Early Detection of Tomato Gray Mold Based on Multispectral Imaging and Machine Learning
Abstract
1. Introduction
- (1)
- Acquire multispectral fluorescence and reflectance image data of tomato leaves at different health and disease stages (from the latent period L1 to the symptomatic period L5, corresponding to 0–120 h post-inoculation). Perform data fusion to construct a comprehensive dataset for early disease detection.
- (2)
- Apply Partial Least Squares Regression (PLSR) to quantify the correlations between spectral features and key physiological and biochemical indicators (CHL, SOD, MDA, CAT, WC), thereby identifying spectral response patterns that can characterize the early progression of the disease.
- (3)
- Employ a WGAN-GP (Wasserstein Generative Adversarial Network with Gradient Penalty) model for data augmentation to expand the dataset and optimize model performance, thereby addressing the challenge of limited sample size.
- (4)
- Develop a tomato gray mold detection model based on Random Forest to achieve precise grading of the disease severity in tomatoes.
2. Materials and Methods
2.1. Sample Collection and Disease Induction
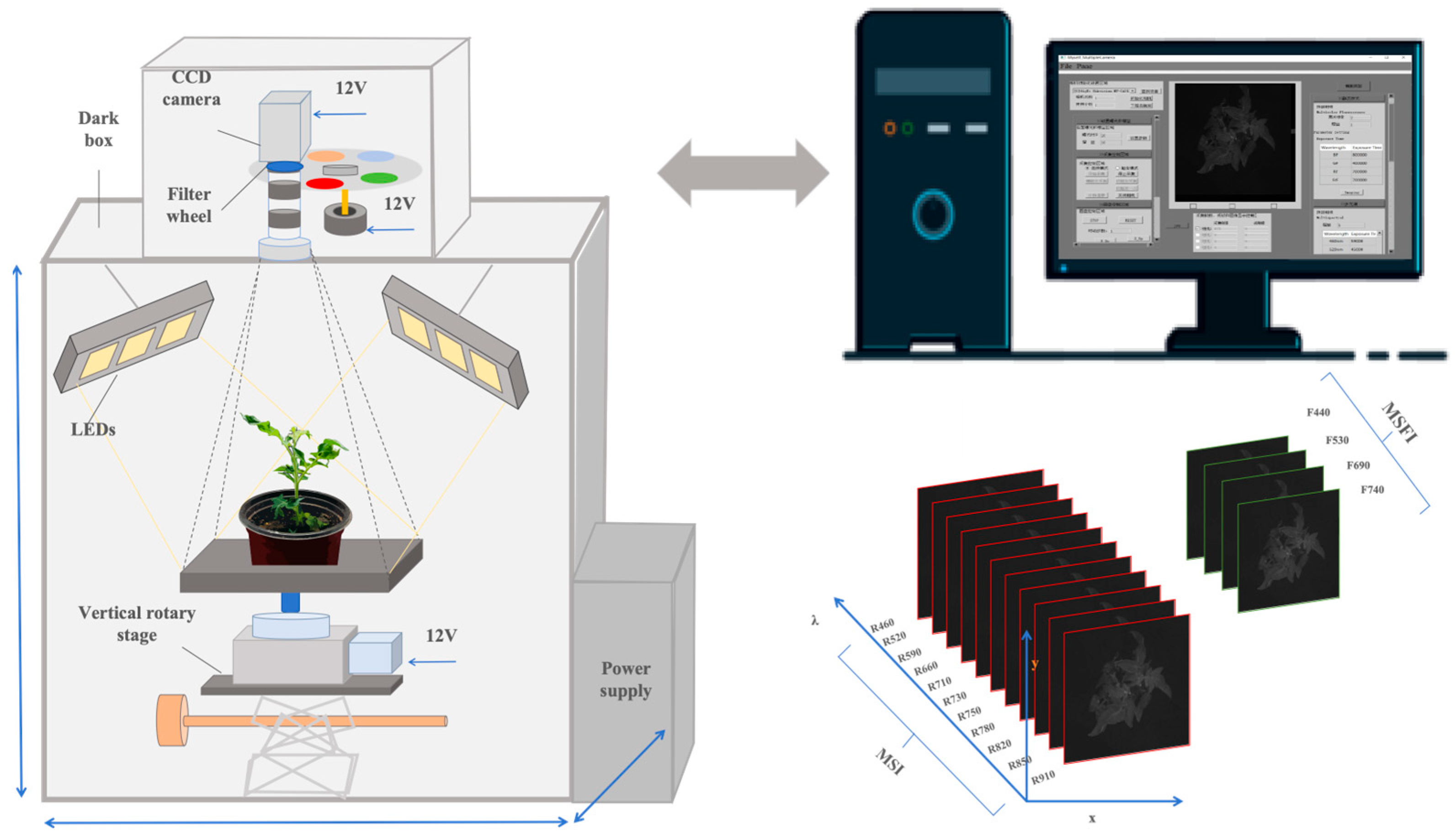
2.2. Multispectral Image Acquisition
2.3. Measurement of Physiological and Biochemical Parameters
2.4. Disease Severity Classification
2.5. Data Preprocessing
2.6. Model Development and Evaluation
2.6.1. PLSR Model for Predicting Physiological and Biochemical Parameters
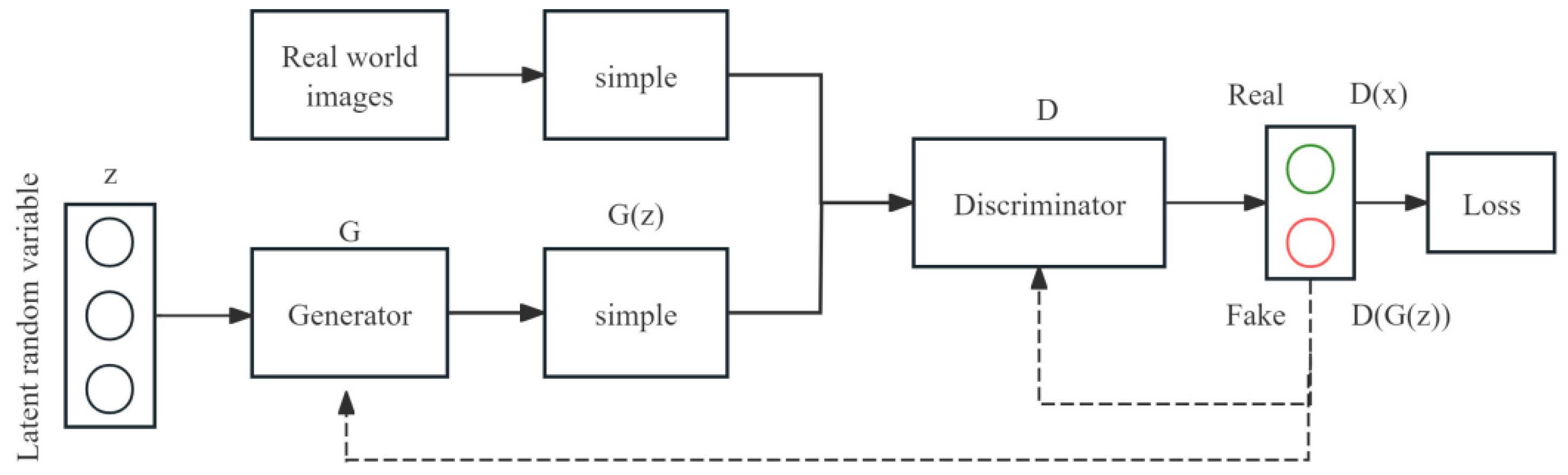
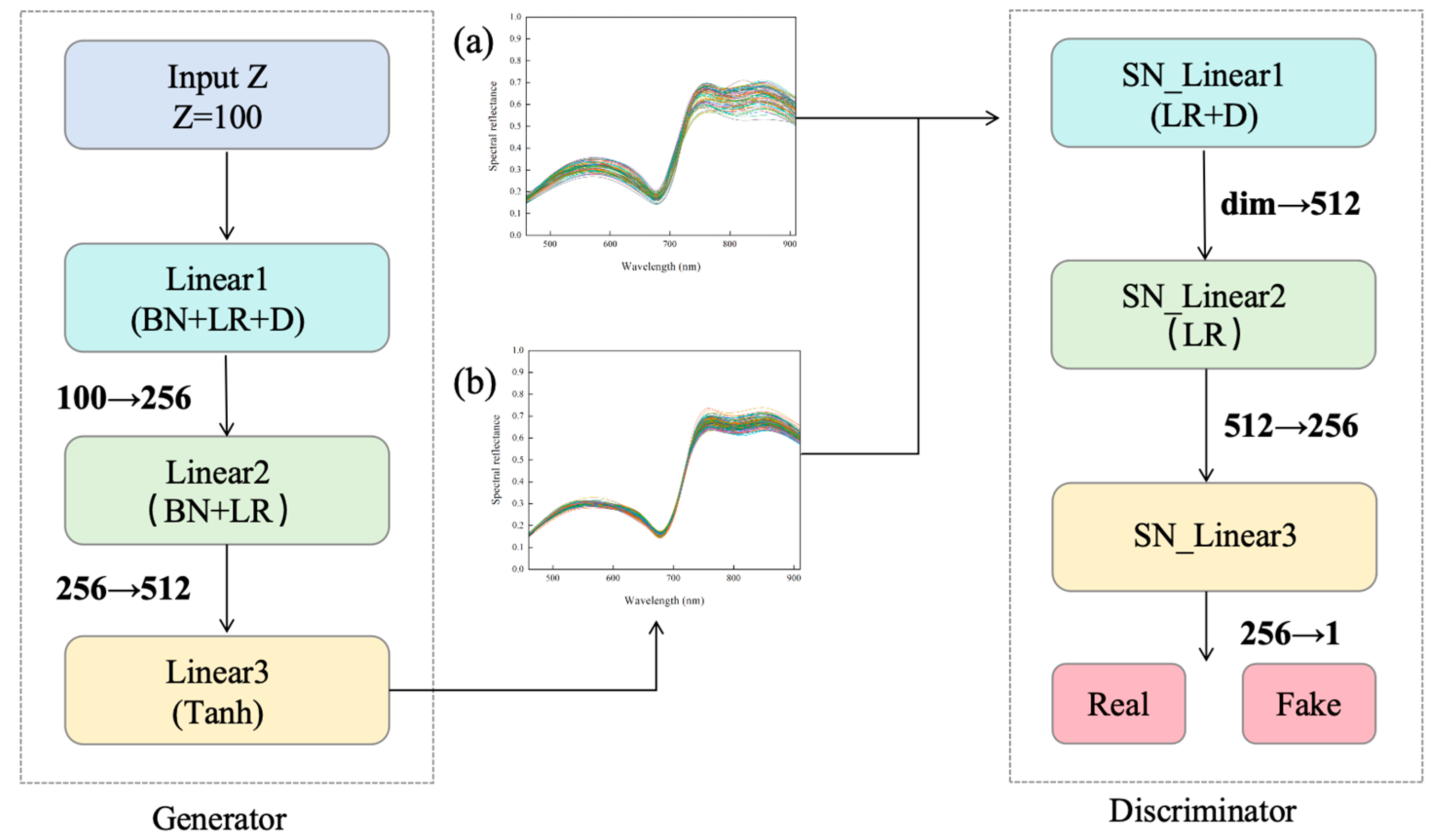
2.6.2. Improved Generative Adversarial Network (WGAN-GP) Model for Data Augmentation
2.6.3. Early Detection Model
2.6.4. Model Evaluation Method
2.7. Statistical Analysis
3. Results and Analysis
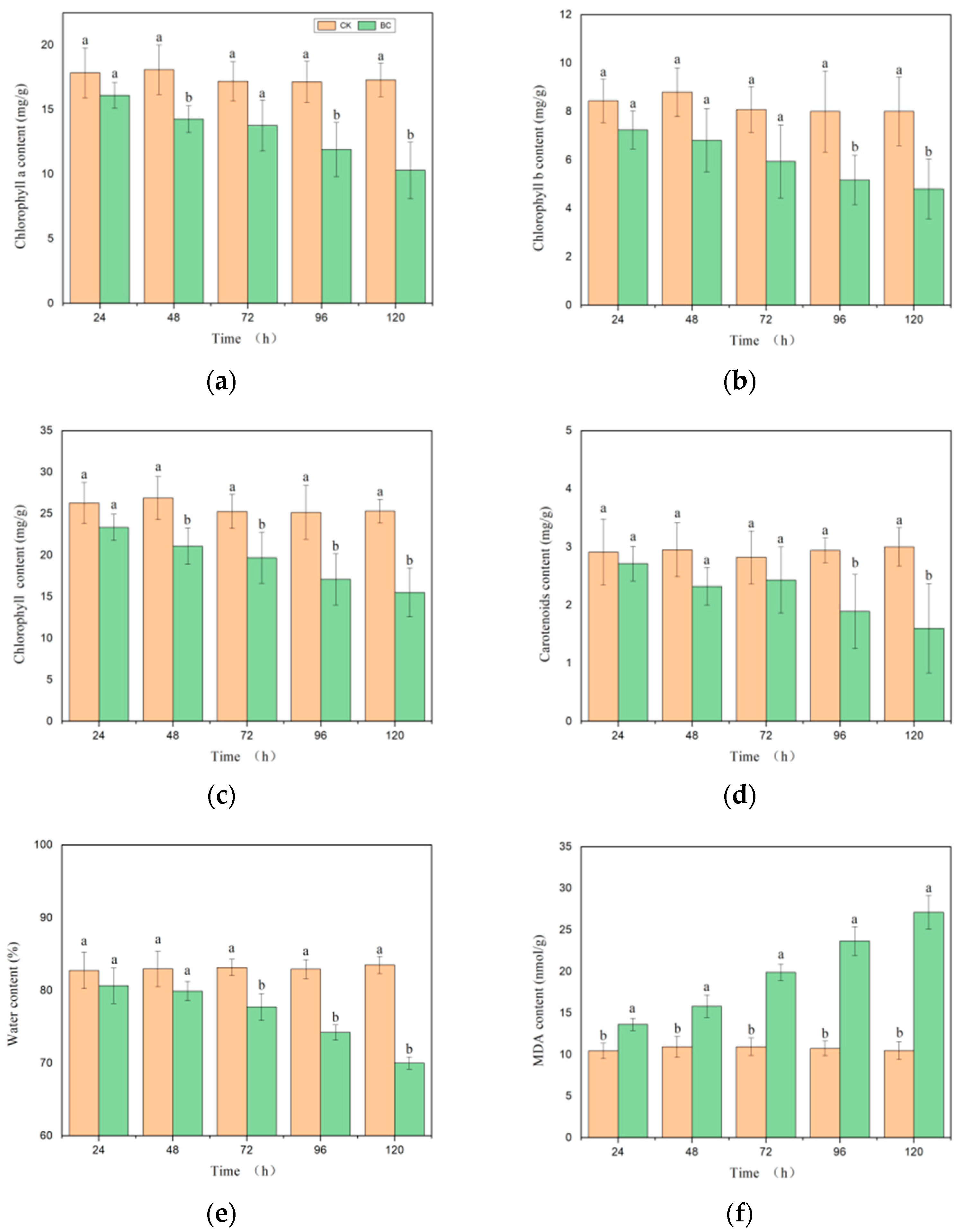
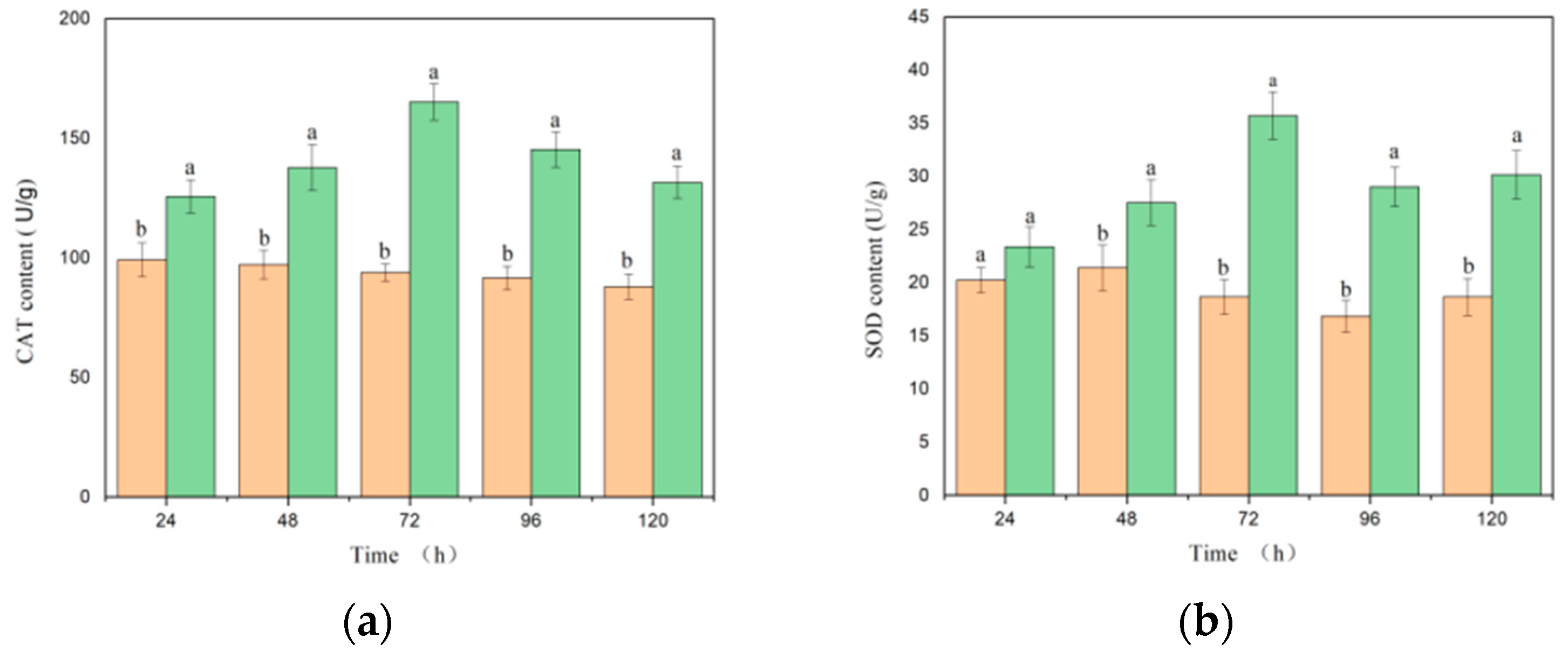
3.1. Effects of Botrytis cinerea Infection on Tomato Physiological Characteristics
3.2. Effects of Botrytis cinerea Infection on Tomato Biochemical Characteristics
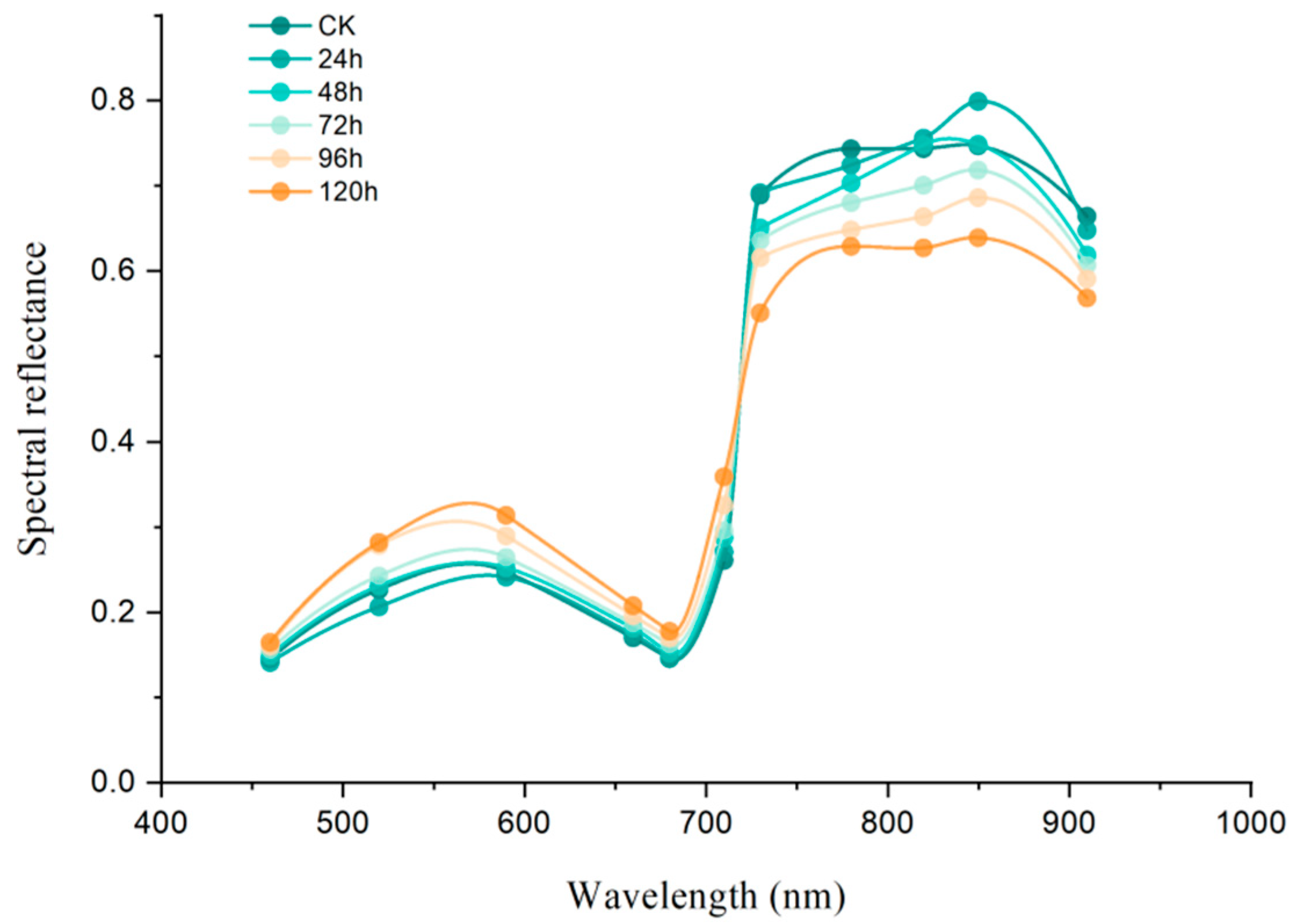
3.3. Response of Tomato Leaves to Multispectral Reflectance Under Botrytis cinerea Infection
3.3.1. Multispectral Reflectance and Vegetation Index Analysis of Tomato Leaves
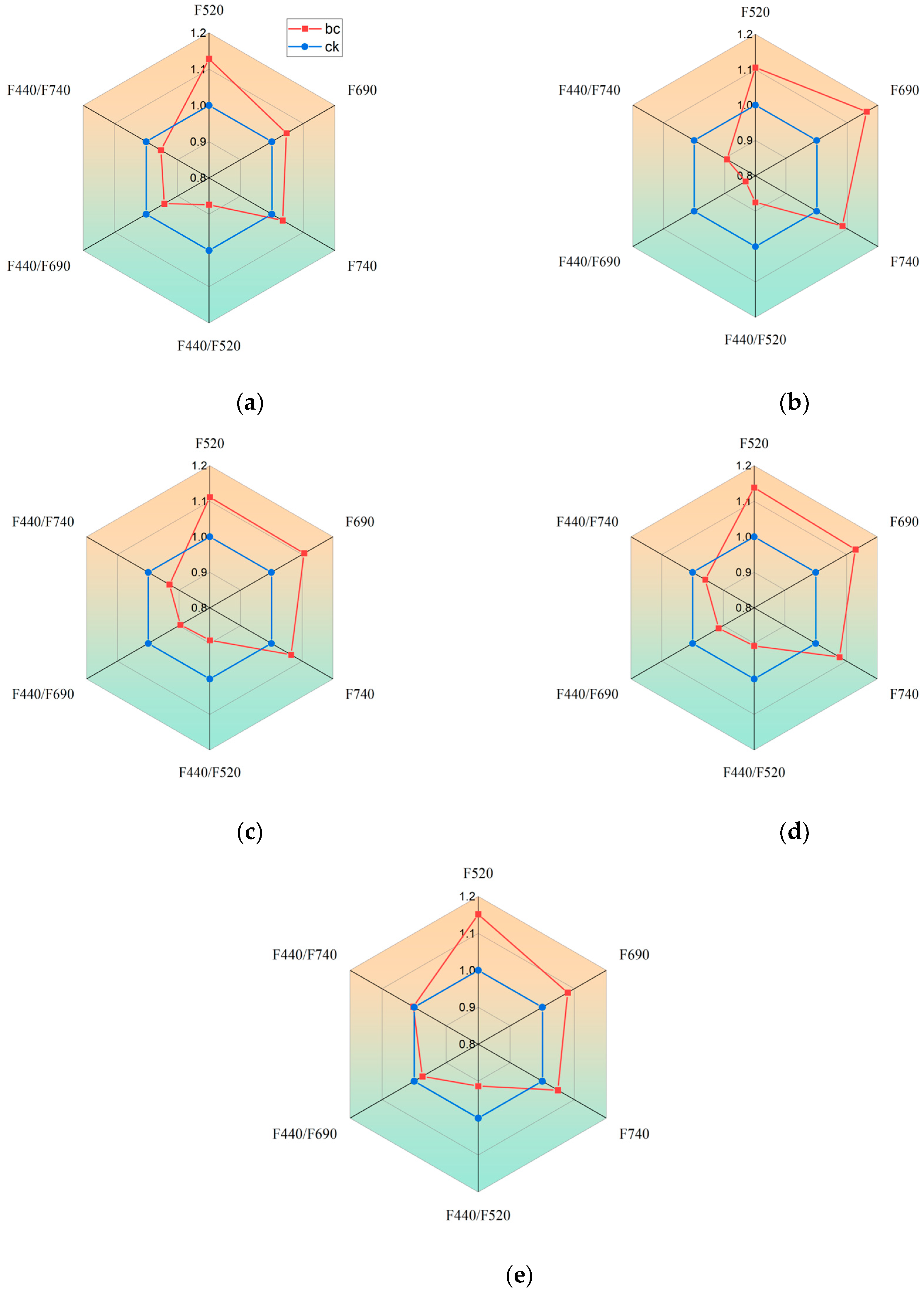
3.3.2. Changes in Multispectral Fluorescence of Tomato Leaves Under Gray Mold Infection
3.4. Augmentation of Tomato Gray Mold Spectra Using the WGAN-GP Generative Adversarial Network
3.5. PLSR Predictive Model for Physiological and Biochemical Indicators
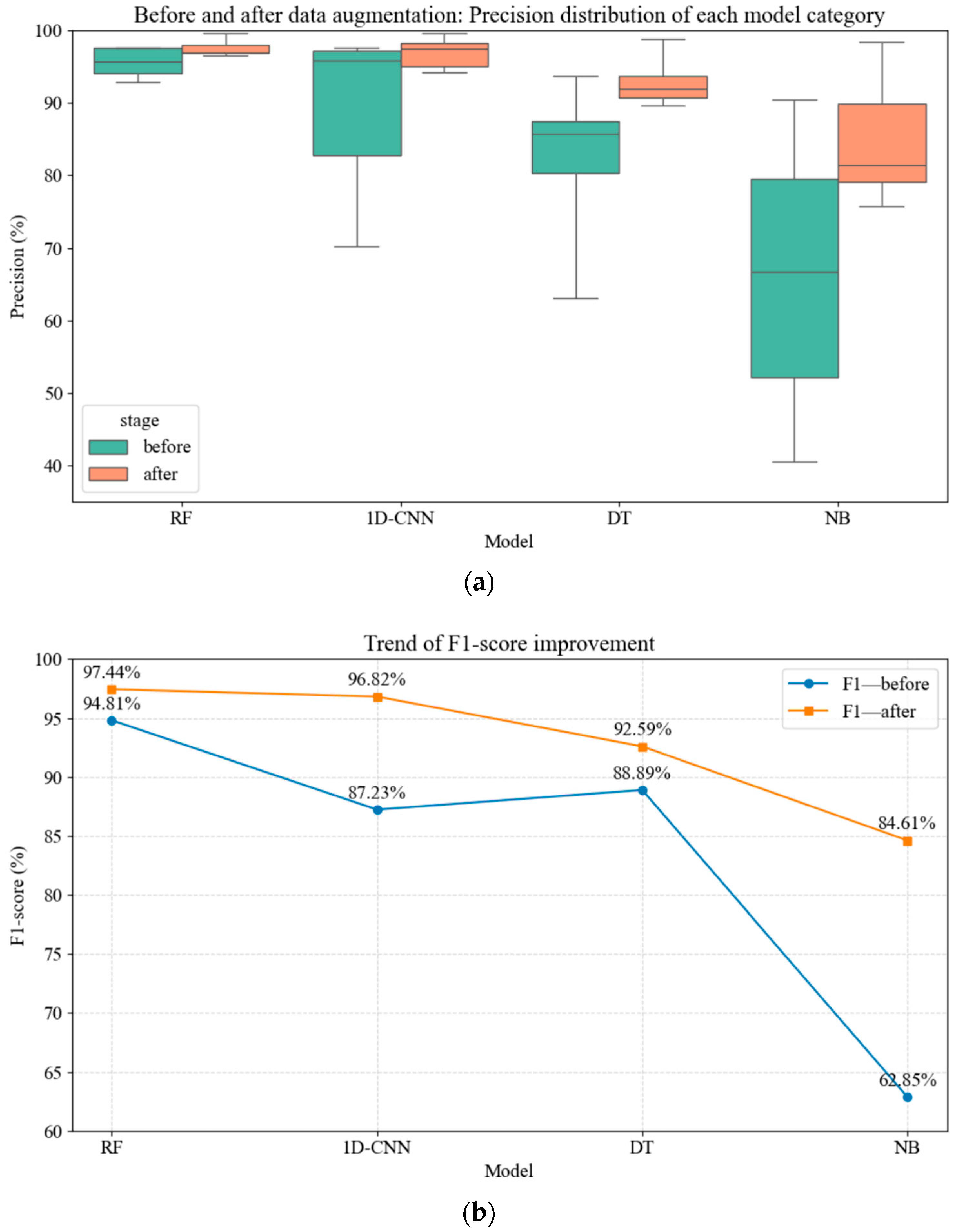
3.6. Model Comparison
4. Discussion
5. Conclusions
- (1)
- The synergistic integration of fluorescence and reflectance imaging can sensitively capture early physiological damage, and their complementarity markedly enhances the sensitivity of disease detection at the initial stage.
- (2)
- The development of the disease stages in tomatoes is mainly related to changes in malondialdehyde (MDA) and water content, and there is a certain correlation with changes in chlorophyll, superoxide dismutase (SOD), and catalase (CAT) levels. Physiological and biochemical indicators show strong correlations with all spectral parameters.
- (3)
- The Random Forest (RF) model with data augmentation achieved the best performance, with an average accuracy of 97.56% and an F1 score of 97.44%. Its overall recognition rate for early-stage diseased plants (L1–L4) reached 97.21%, significantly outperforming the 1D-CNN, DT, and NB models.
- (4)
- Data augmentation significantly enhanced the generalization ability of all models. Specifically, the NB model exhibited an 18.88% improvement in average accuracy, while the 1D-CNN achieved over 15% higher recognition rates for weaker classes (L2/L3). In addition, inter-class performance disparities among models were reduced, and the variability (standard deviation) of both RF and 1D-CNN decreased by more than 40%.
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ambresh, L.; Lingaiah, H.B.; Renuka, M.; Bhat, B.A. Development and characterization of recombinant inbreed lines for segregating bacterial wilt disease in tomato. Int. J. Curr. Microbiol. Appl. Sci. 2017, 6, 1050–1054. [Google Scholar] [CrossRef]
- Dean, R.; van Kan, J.A.L.; Pretorius, Z.A.; Hammond-Kosack, K.E.; Di Pietro, A.; Spanu, P.D.; Rudd, J.J.; Dickman, M.; Kahmann, R.; Ellis, J.; et al. The top 10 fungal pathogens in molecular plant pathology. Mol. Plant Pathol. 2012, 13, 414–430. [Google Scholar] [CrossRef]
- Sánchez-Sánchez, M.; Aispuro-Hernández, E.; Quintana-Obregón, E.A.; Vargas-Arispuro, I.C.; Martínez-Téllez, M.Á. Estimating tomato production losses due to plant viruses, a look at the past and new challenges. Comun. Sci. 2024, 15, e4247. [Google Scholar] [CrossRef]
- Singh, R.; Caseys, C.; Kliebenstein, D.J. Genetic and molecular landscapes of the generalist phytopathogen Botrytis cinerea. Mol. Plant Pathol. 2024, 25, e13404. [Google Scholar] [CrossRef] [PubMed]
- Hahn, M. The rising threat of fungicide resistance in plant pathogenic fungi: Botrytis as a case study. J. Chem. Biol. 2014, 7, 133–141. [Google Scholar] [CrossRef] [PubMed]
- Asselbergh, B.; Curvers, K.; França, S.C.; Audenaert, K.; Vuylsteke, M.; Van Breusegem, F.; Höfte, M. Resistance to Botrytis cinerea in sitiens, an abscisic acid-deficient tomato mutant, involves timely production of hydrogen peroxide and cell wall modifications in the epidermis. Plant Physiol. 2007, 144, 1863–1877. [Google Scholar] [CrossRef]
- Moter, A.; Göbel, U.B. Fluorescence in situ hybridization (FISH) for direct visualization of microorganisms. J. Microbiol. Methods 2000, 41, 85–112. [Google Scholar] [CrossRef]
- Liu, S.; Xu, H.; Deng, Y.; Cai, Y.; Wu, Y.; Zhong, X.; Zheng, J.; Lin, Z.; Ruan, M.; Chen, J.; et al. YOLOv8-LSW: A Lightweight Bitter Melon Leaf Disease Detection Model. Agriculture 2025, 15, 1281. [Google Scholar] [CrossRef]
- De Silva, M.; Brown, D. Multispectral plant disease detection with vision transformer–convolutional neural network hybrid approaches. Sensors 2023, 23, 8531. [Google Scholar] [CrossRef]
- Duan, Z.; Li, H.; Li, C.; Zhang, J.; Zhang, D.; Fan, X.; Chen, X. A CNN model for early detection of pepper Phytophthora blight using multispectral imaging, integrating spectral and textural information. Plant Methods 2024, 20, 115. [Google Scholar] [CrossRef]
- Guo, Z.; Sun, X.; Qin, L.; Dong, L.; Xiong, L.; Xie, F.; Qin, D.; Chen, Y. Identification of Golovinomyces artemisiae causing powdery mildew, changes in chlorophyll fluorescence parameters, and antioxidant levels in Artemisia selengensis. Front. Plant Sci. 2022, 13, 876050. [Google Scholar] [CrossRef]
- Matorin, D.N.; Timofeev, N.P.; Glinushkin, A.P.; Bratkovskaja, L.B.; Zayadan, B.K. Effect of fungal infection with Bipolaris sorokiniana on photosynthetic light reactions in wheat analyzed by fluorescence spectroscopy. Mosc. Univ. Biol. Sci. Bull. 2018, 73, 203–208. [Google Scholar] [CrossRef]
- Sapoukhina, N.; Boureau, T.; Rousseau, D. Plant disease symptom segmentation in chlorophyll fluorescence imaging with a synthetic dataset. Front. Plant Sci. 2022, 13, 969205. [Google Scholar] [CrossRef] [PubMed]
- Qiu, X.; Chen, H.; Huang, P.; Zhong, D.; Guo, T.; Pu, C.; Li, Z.; Liu, Y.; Chen, J.; Wang, S. Detection of citrus diseases in complex backgrounds based on image–text multimodal fusion and knowledge assistance. Front. Plant Sci. 2023, 14, 1280365. [Google Scholar] [CrossRef]
- Chen, Y.; Pan, J.; Wu, Q. Apple leaf disease identification via improved CycleGAN and convolutional neural network. Soft Comput. 2022, 27, 9773–9786. [Google Scholar] [CrossRef]
- Kanda, P.S.; Xia, K.; Sanusi, O.H. A deep learning-based recognition technique for plant leaf classification. IEEE Access 2021, 9, 162590–162613. [Google Scholar] [CrossRef]
- Min, B.; Kim, T.; Shin, D.; Shin, D. Data augmentation method for plant leaf disease recognition. Appl. Sci. 2023, 13, 1465. [Google Scholar] [CrossRef]
- Li, J.; Zeng, H.; Huang, C.; Wu, L.; Ma, J.; Zhou, B.; Ye, D.; Weng, H. Noninvasive detection of salt stress in cotton seedlings by combining multicolor fluorescence–multispectral reflectance imaging with efficientnet-OB2. Plant Phenomics 2023, 5, 0125. [Google Scholar] [CrossRef]
- Xu, F. Indirect photometric determination of chlorophyll in plant leaves. Phys. Test. Chem. Anal. B 2005, 41, 661–662. [Google Scholar] [CrossRef]
- Wang, J.-H.; Huang, W.-J.; Zhao, C.-J.; Yang, M.-H.; Wang, Z.-J. The inversion of leaf biochemical components and grain quality indicators of winter wheat with spectral reflectance. J. Remote Sens. 2003, 7, 277–284. [Google Scholar] [CrossRef]
- Senthilkumar, M.; Amaresan, N.; Sankaranarayanan, A. Estimation of malondialdehyde (MDA) by thiobarbituric acid (TBA) assay. In Plant-Microbe Interactions: Laboratory Techniques; Springer Science+Business Media: New York, NY, USA, 2021; pp. 103–104. [Google Scholar] [CrossRef]
- Li, C.; Kang, J.H.; Jung, K.I.; Park, M.H.; Kim, M. Effects of Haskap (Lonicera caerulea L.) extracts against oxidative stress and inflammation in RAW 264.7 cells. Prev. Nutr. Food Sci. 2024, 29, 146–153. [Google Scholar] [CrossRef]
- Zou, P.; Lu, X.; Jing, C.; Yuan, Y.; Lu, Y.; Zhang, C.; Meng, L.; Zhao, H.; Li, Y. Low-molecular-weight polysaccharides from Pyropia yezoensis enhance tolerance of wheat seedlings (Triticum aestivum L.) to salt stress. Front. Plant Sci. 2018, 9, 427. [Google Scholar] [CrossRef] [PubMed]
- Xie, C.; Yang, C.; He, Y. Hyperspectral Imaging for Classification of Healthy and Gray Mold Diseased Tomato Leaves with Different Infection Severities. Comput. Electron. Agric. 2017, 135, 154–162. [Google Scholar] [CrossRef]
- Wang, X.Q.; Chen, S.Y.; Zheng, W.C. Traffic Incident Duration Prediction Based on Partial Least Squares Regression. Procedia Soc. Behav. Sci. 2013, 96, 425–432. [Google Scholar] [CrossRef]
- Koumoutsou, D.; Siolas, G.; Charou, E.; Stamou, G. Generative Adversarial Networks for Data Augmentation in Hyperspectral Image Classification. In Generative Adversarial Learning: Architectures and Applications; Springer: Cham, Switzerland, 2022; pp. 115–136. [Google Scholar] [CrossRef]
- Li, Y.; Wang, B.; Yang, Z.; Li, J.; Chen, C. Hierarchical Stochastic Scheduling of Multi-Community Integrated Energy Systems in Uncertain Environments via Stackelberg Game. Energy Convers. Manag. 2022, 235, 113996. [Google Scholar] [CrossRef]
- Zhang, F.F.; Li, J.S.; Wang, C.; Wang, S.L. Estimation of water quality parameters of GF-1 WFV in turbid water based on soft classification. Natl. Remote Sens. Bull. 2023, 27, 769–779. [Google Scholar] [CrossRef]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Kiranyaz, S.; Avci, O.; Abdeljaber, O.; Ince, T.; Gabbouj, M.; Inman, D.J. 1D convolutional neural networks and applications: A survey. Mech. Syst. Signal Process. 2021, 151, 107398. [Google Scholar] [CrossRef]
- Zhang, Z.H. Naïve Bayes classification in R. Ann. Transl. Med. 2016, 4, 241. [Google Scholar] [CrossRef]
- Silva Júnior, A.C.; Moura, W.M.; Bhering, L.L.; Siqueira, M.J.S.; Costa, W.G.; Nascimento, M.; Cruz, C.D. Prediction and importance of predictors in approaches based on computational intelligence and machine learning. Agron. Sci. Biotechnol. 2023, 9, 1–24. [Google Scholar] [CrossRef]
- Arora, S.; Hu, W.; Kothari, P.K. An Analysis of the t-SNE Algorithm for Data Visualization. In Proceedings of the Conference on Learning Theory (COLT), Stockholm, Sweden, 6–9 July 2018; Volume 75, pp. 1–32. [Google Scholar]
- DeVries, T.; Taylor, G.W.; Romero, A.; Pineda, L.; Drozdzal, M. On the Evaluation of Conditional GANs. arXiv 2019, arXiv:1907.08175 2019. [Google Scholar] [CrossRef]
- Liu, Y.; Gao, Y.; Yue, L.; Zhang, H.; Sun, J.; Wu, X. A Real-Time Detection of Pilot Workload Using Low-Interference Devices. Appl. Sci. 2024, 14, 6521. [Google Scholar] [CrossRef]
- Muningsih, E. Kombinasi Metode K-Means dan Decision Tree dengan Perbandingan Kriteria dan Split Data. J. Teknonfo 2022, 16, 113–118. [Google Scholar] [CrossRef]
- Kim, T.K. T test as a parametric statistic. Korean J. Anesthesiol. 2015, 68, 540–546. [Google Scholar] [CrossRef]
- Franzese, M.; Iuliano, A. Correlation Analysis. Encycl. Bioinform. Comput. Biol. 2019, 1, 706–721. [Google Scholar] [CrossRef]
- Zhang, Z.; Ma, L.; Wei, C.; Yang, M.; Qin, S.; Lv, X.; Zhang, Z. Cotton Fusarium wilt diagnosis based on generative adversarial networks in small samples. Front. Plant Sci. 2023, 14, 1290774. [Google Scholar] [CrossRef]
- Cho, S.B.; Soleh, H.M.; Choi, J.W.; Hwang, W.-H.; Lee, H.; Cho, Y.-S.; Cho, B.-K.; Kim, M.S.; Baek, I.; Kim, G. Recent Methods for Evaluating Crop Water Stress Using AI Techniques: A Review. Sensors 2024, 24, 6313. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Xiang, J.; Jin, Y.; Liu, R.; Yan, J.; Wang, L. Boost Precision Agriculture with Unmanned Aerial Vehicle Remote Sensing and Edge Intelligence: A Survey. Remote Sens. 2021, 13, 4387. [Google Scholar] [CrossRef]
- Popescu, D.; Stoican, F.; Stamatescu, G.; Ichim, L.; Dragana, C. Advanced UAV–WSN System for Intelligent Monitoring in Precision Agriculture. Sensors 2020, 20, 817. [Google Scholar] [CrossRef]





| Time | NDVI | PRI | WI | GNDVI | OSAVI |
|---|---|---|---|---|---|
| CK | 0.61 | −0.08 | 0.81 | 0.56 | 0.52 |
| 24 h | 0.59 | −0.04 | 0.83 | 0.51 | 0.50 |
| 48 h | 0.57 | −0.04 | 0.85 | 0.48 | 0.48 |
| 72 h | 0.54 | −0.02 | 0.86 | 0.40 | 0.45 |
| 96 h | 0.50 | −0.05 | 0.89 | 0.38 | 0.42 |
| 120 h | 0.64 | −0.09 | 0.85 | 0.59 | 0.54 |
| Category | FID Score | JS Divergence |
|---|---|---|
| C0 | 0.05 | 0.03 |
| L1 | 0.08 | 0.04 |
| L2 | 0.05 | 0.03 |
| L3 | 0.05 | 0.02 |
| L4 | 0.08 | 0.02 |
| L5 | 0.03 | 0.02 |
| Category | Indicators | Equation | Rp2 | RMSEP |
|---|---|---|---|---|
| Physiological | MDA | MDA = −0.64*R730 + 0.91*R710 + 0.48*GNDVI + 0.21*PRI | 0.82 | 0.42 |
| WC | WC = +0.52*R730 − 0.65*R710 − 0.17*GNDVI − 0.08*R590 | 0.73 | 0.47 | |
| TCHL | TCHL = 0.44*R730 − 0.19*R710 − 0.17*R460 − 0.16*R520 | 0.42 | 0.72 | |
| Biochemical | SOD | SOD = +0.34*F520 + 0.28*F690/F740 + 0.38*R460 − 0.22*R730 | 0.32 | 0.40 |
| CAT | CAT = +0.51*F520 + 0.25*F690/F740 + 0.27*R850 − 0.48*GNDVI | 0.25 | 0.76 |
| Models | Precision | F1-Score | ||||||
|---|---|---|---|---|---|---|---|---|
| C0 | L1 | L2 | L3 | L4 | L5 | Average | ||
| RF | 94.05 | 97.62 | 94.05 | 92.86 | 97.62 | 97.50 | 95.60 | 94.81 |
| 1D-CNN | 95.24 | 97.62 | 78.57 | 70.24 | 96.43 | 97.50 | 89.20 | 87.23 |
| DT | 85.71 | 85.71 | 78.57 | 63.10 | 88.10 | 93.75 | 82.40 | 88.89 |
| NB | 90.48 | 79.76 | 54.76 | 40.48 | 51.19 | 78.75 | 65.80 | 62.85 |
| Models | Precision | F1-Score | ||||||
|---|---|---|---|---|---|---|---|---|
| C0 | L1 | L2 | L3 | L4 | L5 | Average | ||
| RF | 96.92 | 96.54 | 96.92 | 96.92 | 98.46 | 99.62 | 97.56 | 97.44 |
| 1D-CNN | 97.31 | 97.69 | 94.23 | 94.23 | 98.46 | 99.62 | 96.92 | 96.82 |
| DT | 89.62 | 91.54 | 90.38 | 92.31 | 94.23 | 98.85 | 92.82 | 92.59 |
| NB | 92.69 | 75.77 | 78.46 | 81.15 | 81.54 | 98.46 | 84.68 | 84.61 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhong, X.; Li, H.; Cai, Y.; Deng, Y.; Xu, H.; Tian, J.; Liu, S.; Hou, M.; Weng, H.; Wang, L.; et al. Early Detection of Tomato Gray Mold Based on Multispectral Imaging and Machine Learning. Horticulturae 2025, 11, 1073. https://doi.org/10.3390/horticulturae11091073
Zhong X, Li H, Cai Y, Deng Y, Xu H, Tian J, Liu S, Hou M, Weng H, Wang L, et al. Early Detection of Tomato Gray Mold Based on Multispectral Imaging and Machine Learning. Horticulturae. 2025; 11(9):1073. https://doi.org/10.3390/horticulturae11091073
Chicago/Turabian StyleZhong, Xiaohao, Huicheng Li, Yixin Cai, Ying Deng, Haobin Xu, Jun Tian, Shuang Liu, Maomao Hou, Haiyong Weng, Lijing Wang, and et al. 2025. "Early Detection of Tomato Gray Mold Based on Multispectral Imaging and Machine Learning" Horticulturae 11, no. 9: 1073. https://doi.org/10.3390/horticulturae11091073
APA StyleZhong, X., Li, H., Cai, Y., Deng, Y., Xu, H., Tian, J., Liu, S., Hou, M., Weng, H., Wang, L., Ruan, M., Zhong, F., Zhu, C., & Xu, L. (2025). Early Detection of Tomato Gray Mold Based on Multispectral Imaging and Machine Learning. Horticulturae, 11(9), 1073. https://doi.org/10.3390/horticulturae11091073





