Insights into Loquat Flowering Regulation Through Analysis of Alternative Splicing of Flowering-Time Genes and Functions of EjCO1 Isoforms
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. RNA Extraction and qRT-PCR
2.3. PacBio SMRT Sequencing and Data Analysis
2.4. Alternative Splicing Analysis and Validation
2.5. Prediction of LncRNA
2.6. Subcellular Localization
2.7. Genetic Transformation of Arabidopsis
2.8. Yeast Two-Hybrid Assay
2.9. Dual LUC Assay
3. Results
3.1. Summary of SMRT Sequencing
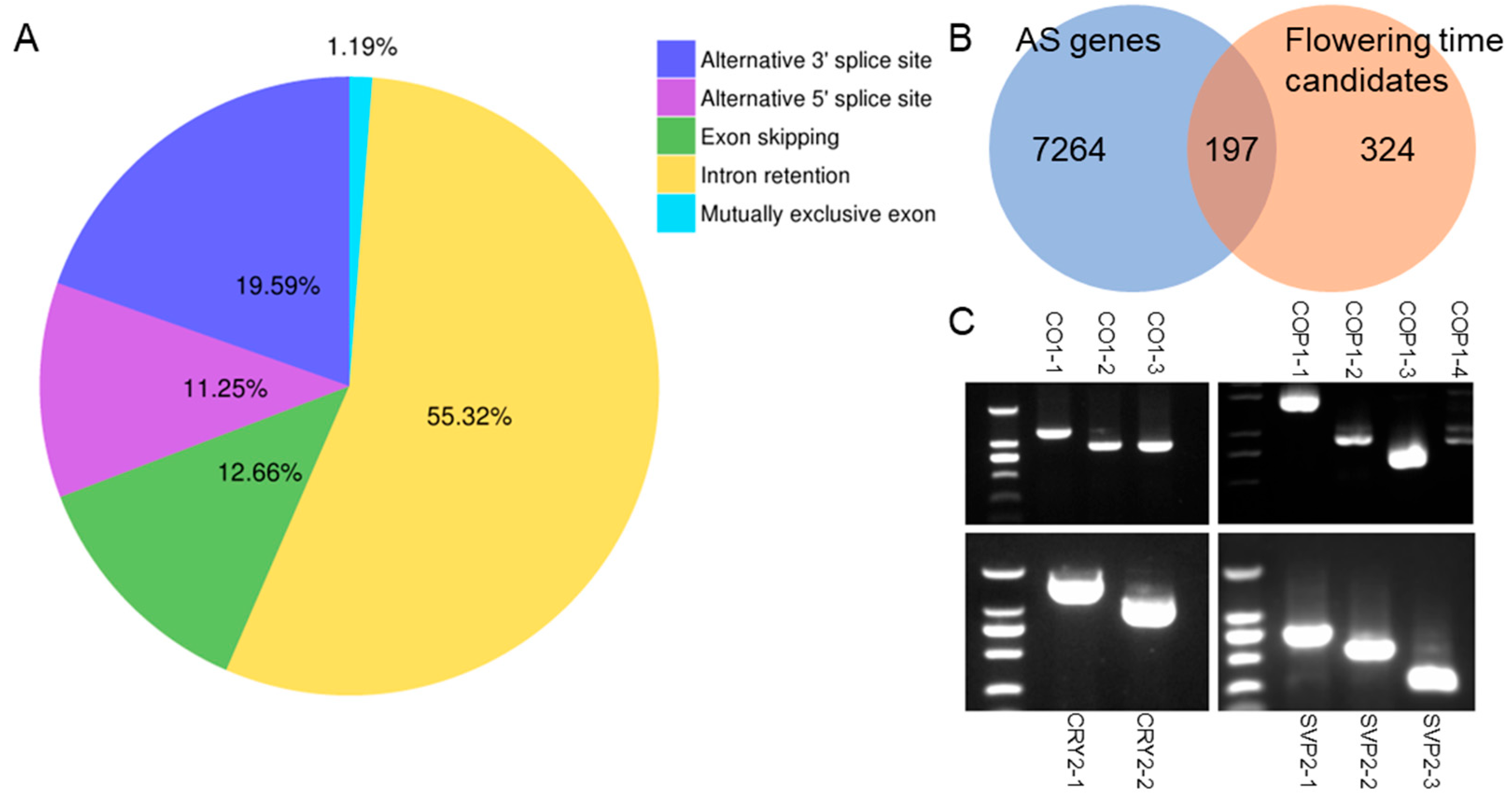
3.2. Alternative Splicing of Flowering Time Homologs and Validation
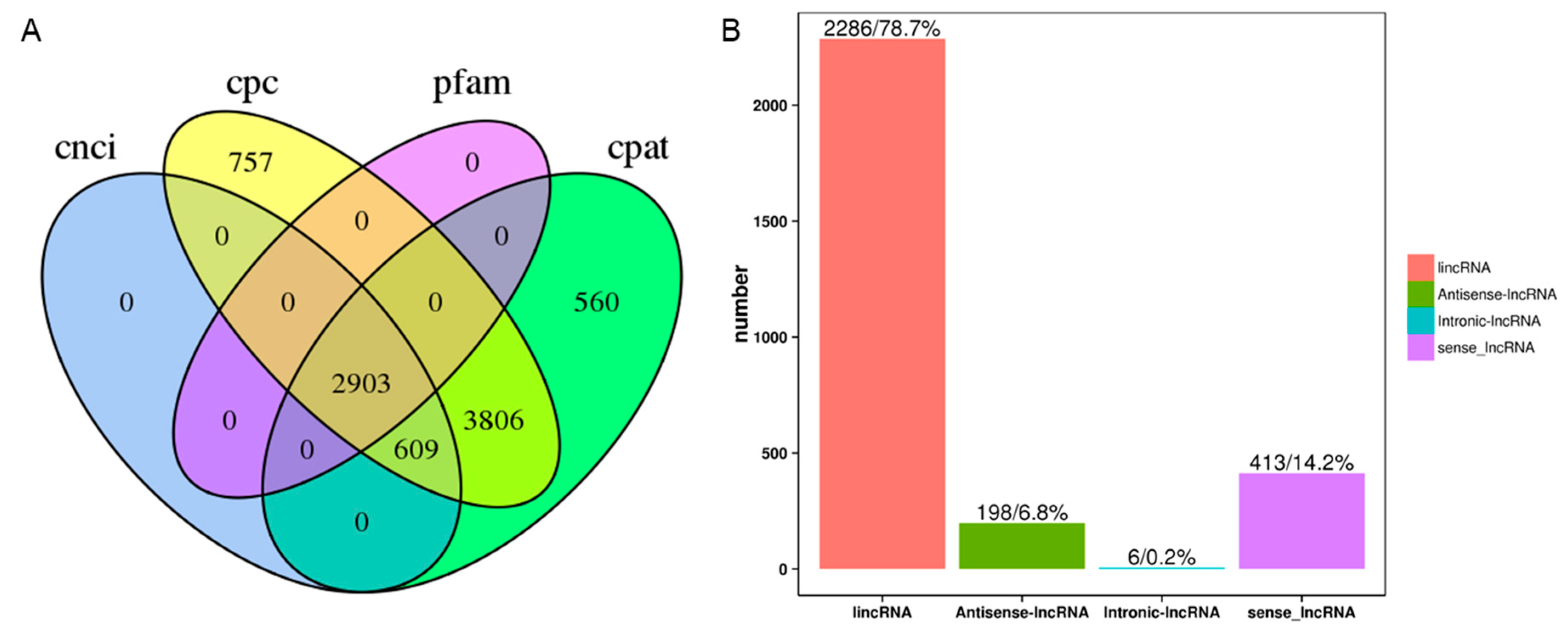
3.3. LncRNA Prediction
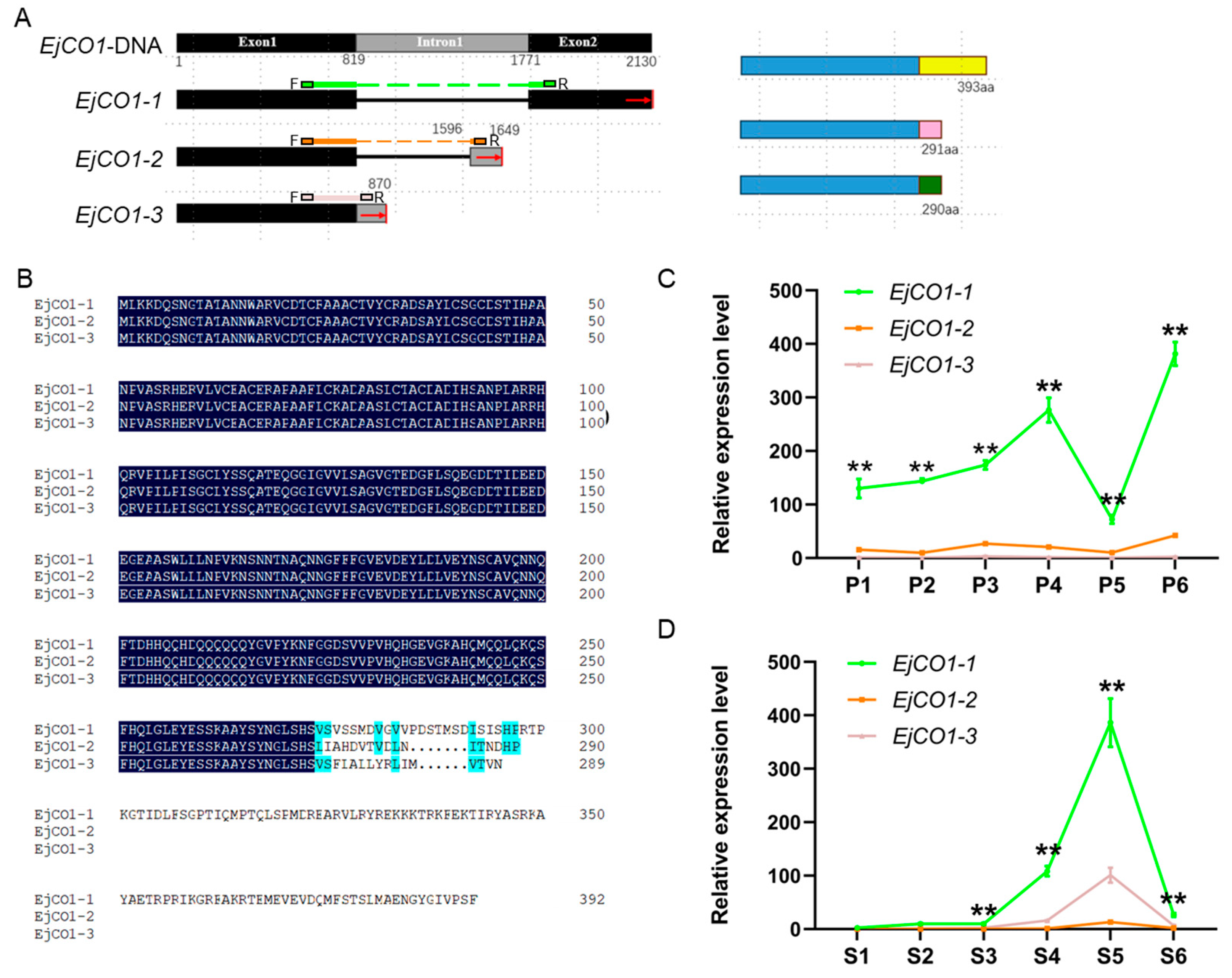
3.4. Comparison and Interaction Among Three Isoforms of EjCO1
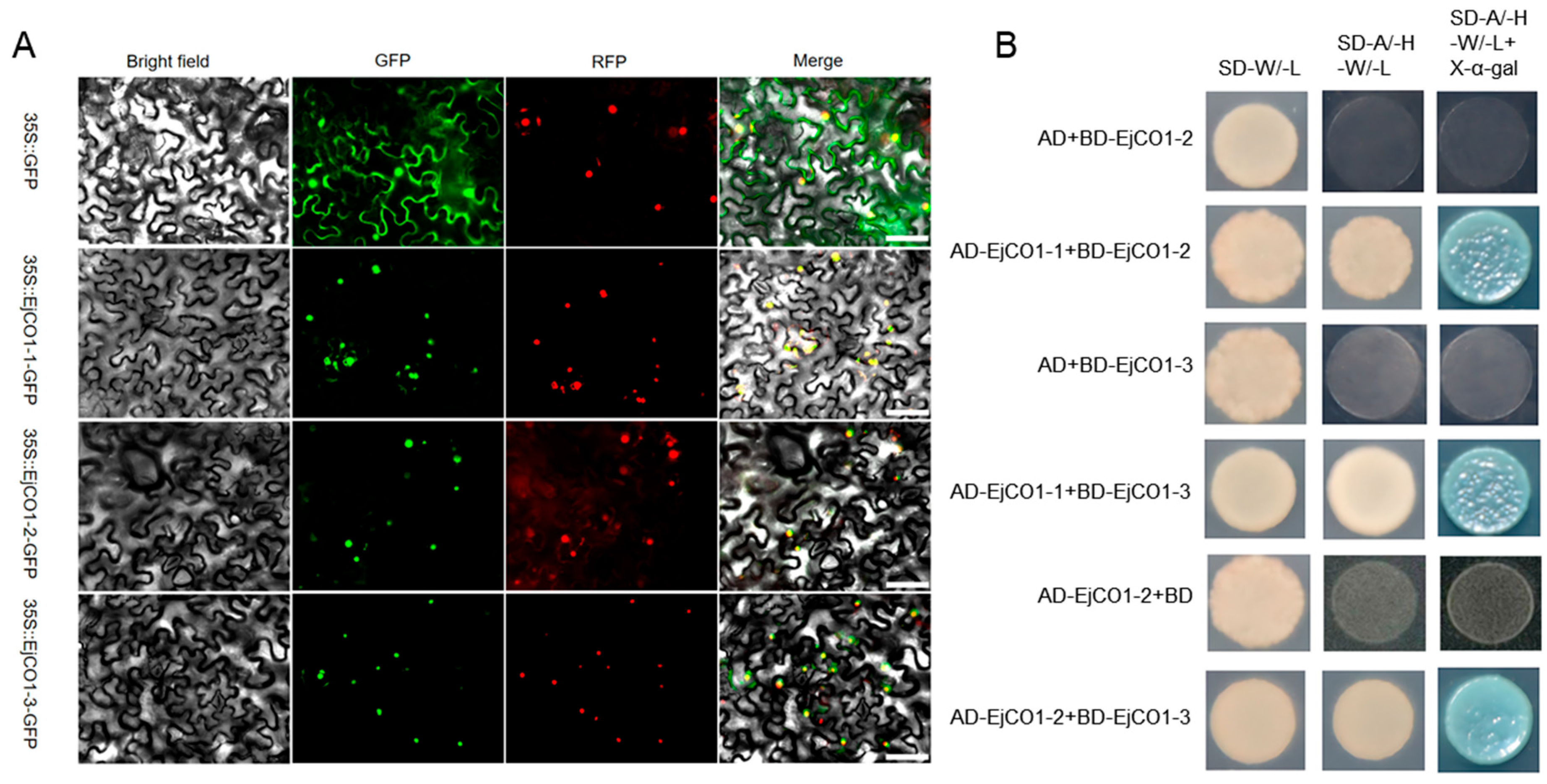
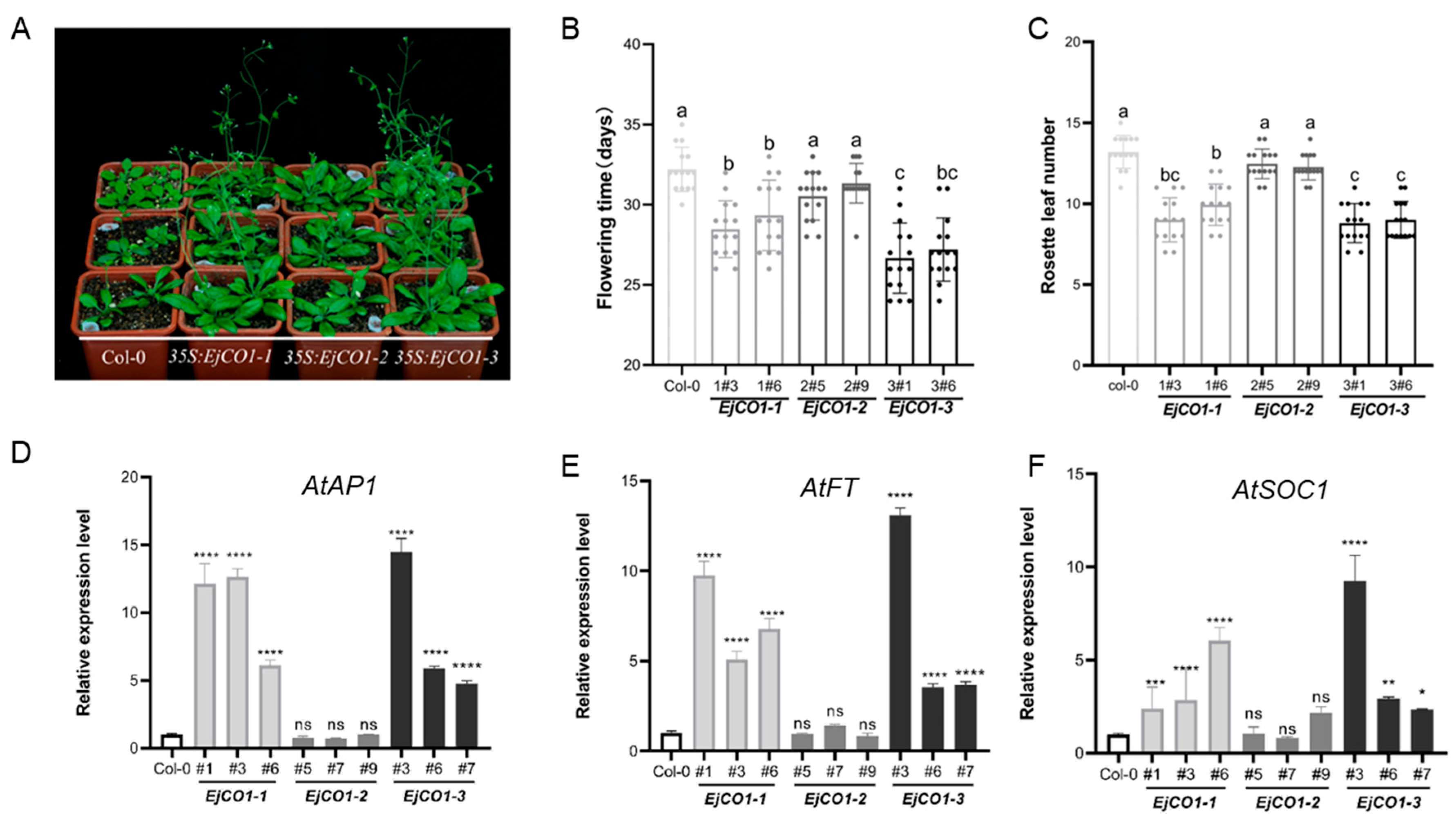
3.5. Functional Differentiation Among EjCO1 Isoforms
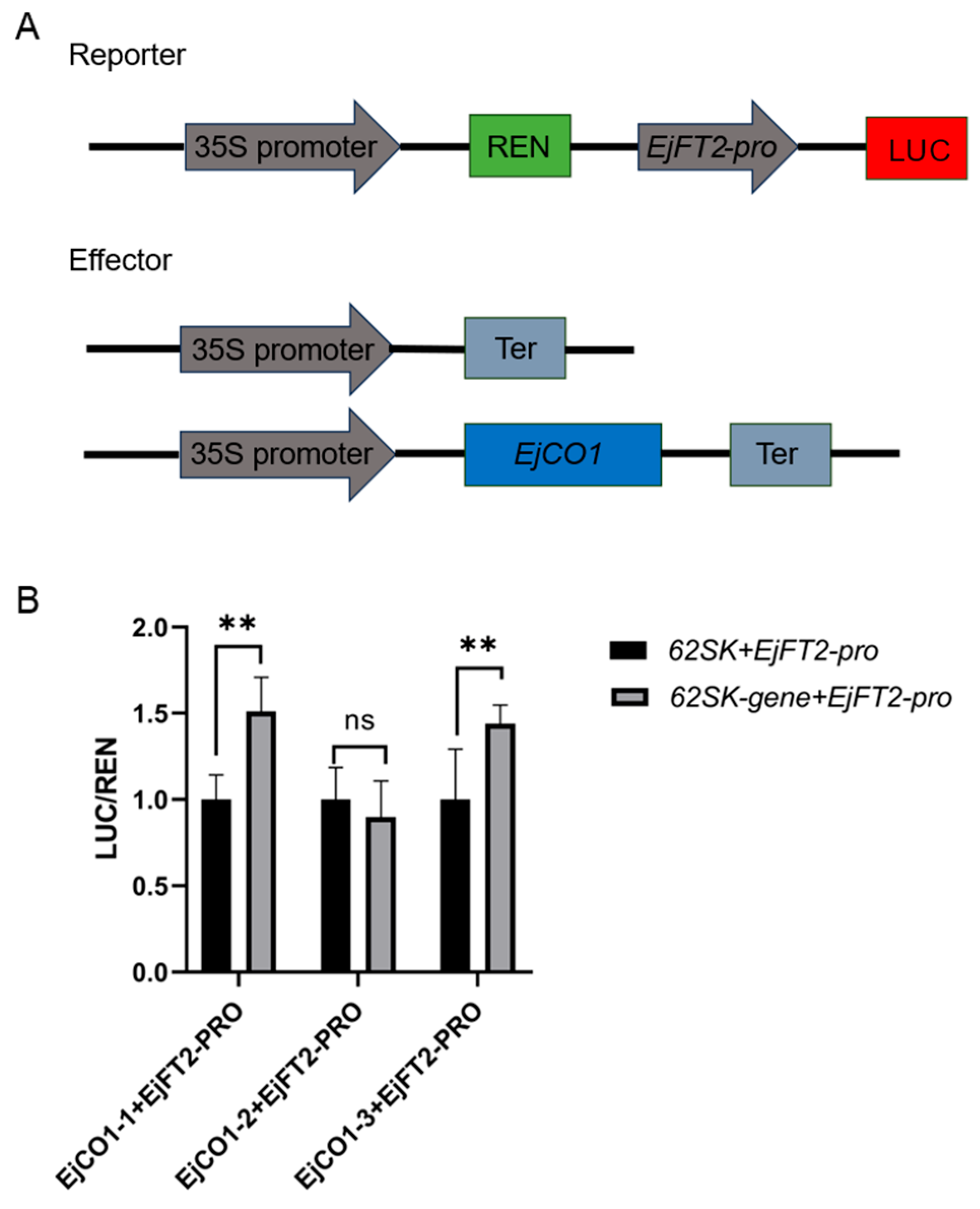
3.6. Differential Regulation of EjFT2 Promoter Activity by EjCO1 Isoforms
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Bouché, F.; Lobet, G.; Tocquin, P.; Périlleux, C. FLOR-ID: An interactive database of flowering-time gene networks in Arabidopsis thaliana. Nucleic Acids Res. 2016, 44, D1167–D1171. [Google Scholar] [CrossRef]
- Fornara, F.; de Montaigu, A.; Coupland, G. SnapShot: Control of flowering in Arabidopsis. Cell 2010, 141, 550. [Google Scholar] [CrossRef]
- Song, S.; Hao, Q.; Su, L.; Xia, S.; Zhang, R.; Liu, Y.; Li, Y.; Zhu, Y.; Luo, Q.; Lai, Y. FLOWERING LOCUS T (FT) gene regulates short-day flowering in low latitude Xishuangbanna cucumber (Cucumis sativus var. Xishuangbannanesis). Veg. Res. 2023, 3, 15. [Google Scholar] [CrossRef]
- Xu, P.; Wang, X.; Luo, S.; Cheng, A.; Xu, J.; Ma, H.; Zhang, Y.; Zhang, H. MAPK and hormone signaling and carotenoid biosynthesis pathways are involved in regulating flower opening of pear tree by response to temperature mediated by melatonin. Fruit Res. 2024, 4, e26. [Google Scholar] [CrossRef]
- Andrés, F.; Coupland, G. The genetic basis of flowering responses to seasonal cues. Nat. Rev. Genet. 2012, 13, 627–639. [Google Scholar] [CrossRef]
- Kurokura, T.; Mimida, N.; Battey, N.H.; Hytönen, T. The regulation of seasonal flowering in the Rosaceae. J. Exp. Bot. 2013, 64, 4131–4141. [Google Scholar] [CrossRef]
- Lin, S. World loquat production and research with special reference to China. Acta Hortic. 2007, 750, 37–43. [Google Scholar] [CrossRef]
- Jiang, Y.; Peng, J.; Zhu, Y.; Su, W.; Zhang, L.; Jing, Y.; Lin, S.; Gao, Y. The Role of EjSOC1s in Flower Initiation in Eriobotrya japonica. Front. Plant Sci. 2019, 10, 253. [Google Scholar] [CrossRef]
- Peng, Z.; Wang, M.; Zhang, L.; Jiang, Y.; Zhao, C.; Shahid, M.Q.; Bai, Y.; Hao, J.; Peng, J.; Gao, Y.; et al. EjRAV1/2 delay flowering through transcriptional repression of EjFTs and EjSOC1s in loquat. Front. Plant Sci. 2021, 12, 816086. [Google Scholar] [CrossRef]
- Chen, Q.; Yong, S.; Xu, F.; Fu, H.; Dang, J.; He, Q.; Jing, D.; Wu, D.; Liang, G.; Guo, Q. EjGASA6 promotes flowering and root elongation by enhancing gibberellin biosynthesis. J. Integr. Agr. 2024, 23, 1568–1579. [Google Scholar] [CrossRef]
- Reig, C.; Gil-Muñoz, F.; Vera-Sirera, F.; García-Lorca, A.; Martínez-Fuentes, A.; Mesejo, C.; Pérez-Amador, M.A.; Agustí, M. Bud sprouting and floral induction and expression of FT in loquat [Eriobotrya japonica (Thunb.) Lindl.]. Planta 2017, 246, 915–925. [Google Scholar] [CrossRef]
- Jiang, Y.; Zhu, Y.; Peng, Z.; Su, W.; Peng, J.; Yuan, Y.; Zhang, L.; Zhang, Z.; Yang, X.; Gao, Y.; et al. Two FT genes synergistically regulate the reproductive transition of loquat. Hortic. Plant J. 2025, 11, 548–563. [Google Scholar] [CrossRef]
- Song, Y.H.; Shim, J.S.; Kinmonth-Schultz, H.A.; Imaizumi, T. Photoperiodic flowering: Time measurement mechanisms in leaves. Annu. Rev. Plant Biol. 2015, 66, 441–464. [Google Scholar] [CrossRef]
- Shim, J.S.; Kubota, A.; Imaizumi, T. Circadian clock and photoperiodic flowering in Arabidopsis: CONSTANS is a hub for signal integration. Plant Physiol. 2017, 173, 5–15. [Google Scholar] [CrossRef] [PubMed]
- Gil, K.E.; Park, M.J.; Lee, H.J.; Park, Y.J.; Han, S.H.; Kwon, Y.J.; Seo, P.J.; Jung, J.H.; Park, C.M. Alternative splicing provides a proactive mechanism for the diurnal CONSTANS dynamics in Arabidopsis photoperiodic flowering. Plant J. 2017, 89, 128–140. [Google Scholar] [CrossRef]
- Capovilla, G.; Pajoro, A.; Immink, R.G.; Schmid, M. Role of alternative pre-mRNA splicing in temperature signaling. Curr. Opin. Plant Biol. 2015, 27, 97–103. [Google Scholar] [CrossRef]
- Cheng, Y.; Tu, S. Alternative splicing and Cross-Talk with light signaling. Plant Cell Physiol. 2018, 59, 1104–1110. [Google Scholar] [CrossRef]
- Wang, Y.Y.; Xiong, F.; Ren, Q.P.; Wang, X.L. Regulation of flowering transition by alternative splicing: The role of the U2 auxiliary factor. J. Exp. Bot. 2020, 71, 751–758. [Google Scholar] [CrossRef]
- Huang, C.K.; Lin, W.D.; Wu, S.H. An improved repertoire of splicing variants and their potential roles in Arabidopsis photomorphogenic development. Genome Biol. 2022, 23, 50. [Google Scholar] [CrossRef]
- An, Y.; Wu, J.; Chen, Y.; Li, S. Comprehensive analysis of alternative splicing in Rosa roxburghii Tratt reveals its role in flavonoid synthesis. Front. Plant Sci. 2025, 16, 1627126. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Dai, C.; Hu, C.; Liu, Z.; Kang, C. Global identification of alternative splicing via comparative analysis of SMRT- and Illumina-based RNA-seq in strawberry. Plant J. 2017, 90, 164–176. [Google Scholar] [CrossRef]
- Zhou, T.; He, Y.; Zeng, X.; Cai, B.; Qu, S.; Wang, S. Comparative Analysis of Alternative Splicing in Two Contrasting Apple Cultivars Defense against Alternaria alternata Apple Pathotype Infection. Int. J. Mol. Sci. 2022, 23, 14202. [Google Scholar] [CrossRef] [PubMed]
- Kagaya, H.; Ito, N.; Shibuya, T.; Komori, S.; Kato, K.; Kanayama, Y. Characterization of FLOWERING LOCUS C homologs in apple as a model for fruit trees. Int. J. Mol. Sci. 2020, 21, 4562. [Google Scholar] [CrossRef]
- Li, J.; Yan, X.; Ahmad, M.; Yu, W.; Song, Z.; Ni, J.; Yang, Q.; Teng, Y.; Zhang, H.; Bai, S. Alternative splicing of the dormancy-associated MADS-box transcription factor gene PpDAM1 is associated with flower bud dormancy in ‘Dangshansu’ pear (Pyrus pyrifolia white pear group). Plant Physiol. Bioch. 2021, 166, 1096–1108. [Google Scholar] [CrossRef] [PubMed]
- Kurokawa, K.; Kobayashi, J.; Nemoto, K.; Nozawa, A.; Sawasaki, T.; Nakatsuka, T.; Yamagishi, M. Expression of LhFT1, the flowering inducer of asiatic hybrid lily, in the bulb scales. Front. Plant Sci. 2020, 11, 570915. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Zhang, H.; Li, L. Alternative mRNA processing increases the complexity of microRNA-based gene regulation in Arabidopsis. Plant J. 2012, 70, 421–431. [Google Scholar] [CrossRef]
- Wang, P.; Li, Y.; Liu, Z.; Li, X.; Wang, Y.; Liu, W.; Li, X.; Hu, J.; Zhu, W.; Wang, C.; et al. Reciprocal regulation of flower induction by ELF3α and ELF3β generated via alternative promoter usage. Plant Cell 2023, 35, 2095–2113. [Google Scholar] [CrossRef]
- Wang, X.; Zhao, F.; Wu, Q.; Xing, S.; Yu, Y.; Qi, S. Physiological and transcriptome analyses to infer regulatory networks in flowering transition of Rosa rugosa. Ornam. Plant Res. 2023, 3, 1–12. [Google Scholar] [CrossRef]
- Peng, L.; Song, W.; Tan, W.; Liu, Z.; Wang, X.; Li, Y.; Shu, Q. Integration of genome-wide identification, transcriptome and association analysis of HSP20 gene family to revealing genetic basis of floral organ number-related traits in tree peony. Ornam. Plant Res. 2023, 3, 22. [Google Scholar] [CrossRef]
- Wu, M.; Liu, K.; Li, H.; Li, Y.; Zhu, Y.; Su, D.; Zhang, Y.; Deng, H.; Wang, Y.; Liu, M. Gibberellins involved in fruit ripening and softening by mediating multiple hormonal signals in tomato. Hortic. Res. 2024, 11, d275. [Google Scholar] [CrossRef]
- Arro, J.; Yang, Y.; Song, G.; Cousins, P.; Liu, Z.; Zhong, G. Transcriptome analysis unveils a potential novel role of VvAP1 in regulating the developmental fate of primordia in grapevine. Fruit Res. 2024, 4, e11. [Google Scholar] [CrossRef]
- An, D.; Cao, H.X.; Li, C.; Humbeck, K.; Wang, W. Isoform sequencing and State-of-Art applications for unravelling complexity of plant transcriptomes. Genes 2018, 9, 43. [Google Scholar] [CrossRef]
- Cai, F.; Shao, C.; Sun, Y. The role of alternative splicing in floral transition. Chin. Bull. Bot. 2022, 1, 69–79. [Google Scholar]
- Yan, C.; Zhang, N.; Wang, Q.; Fu, Y.; Zhao, H.; Wang, J.; Wu, G.; Wang, F.; Li, X.; Liao, H. Full-length transcriptome sequencing reveals the molecular mechanism of potato seedlings responding to low-temperature. BMC Plant Biol. 2022, 22, 125. [Google Scholar] [CrossRef]
- Zheng, L.; Zhao, Y.; Gan, Y.; Li, H.; Luo, S.; Liu, X.; Li, Y.; Shao, Q.; Zhang, H.; Zhao, Y.; et al. Full-Length transcriptome sequencing reveals the impact of cold stress on alternative splicing in quinoa. Int. J. Mol. Sci. 2022, 23, 5724. [Google Scholar] [CrossRef]
- Zhou, H.; Sheng, Y.; Qiu, K.; Ren, F.; Shi, P.; Xie, Q.; Guo, J.; Pan, H.; Zhang, J. Improved annotation of the peach (Prunus persica) genome and identification of tissue- or development Stage-Specific alternative splicing through the integration of Iso-Seq and RNA-Seq data. Horticulturae 2023, 2, 175. [Google Scholar] [CrossRef]
- Pan, C.; Wang, Y.; Tao, L.; Zhang, H.; Deng, Q.; Yang, Z.; Chi, Z.; Yang, Y. Single-molecule real-time sequencing of the full-length transcriptome of loquat under low-temperature stress. PLoS ONE 2020, 15, e238942. [Google Scholar] [CrossRef]
- Jiang, Y.; Zhu, Y.; Zhang, L.; Su, W.; Peng, J.; Yang, X.; Song, H.; Gao, Y.; Lin, S. EjTFL1 genes promote growth but inhibit flower bud differentiation in loquat. Front. Plant Sci. 2020, 11, 576. [Google Scholar] [CrossRef]
- Su, W.; Jing, Y.; Lin, S.; Yue, Z.; Yang, X.; Xu, J.; Wu, J.; Zhang, Z.; Xia, R.; Zhu, J.; et al. Polyploidy underlies co-option and diversification of biosynthetic triterpene pathways in the apple tribe. Proc. Natl. Acad. Sci USA 2021, 118, e2101767118. [Google Scholar] [CrossRef]
- Su, W.; Yuan, Y.; Zhang, L.; Jiang, Y.; Gan, X.; Bai, Y.; Peng, J.; Wu, J.; Liu, Y.; Lin, S. Selection of the optimal reference genes for expression analyses in different materials of Eriobotrya japonica. Plant Methods 2019, 15, 7. [Google Scholar] [CrossRef]
- Ferreira, M.J.; Silva, J.; Pinto, S.C.; Coimbra, S. I choose you: Selecting accurate reference genes for qPCR expression analysis in reproductive tissues in Arabidopsis thaliana. Biomolecules 2023, 13, 463. [Google Scholar] [CrossRef]
- Foissac, S.; Sammeth, M. ASTALAVISTA: Dynamic and flexible analysis of alternative splicing events in custom gene datasets. Nucleic Acids Res. 2007, 35, W297–W299. [Google Scholar] [CrossRef]
- Yao, J.L.; Kang, C.; Gu, C.; Gleave, A.P. The roles of floral organ genes in regulating Rosaceae fruit development. Front. Plant Sci. 2021, 12, 644424. [Google Scholar] [CrossRef]
- Kong, L.; Zhang, Y.; Ye, Z.Q.; Liu, X.Q.; Zhao, S.Q.; Wei, L.; Gao, G. CPC: Assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res. 2007, 35, W345–W349. [Google Scholar] [CrossRef]
- Sun, L.; Luo, H.; Bu, D.; Zhao, G.; Yu, K.; Zhang, C.; Liu, Y.; Chen, R.; Zhao, Y. Utilizing sequence intrinsic composition to classify protein-coding and long non-coding transcripts. Nucleic Acids Res. 2013, 41, e166. [Google Scholar] [CrossRef]
- El-Gebali, S.; Mistry, J.; Bateman, A.; Eddy, S.R.; Luciani, A.; Potter, S.C.; Qureshi, M.; Richardson, L.J.; Salazar, G.A.; Smart, A.; et al. The Pfam protein families database in 2019. Nucleic Acids Res. 2019, 47, D427–D432. [Google Scholar] [CrossRef]
- Wang, L.; Park, H.J.; Dasari, S.; Wang, S.; Kocher, J.P.; Li, W. CPAT: Coding-Potential Assessment Tool using an alignment-free logistic regression model. Nucleic Acids Res. 2013, 41, e74. [Google Scholar] [CrossRef]
- Sparkes, I.A.; Runions, J.; Kearns, A.; Hawes, C. Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nat. Protoc. 2006, 1, 2019–2025. [Google Scholar] [CrossRef]
- Hou, X.; Zhou, J.; Liu, C.; Liu, L.; Shen, L.; Yu, H. Nuclear factor Y-mediated H3K27me3 demethylation of the SOC1 locus orchestrates flowering responses of Arabidopsis. Nat. Commun. 2014, 5, 4601. [Google Scholar] [CrossRef]
- Zhang, X.; Henriques, R.; Lin, S.S.; Niu, Q.W.; Chua, N.H. Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat. Protoc. 2006, 1, 641–646. [Google Scholar] [CrossRef]
- Hellens, R.P.; Allan, A.C.; Friel, E.N.; Bolitho, K.; Grafton, K.; Templeton, M.D.; Karunairetnam, S.; Gleave, A.P.; Laing, W.A. Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods 2005, 1, 13. [Google Scholar] [CrossRef]
- Yoo, S.; Chung, K.S.; Mondes, E.; Lee, J.H.; Hong, S.; Yoo, S.; Yoo, S.; Lee, J.S.; Ahn, J.H. CONSTANS activates suppressor of overexpression of CONSTANS T through flowering locus T to promote flowering in Arabidopsis. Plant Physiol. 2005, 139, 770–778. [Google Scholar] [CrossRef]
- Wang, S.; Shi, Y.; Zhou, Y.; Hu, W.; Liu, F. Full-length transcriptome sequencing of Arabidopsis plants provided new insights into the autophagic regulation of photosynthesis. Sci. Rep.-UK 2024, 14, 14588. [Google Scholar] [CrossRef]
- Ma, H.; Pei, J.; Zhuo, J.; Tang, Q.; Hou, D.; Lin, X. The CONSTANS-LIKE gene PeCOL13 regulates flowering through intron-retained alternative splicing in Phyllostachys edulis. Int. J. Biol. Macromol. 2024, 274, 133393. [Google Scholar] [CrossRef]
- Palos, K.; Yu, L.; Railey, C.E.; Nelson, D.A.; Nelson, A. Linking discoveries, mechanisms, and technologies to develop a clearer perspective on plant long noncoding RNAs. Plant Cell 2023, 35, 1762–1786. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Li, F.; Ali, M.; Li, X.; Fu, X.; Zhang, X. Emerging roles and mechanisms of lncRNAs in fruit and vegetables. Hortic. Res. 2024, 11, e46. [Google Scholar] [CrossRef] [PubMed]
- Heo, J.B.; Sung, S. Vernalization-mediated epigenetic silencing by a long intronic noncoding RNA. Science 2011, 331, 76–79. [Google Scholar] [CrossRef]
- Kim, D.H.; Sung, S. Vernalization-Triggered intragenic chromatin loop formation by long noncoding RNAs. Dev. Cell 2017, 40, 302–312. [Google Scholar] [CrossRef]
- Robson, F.; Costa, M.M.; Hepworth, S.R.; Vizir, I.; Piñeiro, M.; Reeves, P.H.; Putterill, J.; Coupland, G. Functional importance of conserved domains in the flowering-time gene CONSTANS demonstrated by analysis of mutant alleles and transgenic plants. Plant J. 2001, 28, 619–631. [Google Scholar] [CrossRef]
| Item | Information |
|---|---|
| Subreads base (bp) | 41,973,500,803 |
| Number of subreads | 30,346,772 |
| Average subreads length (bp) | 1383 |
| Number of CCS reads | 516,184 |
| Average CCS read Length (bp) | 1683 |
| Number of full-length non-chimeric reads | 418,868 |
| Number of consensus isoforms | 172,158 |
| Average read length of consensus isoforms (bp) | 1598 |
| Number of high-quality isoforms | 172,106 |
| Number of non-redundant high-quality isoforms | 94,194 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, W.; Zhao, C.; Jiang, J.; Li, H.; Su, W.; Jiang, Y.; Yang, X.; Peng, Z. Insights into Loquat Flowering Regulation Through Analysis of Alternative Splicing of Flowering-Time Genes and Functions of EjCO1 Isoforms. Horticulturae 2025, 11, 1064. https://doi.org/10.3390/horticulturae11091064
Wu W, Zhao C, Jiang J, Li H, Su W, Jiang Y, Yang X, Peng Z. Insights into Loquat Flowering Regulation Through Analysis of Alternative Splicing of Flowering-Time Genes and Functions of EjCO1 Isoforms. Horticulturae. 2025; 11(9):1064. https://doi.org/10.3390/horticulturae11091064
Chicago/Turabian StyleWu, Wendong, Chongbin Zhao, Jie Jiang, Huijie Li, Wenbing Su, Yuanyuan Jiang, Xianghui Yang, and Ze Peng. 2025. "Insights into Loquat Flowering Regulation Through Analysis of Alternative Splicing of Flowering-Time Genes and Functions of EjCO1 Isoforms" Horticulturae 11, no. 9: 1064. https://doi.org/10.3390/horticulturae11091064
APA StyleWu, W., Zhao, C., Jiang, J., Li, H., Su, W., Jiang, Y., Yang, X., & Peng, Z. (2025). Insights into Loquat Flowering Regulation Through Analysis of Alternative Splicing of Flowering-Time Genes and Functions of EjCO1 Isoforms. Horticulturae, 11(9), 1064. https://doi.org/10.3390/horticulturae11091064







