Plant Growth Regulators Promote Petaloidy and Modulate Related Gene Expression in Ornamental Pomegranate
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Site and Plant Materials
2.2. Plant Growth Regulator Treatment Methods
2.3. Phenotypic Indicator Statistics
2.4. qRT-PCR Analysis
2.5. Data Processing
3. Results
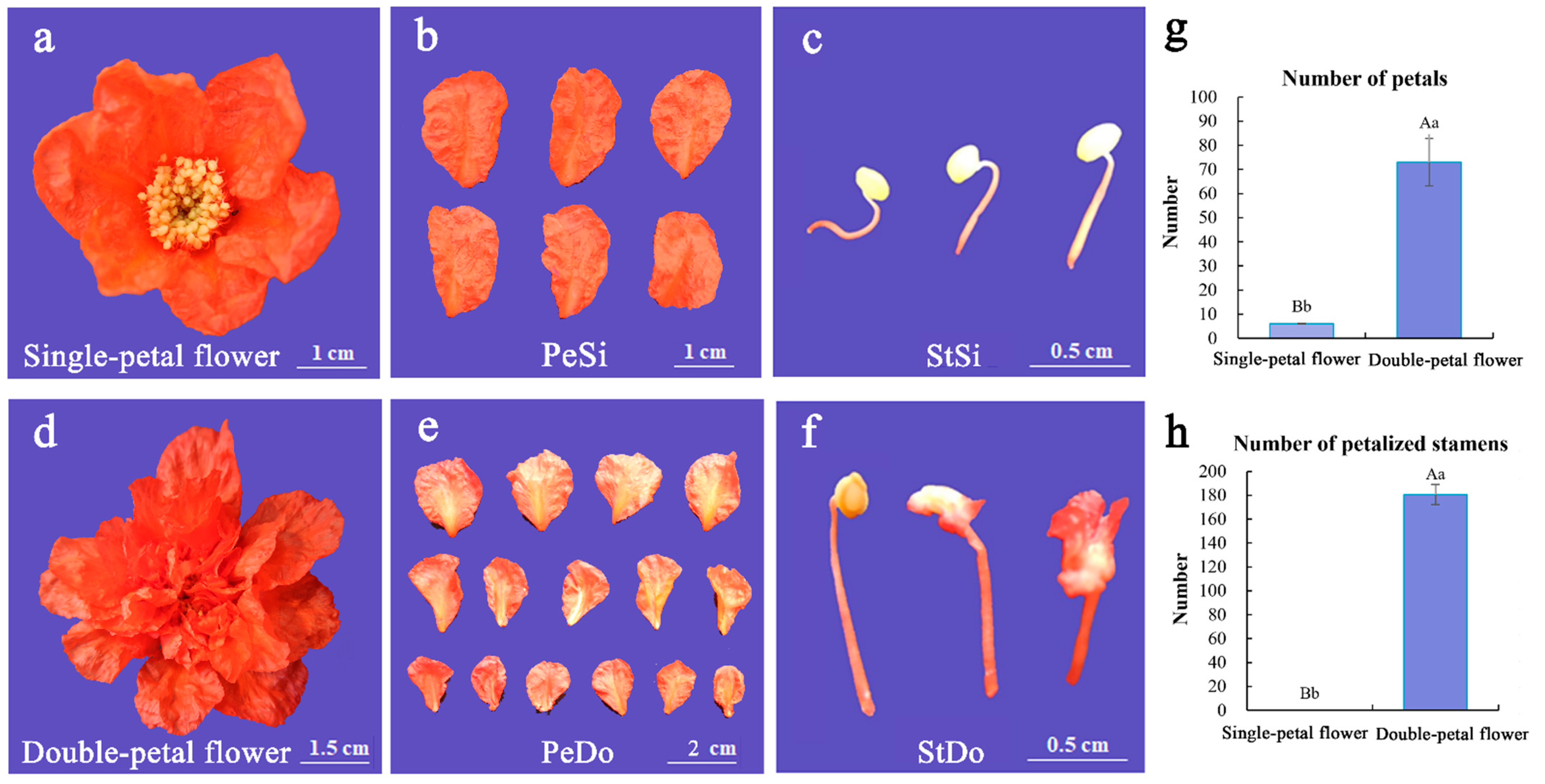
3.1. Effect of Growth Regulator Treatment on Petaloidy Phenotype
3.2. Effects of NAA on Hormone-Related Genes Involved in Petaloidy
3.3. Effects of MeJA on Hormone-Related Genes Involved in Petaloidy
4. Discussion
4.1. Plant Growth Regulator-Induced Petaloidy in Ornamental Pomegranate
4.2. Response Patterns of Hormone-Related Genes in Petaloidy of Ornamental Pomegranate Under NAA Induction
4.3. Response Patterns of Hormone-Related Genes in Petaloidy of Ornamental Pomegranate Under MeJA Induction
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| NAA | Naphthaleneacetic acid |
| MeJA | Methyl jasmonate |
| ABA | Abscisic acid |
| ETH | Ethephon |
| NOPSs | Number of petalized stamens |
| NOPs | Number of petals |
| PeSi | Petals of the single-petal flowers |
| PeDo | Petals of the double-petal flowers, including the transitional petals |
| StSi | Stamens of the single-petal flowers |
| StDo | Stamens of the double-petal flowers, including the petalized stamens |
| YUC | Flavin-containing monooxygenase |
| ILR1 | IAA-amino acid hydrolase 1 |
| GH3.17 | Indole-3-acetic acid-amido synthetase |
| LAX2 | Auxin transporter 2 |
| ARF | Auxin response factor |
| AIR12 | Auxin-induced in root cultures protein 12 |
| JAR1 | Jasmonic acid-amido synthetase 1 |
| ASR | ABA stress ripening-induced protein |
| ANOVA | Analysis of variance |
| PTL | PETAL LOSS |
| PIN | Efflux carrier |
References
- Xiao, Y.G.; Chen, Y.; Charnikhova, T.; Mulder, P.P.; Heijmans, J.; Hoogenboom, A.; Agalou, A.; Michel, C.; Morel, J.-B.; Dreni, L.; et al. OsJAR1 is required for JA–regulated floret opening and anther dehiscence in rice. Plant Mol. Biol. 2014, 86, 19–33. [Google Scholar] [CrossRef]
- Aloni, R.; Aloni, E.; Langhans, M.; Cornelia, I.U. Role of auxin in regulating Arabidopsis flower development. Planta 2006, 223, 315–328. [Google Scholar] [CrossRef]
- Cecchetti, V.; Brunetti, P.; Napoli, N.; Fattorini, L.; Altamura, M.M.; Costantino, P.; Cardarelli, M. ABCB1 and ABCB19 auxin transporters have synergistic effects on early and late Arabidopsis anther development. J. Integr. Plant Biol. 2015, 57, 1089–1098. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Jin, M.; Wu, C.Y.; Bao, J.P. Effects of plant growth regulators on the endogenous hormone content of calyx development in ‘Korla’ fragrant pear. Hortscience 2022, 57, 497–503. [Google Scholar] [CrossRef]
- Pan, J.; Wang, G.; Wen, H.F.; Du, H.; Lian, H.L.; He, H.L.; Pan, J.S.; Cai, R. Differential gene expression caused by the F and M loci provides insight into ethylene–mediated female flower differentiation in cucumber. Front. Plant Sci. 2018, 9, 1091. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Y.Y. Study on the Double Petal Formation Mechanism of Grandiflora Clematis. Master’s Thesis, Southwest Forestry University, Kunming, China, 4 June 2019. [Google Scholar]
- Wei, J.; Yang, Q.S.; Ni, J.B.; Gao, Y.H.; Tang, Y.X.; Bai, S.L.; Teng, Y.W. Early defoliation induces auxin redistribution, promoting paradormancy release in pear buds. Plant Physiol. 2022, 190, 2739–2756. [Google Scholar] [CrossRef]
- Chen, L.N.; Zhang, J.; Li, H.X.; Niu, J.; Xue, H.; Liu, B.B.; Wang, Q.; Luo, X.; Zhang, F.H.; Zhao, D.G.; et al. Transcriptomic analysis reveals candidate genes for female sterility in pomegranate flowers. Front. Plant Sci. 2017, 8, 1430. [Google Scholar] [CrossRef]
- Xia, S.S.; Niu, J.; Chen, L.N.; Cao, S.Y. Effect of different reagents on the proportion of fertile flower and fruit yield and quality in pomegranate. J. Northwest For. Univ. 2017, 32, 111–116. [Google Scholar]
- Lin, Z.; Damaris, R.N.; Shi, T.; Li, J.; Yang, P. Transcriptomic analysis identifies the key genes involved in stamen petaloid in lotus (Nelumbo nucifera). BMC Genom. 2018, 19, 554. [Google Scholar] [CrossRef]
- Hera, G. Morphology of Single-Petaled/Double-Petaled Flower Traits in ‘Malus’ Through Anatomical and Transcriptome Analyses. Ph.D. Thesis, Chinese Academy of Agricultural Sciences, Beijing, China, 31 May 2013. [Google Scholar]
- Huo, Y.; Yang, H.; Ding, W.J.; Yuan, Z.H.; Zhu, Z.L. Exploring the relationship between genomic variation and phenotype in ornamental pomegranate. Horticulturae 2023, 9, 361. [Google Scholar] [CrossRef]
- Huo, Y.; Yang, H.; Ding, W.J.; Huang, T.; Yuan, Z.H.; Zhu, Z.L. Combined transcriptome and proteome analysis provides insights into petaloidy in ornamental pomegranate. Plants 2023, 12, 2402. [Google Scholar] [CrossRef]
- Yang, J.; Tian, L.; Sun, M.X.; Huang, X.Y.; Zhu, J.; Guan, Y.F.; Jia, Q.S.; Yang, Z.N. AUXIN RESPONSE FACTOR17 is essential for pollen wall pattern formation in Arabidopsis. Plant Physiol. 2013, 162, 720–731. [Google Scholar] [CrossRef]
- Hagen, G.; Guilfoyle, T. Auxin–responsive gene expression: Genes, promoters and regulatory factors. Plant Mol. Biol. 2002, 49, 373–385. [Google Scholar] [CrossRef]
- Schruff, M.C.; Spielman, M.; Tiwari, S.; Adams, S.; Fenby, N.; Scott, R.J. The AUXIN RESPONSE FACTOR 2 gene of Arabidopsis links auxin signalling, cell division, and the size of seeds and other organs. Development 2006, 133, 251–261. [Google Scholar] [CrossRef]
- Stintzi, A.; Browse, J. The Arabidopsis male–sterile mutant, opr3, lacks the 12–oxophytodienoic acid reductase required for jasmonate synthesis. Proc Natl. Acad. Sci USA 2000, 97, 10625–10630. [Google Scholar] [CrossRef]
- Xiang, Q.J.; Yang, H.Y.; Wu, J.; Peng, Z.; Shen, F.; Zhou, L.Y.; Wang, Y.Y. Identification and MeJA sensitivity analysis of Arabidopsis mutant jar1. J. Southwest For. Univ. 2018, 38, 69–73. [Google Scholar]
- Cirilli, M.; Rossini, L.; Chiozzotto, R.; Baccichet, I.; Florio, F.E.; Mazzaglia, A.; Turco, S.; Bassi, D.; Gattolin, S. Less is more: Natural variation disrupting a miR172 gene at the di locus underlies the recessive double-flower trait in peach (P. persica L. Batsch). BMC Plant Biol. 2022, 22, 318. [Google Scholar] [CrossRef] [PubMed]
- Hu, L.; Zheng, T.; Cai, M.; Pan, H.; Wang, J.; Zhang, Q. Transcriptome analysis during floral organ development provides insights into stamen petaloidy in Lagerstroemia speciosa. Plant Physiol. Biochem. 2019, 142, 510–518. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Huang, X.C.; Wang, Y.L.; Zhang, R.J.; Shi, D.Y.; Li, T.F.; Zhou, G.C.; Xue, J.Y. Plastid phylogenomics of Paeonia and the evolution of ten flower types in tree peony. Genes 2022, 13, 2229. [Google Scholar] [CrossRef]
- Lou, P.; Kang, J.G.; Zhang, G.Y.; Bonnema, G.; Fang, Z.Y.; Wang, X.W. Transcript profiling of a dominant male sterile mutant (Ms-cd1) in cabbage during flower bud development. Plant Sci. 2007, 172, 111–119. [Google Scholar] [CrossRef]
- Peng, S.; Huang, S.; Liu, Z.; Feng, H. Mutation of ACX1, a jasmonic acid biosynthetic enzyme, leads to petal degeneration in chinese cabbage (Brassica campestris ssp. pekinensis). Int. J. Mol. Sci. 2019, 20, 2310. [Google Scholar] [CrossRef]
- Wang, W.B.; He, X.F.; Li, X.Y.; Wang, W.H. Transcriptome profiling during double–flower development provides insight into stamen petaloid in cultivated Lilium. Ornam. Plant Res. 2022, 2, 10. [Google Scholar] [CrossRef]
- Lampugnani, E.R.; Kilinc, A.; Smyth, D.R. Auxin controls petal initiation in Arabidopsis. Development 2013, 140, 185–194. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.Y.; Wu, K.; Yang, M.M.; Zhou, X.A.; Zhao, Y.Z. Variation of soluble sugar, starch and plant hormones contents in sesame dominat genic male sterile line during bud development. Chin. J. Oil Crop Sci. 2014, 36, 175–180. [Google Scholar]
- Song, S.S.; Qi, T.C.; Huang, H.; Xie, D.X. Regulation of stamen development by coordinated actions of jasmonate, auxin, and gibberellin in Arabidopsis. Mol. Plant 2013, 6, 1065–1073. [Google Scholar] [CrossRef]
- Jiang, Y.J.; Liang, G.; Yang, S.Z.; Yu, D.Q. Arabidopsis WRKY57 functions as a node of convergence for jasmonic acid–and auxinmediated signaling in jasmonic acid–induced leaf senescence. Plant Cell 2014, 26, 230–245. [Google Scholar] [CrossRef]
- Cheng, Y.; Zhao, Y. A role for auxin in flower development. J. Integr. Plant Biol. 2007, 49, 99–104. [Google Scholar] [CrossRef]
- Staswick, P.E. The tryptophan conjugates of jasmonic and indole–3–acetic acids are endogenous auxin inhibitors. Plant Physiol. 2009, 150, 1310–1321. [Google Scholar] [CrossRef]
- Yang, Y.; Wang, J.Y.; Xu, Y.; Abbas, F.; Xu, D.B.; Tao, S.C.; Xie, X.T.; Song, F.; Huang, Q.Y.; Sharma, A.; et al. Genome-wide identification and expression analysis of AUX/LAX family genes in Chinese hickory (Carya cathayensis Sarg.) under various abiotic stresses and grafting. Front Plant Sci. 2023, 13, 1060965. [Google Scholar] [CrossRef]
- Chen, J.W.; Li, Y.; Li, Y.H.; Li, Y.Q.; Wang, Y.; Jiang, C.Y.; Choisy, P.; Xu, T.; Cai, Y.M.; Pei, D.; et al. Auxin response factor 18–histone deacetylase 6 module regulates floral organ identity in rose (Rosa hybrida). Plant Physiol. 2021, 186, 1074–1087. [Google Scholar] [CrossRef]
- Yanofsk, M.F. Floral meristems to floral organs: Gens controlling early events in Arabidopsis flower development. Annu. Rev. Plant Physiol. 1995, 46, 167–188. [Google Scholar] [CrossRef]
- Davies, B.; Motte, P.; Keck, E.; Saedler, H.; Sommer, H.; Schwarz-Sommer, Z. Plena and Farinelli: Redundancy and regulatory interactions between two Antirrhinum MADS–box factors controlling flower development. EMBO J. 1999, 18, 4023–4034. [Google Scholar] [CrossRef]
- Nakamura, T.; Fukuda, T.; Nakana, M.; Hasebe, M.; Kameya, T.; Kanno, A. The modified ABC model explains the development of the petaloid perianth of Agapanthus praecox ssp. Orientalis (Agapanthaceae) flower. Plant Mol. Biol. 2005, 58, 435–445. [Google Scholar] [CrossRef] [PubMed]
- Dubois, A.; Raymond, O.; Maene, M.; Baudino, S.; Langlde, B.N.; Boltz, V.; Vergne, P.; Bendahmane, M. Tinkering with the C–function: A molecular frame for the selection of double flowers in cultivated roses. PLoS ONE 2010, 5, e9288. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.X.; Zhang, D.D.; Liu, D.; Li, F.L.; Lu, H. Exon skipping of AGAMOUS homolog PrseAG in developing double flowers of Prunus lannesiana (Rosaceae). Plant Cell Rep. 2013, 32, 227–237. [Google Scholar] [CrossRef] [PubMed]
- Ellis, C.M.; Nagpal, P.; Young, J.C.; Hagen, G.; Guifoyle, J.T.; Reed, W.J. AUXIN RESPONSE FACTOR1 and AUXIN RESPONSE FACTOR2 regulate senescence and floral organ abscission in Arabidopsis thaliana. Development 2005, 132, 4563–4574. [Google Scholar] [CrossRef]
- Pekker, I.; Alvarez, J.P.; Eshed, Y. Auxin response factors mediate Arabidopsis organ asymmetry via modulation of KANADI activity. Plant Cell 2005, 17, 2899–2910. [Google Scholar] [CrossRef]
- Sessions, A.; Nemhauser, J.L.; McColl, A.; Roe, J.L.; Feldmann, K.A.; Zambryski, P.C. ETTIN patterns the Arabidopsis floral meristerm and reproductive organs. Development 1997, 134, 4481–4491. [Google Scholar] [CrossRef]
- Yang, Y. Molecular Mechanism of ARF19-WOX9/13 Mediated Exogenous Auxin Inducing the Embryogenic Cell Formation in Lilium. Ph.D. Thesis, Shenyang Agricultural University, Shenyang, China, 1 June 2022. [Google Scholar]
- Li, X.N.; Xiao, H.Z.; Wan, S.L.; Zhang, D.; Zhang, Y. Gene cloning and expression analysis of auxin responsive HbJAR1 in rubber tree. Chin. J. Trop. Crops 2017, 38, 1478–1484. [Google Scholar]
- Wang, J.; Song, L.; Gong, X.; Xu, J.F.; Li, M.H. Functions of jasmonic acid in plant regulation and response to abiotic stress. Int. J. Mol. Sci. 2020, 21, 1446. [Google Scholar] [CrossRef]
- Sheard, L.B.; Tan, X.; Mao, H.; Withers, J.; Ben-Nissan, G.; Hinds, T.R.; Kobayashi, Y.; Hsu, F.F.; Sharon, M.; Browse, J.; et al. Jasmonate perception by inositol–phosphate–potentiated COI1–JAZ co–receptor. Nature 2010, 468, 400–405. [Google Scholar] [CrossRef]
- Zhang, C. Identification of jasmonic acid regulatory gene GH3 family and analysis of disease resistance and wounding in potato. Ph.D. Thesis, Northwest Agriculture & Forestry University, Xianyang, China, 1 April 2021. [Google Scholar]
- Zhao, Y.J.; Wang, Y.Y.; Zhao, X.Q.; Yan, M.; Ren, Y.; Yuan, Z.H. ARF6s identification and function analysis provide insights into flower development of Punica granatum L. Front. Plant Sci. 2022, 13, 833747. [Google Scholar]
- Sun, J.Q.; Chen, Q.; Qi, L.L.; Jiang, H.L.; Li, S.U.; Xu, Y.X.; Liu, F.; Zhou, W.K.; Pan, J.W.; Li, X.G.; et al. Jasmonate modulates endocytosis and plasma membrane accumulation of the Arabidopsis PIN2 protein. New Phytol. 2011, 191, 360–375. [Google Scholar] [CrossRef]
- Li, J.T.; Ren, M.D.; Chai, M.M.; Zhang, H.X.; Wang, Z.Y.; Gu, C.F.; Tian, Q.; Peng, C.Y.; Li, Y.X.; Fan, H.Y. Research of jasmonic acid modulated auxin biosynthesis and transport in rice root. J. Xinyang Norm. Univ. 2021, 34, 448–451. [Google Scholar]
- Pak, H.; Guo, Y.; Chen, M.X.; Chen, K.M.; Li, Y.L.; Hua, S.J.; Shamsi, I.; Meng, H.B.; Shi, C.G.; Jiang, L.X. The effect of exogenous methyl jasmonate on the flowering time, floral organ morphology, and transcript levels of a group of genes implicated in the development of oilseed rape flowers (Brassica napus L.). Planta 2009, 231, 79–91. [Google Scholar] [CrossRef]




| Pomegranate Variety | Plant Growth Regulator | Concentration (mg/L) |
|---|---|---|
| Single-petal variety ‘Nldbh’ | NAA | 10 |
| Single-petal variety ‘Nldbh’ | NAA | 50 |
| Single-petal variety ‘Nldbh’ | NAA | 100 |
| Single-petal variety ‘Nldbh’ | MeJA | 50 |
| Single-petal variety ‘Nldbh’ | MeJA | 100 |
| Single-petal variety ‘Nldbh’ | MeJA | 150 |
| Single-petal variety ‘Nldbh’ | ABA | 50 |
| Single-petal variety ‘Nldbh’ | ABA | 100 |
| Single-petal variety ‘Nldbh’ | ABA | 150 |
| Single-petal variety ‘Nldbh’ | ETH | 10 |
| Single-petal variety ‘Nldbh’ | ETH | 50 |
| Single-petal variety ‘Nldbh’ | ETH | 100 |
| Single-petal variety ‘Nldbh’ | CK | – |
| Double-petal variety ‘Nlcbh’ | NAA | 10 |
| Double-petal variety ‘Nlcbh’ | NAA | 50 |
| Double-petal variety ‘Nlcbh’ | NAA | 100 |
| Double-petal variety ‘Nlcbh’ | MeJA | 50 |
| Double-petal variety ‘Nlcbh’ | MeJA | 100 |
| Double-petal variety ‘Nlcbh’ | MeJA | 150 |
| Double-petal variety ‘Nlcbh’ | ABA | 50 |
| Double-petal variety ‘Nlcbh’ | ABA | 100 |
| Double-petal variety ‘Nlcbh’ | ABA | 150 |
| Double-petal variety ‘Nlcbh’ | ETH | 10 |
| Double-petal variety ‘Nlcbh’ | ETH | 50 |
| Double-petal variety ‘Nlcbh’ | ETH | 100 |
| Double-petal variety ‘Nlcbh’ | CK | – |
| Treatment of Plant Growth Regulator | Tissue | |||
|---|---|---|---|---|
| StSi | StDo | PeDo | PeSi | |
| NAA 10 mg/L | √ | – | – | √ |
| NAA 50 mg/L | √ | – | – | √ |
| NAA 100 mg/L | √ | – | – | √ |
| CK | √ | – | – | √ |
| MeJA 50 mg/L | – | √ | √ | – |
| MeJA 100 mg/L | – | √ | √ | – |
| MeJA 150 mg/L | – | √ | √ | – |
| NAA 10 mg/L | – | √ | √ | – |
| NAA 50 mg/L | – | √ | √ | – |
| NAA 100 mg/L | – | √ | √ | – |
| CK | – | √ | √ | – |
| Gene Name | Upstream Primer (5′–3′) | Downstream Primer (5′–3′) |
|---|---|---|
| PgActin | AGTCCTCTTCCAGCCATCTC | CACTGAGCACAATGTTTCCA |
| YUC | CTACCCGACCTACCCGACCAAG | CGTTGAACCGAGGCTGGATGTC |
| LAX2 | CCTGCTGGCTACTCTGTATGTGTTC | GCGTTGGCGTGGTTGAGGAG |
| JAR1 | GACCGAAGTGAAGGTTGGAGAAGAG | CGGCGTGGAGTTGTGGAAGC |
| ILR1 | CTGGCGTTGTCGGTTACATCGG | ATGCTCCCATTCCACTCTCTCCTC |
| GH3.17 | CTGGCGGACTAATGGCAAGGC | TCTCATCAGGGCTCGTGACCAC |
| ASR | ACACCACCACCTCTTCCACCAC | AGCCGACAGCCATCACCTCAG |
| ARF | GAAACATGATGCCGATGCTTTGGG | CAAGGCTTTGAGGAGTTCAGGGTAG |
| AIR12 | AACTGCTCCGACCTCCCGAAG | CAGCGGTGAAGGCAATGGAGAG |
| Treatment | Flower Type | Number of Petalized Stamens (NOPSs) per Flower | Number of Petals (NOPs) per Flower |
|---|---|---|---|
| 10 mg/L NAA | Single-petal flower | 3.10 ± 0.16 Aa | 6.21 ± 0.07 Aa |
| 50 mg/L NAA | Single-petal flower | 1.87 ± 0.21 Bb | 6.17 ± 0.05 ABab |
| 100 mg/L NAA | Single-petal flower | 0.13 ± 0.05 Cc | 6.17 ± 0.05 ABab |
| 50 mg/L MeJA | Single-petal flower | 0.00 ± 0.00 Cc | 6.10 ± 0.08 Abab |
| 100 mg/L MeJA | Single-petal flower | 0.00 ± 0.00 Cc | 6.00 ± 0.05 Bb |
| 150 mg/L MeJA | Single-petal flower | 0.00 ± 0.00 Cc | 6.13 ± 0.05 Abab |
| 50 mg/L ABA | Single-petal flower | 0.00 ± 0.00 Cc | 6.03 ± 0.05 Bb |
| 100 mg/L ABA | Single-petal flower | 0.00 ± 0.00 Cc | 6.03 ± 0.05 Bb |
| 150 mg/L ABA | Single-petal flower | 0.00 ± 0.00 Cc | 6.00 ± 0.00 Bb |
| 10 mg/L ETH | Single-petal flower | 0.00 ± 0.00 Cc | 6.07 ± 0.09 ABb |
| 50 mg/L ETH | Single-petal flower | 0.00 ± 0.00 Cc | 6.03 ± 0.05 Bb |
| 100 mg/L ETH | Single-petal flower | 0.00 ± 0.00 Cc | 6.07 ± 0.05 ABb |
| CK | Single-petal flower | 0.00 ± 0.00 Cc | 6.03 ± 0.05 Bb |
| 10 mg/L NAA | Double-petal flower | 238.47 ± 9.08 ABa | 106.23 ± 4.32 Bb |
| 50 mg/L NAA | Double-petal flower | 205.20 ± 9.40 BCb | 99.97 ± 6.46 BCbc |
| 100 mg/L NAA | Double-petal flower | 174.00 ± 5.08 BCc | 72.133 ± 13.96 BCc |
| 50 mg/L MeJA | Double-petal flower | 207.70 ± 17.68 Bb | 89.50 ± 9.70 BCbc |
| 100 mg/L MeJA | Double-petal flower | 254.93 ± 9.51 ABa | 148.00 ± 23.79 Aa |
| 150 mg/L MeJA | Double-petal flower | 208.00 ± 11.13 Bb | 132.97 ± 12.22 ABa |
| 50 mg/L ABA | Double-petal flower | 154.60 ± 25.77 Cc | 72.57 ± 19.26 BCc |
| 100 mg/L ABA | Double-petal flower | 169.60 ± 20.56 BCc | 89.23 ± 12.55 BCbc |
| 150 mg/L ABA | Double-petal flower | 174.67 ± 13.65 BCc | 66.47 ± 7.92 Cc |
| 10 mg/L ETH | Double-petal flower | 175.57 ± 14.26 BCc | 69.03 ± 14.18 Cc |
| 50 mg/L ETH | Double-petal flower | 167.07 ± 17.64 Cc | 77.10 ± 7.29 BCc |
| 100 mg/L ETH | Double-petal flower | 238.47 ± 9.08 ABa | 106.23 ± 4.32 Bb |
| CK | Double-petal flower | 175.37 ± 3.18 BCc | 71.133 ± 12.75 BCc |
| Treatment | Index | Tissue | Gene Name | Regression Equation | Samples | PCC | F Value | Sig. |
|---|---|---|---|---|---|---|---|---|
| NAA 10 mg/L | NOPS | StSi | YUC | y = 0.003x + 0.019 | 12 | 0.279 | 0.843 | 0.380 |
| ILR1 | y = 0.022x + 0.047 | 12 | 0.949 | 90.346 | 0.000 | |||
| GH3.17 | y = 0.006x + 0.036 | 12 | 0.397 | 1.867 | 0.202 | |||
| LAX2 | y = 0.053x + 0.161 | 12 | 0.888 | 37.457 | 0.000 | |||
| ARF | y = 0.011x + 0.051 | 12 | 0.799 | 17.601 | 0.002 | |||
| AIR12 | y = 0.039x + 0.125 | 12 | 0.822 | 20.757 | 0.001 | |||
| JAR1 | y = 0.048x + 0.026 | 12 | 0.831 | 22.397 | 0.001 | |||
| ASR | y = −4.409x + 45.400 | 12 | 0.548 | 4.295 | 0.065 | |||
| NAA 10 mg/L | NOPS | StDo | YUC | y = 0.000x − 0.008 | 12 | 0.325 | 1.181 | 0.303 |
| ILR1 | y = 0.003x − 0.444 | 12 | 0.901 | 43.212 | 0.000 | |||
| GH3.17 | y = 0.000x − 0.007 | 12 | 0.354 | 1.430 | 0.259 | |||
| LAX2 | y = 0.008x − 1.041 | 12 | 0.878 | 33.573 | 0.000 | |||
| ARF | y = 0.003x − 0.448 | 12 | 0.908 | 47.229 | 0.000 | |||
| AIR12 | y = 0.002x − 0.141 | 12 | 0.705 | 9.860 | 0.011 | |||
| JAR1 | y = 0.005x − 0.648 | 12 | 0.770 | 14.610 | 0.003 | |||
| ASR | y = −0.133x + 62.257 | 12 | 0.510 | 3.514 | 0.090 | |||
| NAA 10 mg/L | NOP | PeDo | YUC | y = 0.000x + 0.025 | 12 | 0.139 | 0.197 | 0.667 |
| ILR1 | y = 0.003x − 0.109 | 12 | 0.783 | 15.847 | 0.003 | |||
| GH3.17 | y = 0.000x + 0.039 | 12 | 0.180 | 1.334 | 0.576 | |||
| LAX2 | y = 0.042x − 0.917 | 12 | 0.656 | 7.572 | 0.020 | |||
| ARF | y = 0.006x − 0.224 | 12 | 0.668 | 8.041 | 0.018 | |||
| AIR12 | y = 0.002x − 0.008 | 12 | 0.681 | 8.655 | 0.015 | |||
| JAR1 | y = 0.005x − 0.123 | 12 | 0.643 | 7.050 | 0.024 | |||
| ASR | y = −0.033x + 12.360 | 12 | 0.375 | 1.638 | 0.230 | |||
| NAA 10 mg/L | NOP | PeSi | YUC | y = −0.086x + 0.572 | 12 | 0.310 | 1.060 | 0.327 |
| ILR1 | y = 0.146x − 0.825 | 12 | 0.417 | 2.107 | 0.177 | |||
| GH3.17 | y = −0.060x + 0.429 | 12 | 0.253 | 0.686 | 0.427 | |||
| LAX2 | y = 1.048x − 5.979 | 12 | 0.504 | 3.410 | 0.095 | |||
| ARF | y = 0.604x − 3.142 | 12 | 0.559 | 4.535 | 0.059 | |||
| AIR12 | y = 0.069x − 0.345 | 12 | 0.224 | 0.530 | 0.483 | |||
| JAR1 | y = 0.177x − 1.023 | 12 | 0.509 | 3.500 | 0.091 | |||
| ASR | y = 2.114x + 4.438 | 12 | 0.071 | 0.050 | 0.827 |
| Treatment | Index | Tissue | Gene Name | Regression Equation | Samples | PCC | F Value | Sig. |
|---|---|---|---|---|---|---|---|---|
| MeJA 100 mg/L | NOPS | StDo | YUC | y = 0.000x + 0.064 | 12 | 0.491 | 3.17 | 0.105 |
| ILR1 | y = 0.002x − 0.202 | 12 | 0.726 | 11.120 | 0.008 | |||
| GH3.17 | y = 0.0.001x − 0.063 | 12 | 0.613 | 6.011 | 0.034 | |||
| LAX2 | y = 0.007x − 0.971 | 12 | 0.873 | 31.982 | 0.000 | |||
| ARF | y = 0.002x − 0.274 | 12 | 0.824 | 21.093 | 0.001 | |||
| AIR12 | y = 0.001x + 0.022 | 12 | 0.386 | 1.748 | 0.216 | |||
| JAR1 | y = 0.003x − 0.438 | 12 | 0.884 | 35.839 | 0.000 | |||
| ASR | y = 0.118x + 10.258 | 12 | 0.456 | 2.626 | 0.136 | |||
| MeJA 100 mg/L | NOP | PeDo | YUC | y = 0.000x + 0.071 | 12 | 0.449 | 2.520 | 0.144 |
| ILR1 | y = 0.002x − 0.077 | 12 | 0.820 | 20.477 | 0.001 | |||
| GH3.17 | y = 0.001x + 0.013 | 12 | 0.633 | 6.687 | 0.027 | |||
| LAX2 | y = 0.023x − 0.290 | 12 | 0.873 | 31.954 | 0.000 | |||
| ARF | y = 0.003x + 0.004 | 12 | 0.718 | 10.619 | 0.009 | |||
| AIR12 | y = 0.001x + 0.061 | 12 | 0.541 | 4.147 | 0.069 | |||
| JAR1 | y = 0.002x − 0.033 | 12 | 0.766 | 14.179 | 0.004 | |||
| ASR | y = −0.206x + 61.615 | 12 | 0.570 | 4.812 | 0.053 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huo, Y.; Lu, F.; Mu, L.; Yang, H.; Ding, W.; Yuan, Z.; Zhu, Z. Plant Growth Regulators Promote Petaloidy and Modulate Related Gene Expression in Ornamental Pomegranate. Horticulturae 2025, 11, 1059. https://doi.org/10.3390/horticulturae11091059
Huo Y, Lu F, Mu L, Yang H, Ding W, Yuan Z, Zhu Z. Plant Growth Regulators Promote Petaloidy and Modulate Related Gene Expression in Ornamental Pomegranate. Horticulturae. 2025; 11(9):1059. https://doi.org/10.3390/horticulturae11091059
Chicago/Turabian StyleHuo, Yan, Fei Lu, Lili Mu, Han Yang, Wenjie Ding, Zhaohe Yuan, and Zunling Zhu. 2025. "Plant Growth Regulators Promote Petaloidy and Modulate Related Gene Expression in Ornamental Pomegranate" Horticulturae 11, no. 9: 1059. https://doi.org/10.3390/horticulturae11091059
APA StyleHuo, Y., Lu, F., Mu, L., Yang, H., Ding, W., Yuan, Z., & Zhu, Z. (2025). Plant Growth Regulators Promote Petaloidy and Modulate Related Gene Expression in Ornamental Pomegranate. Horticulturae, 11(9), 1059. https://doi.org/10.3390/horticulturae11091059






