates | 3.33 mL | Mixture of BS |
| CHIT | Econatur (La Carlota, Spain) | Chitosan hydrochloride 1% | n/a | β-(1→4)-D-glucosamine and N-acetyl-D-glucosamine with a high degree of deacetylation | 4.16 mL | BS |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2026 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license.
Share and Cite
Llamazares De Miguel, D.; Mena-Petite, A.; Corio-Costet, M.-F.; Nieto, J.; Fernández-Navarro, J.R.; Díez-Navajas, A.M. Modulation of the Genetic Response in Vitis vinifera L. Against the Oomycete Plasmopara viticola, Causing Grapevine Downy Mildew, Through the Action of Different Basic Substances. Horticulturae 2026, 12, 112. https://doi.org/10.3390/horticulturae12010112
Llamazares De Miguel D, Mena-Petite A, Corio-Costet M-F, Nieto J, Fernández-Navarro JR, Díez-Navajas AM. Modulation of the Genetic Response in Vitis vinifera L. Against the Oomycete Plasmopara viticola, Causing Grapevine Downy Mildew, Through the Action of Different Basic Substances. Horticulturae. 2026; 12(1):112. https://doi.org/10.3390/horticulturae12010112
Chicago/Turabian StyleLlamazares De Miguel, Diego, Amaia Mena-Petite, Marie-France Corio-Costet, Juan Nieto, José R. Fernández-Navarro, and Ana M. Díez-Navajas. 2026. "Modulation of the Genetic Response in Vitis vinifera L. Against the Oomycete Plasmopara viticola, Causing Grapevine Downy Mildew, Through the Action of Different Basic Substances" Horticulturae 12, no. 1: 112. https://doi.org/10.3390/horticulturae12010112
APA StyleLlamazares De Miguel, D., Mena-Petite, A., Corio-Costet, M.-F., Nieto, J., Fernández-Navarro, J. R., & Díez-Navajas, A. M. (2026). Modulation of the Genetic Response in Vitis vinifera L. Against the Oomycete Plasmopara viticola, Causing Grapevine Downy Mildew, Through the Action of Different Basic Substances. Horticulturae, 12(1), 112. https://doi.org/10.3390/horticulturae12010112






