RrYUC10 Positively Regulates Adventitious Root Formation in Rosa rugosa Stem Cuttings
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material and Growth Conditions
2.2. Detection of Phytohormones
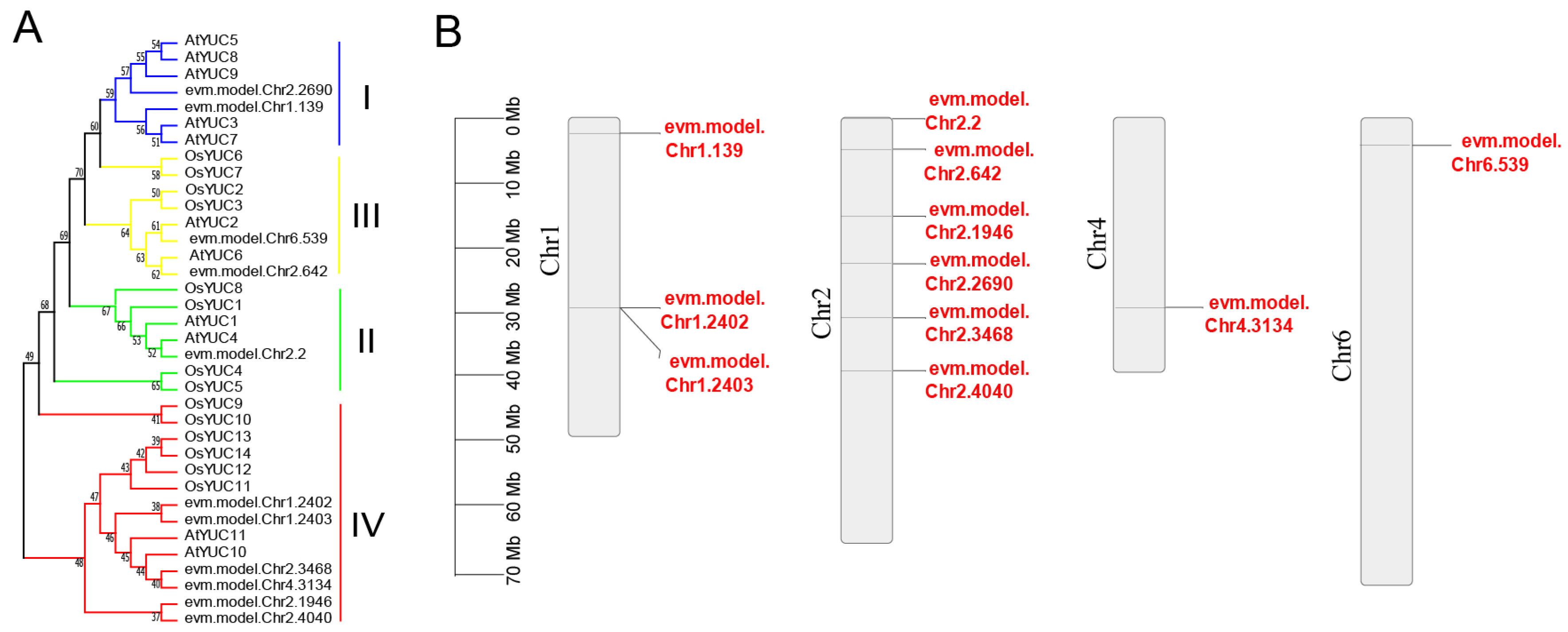
2.3. Identification of RrYUCs and Phylogenetic Analyses
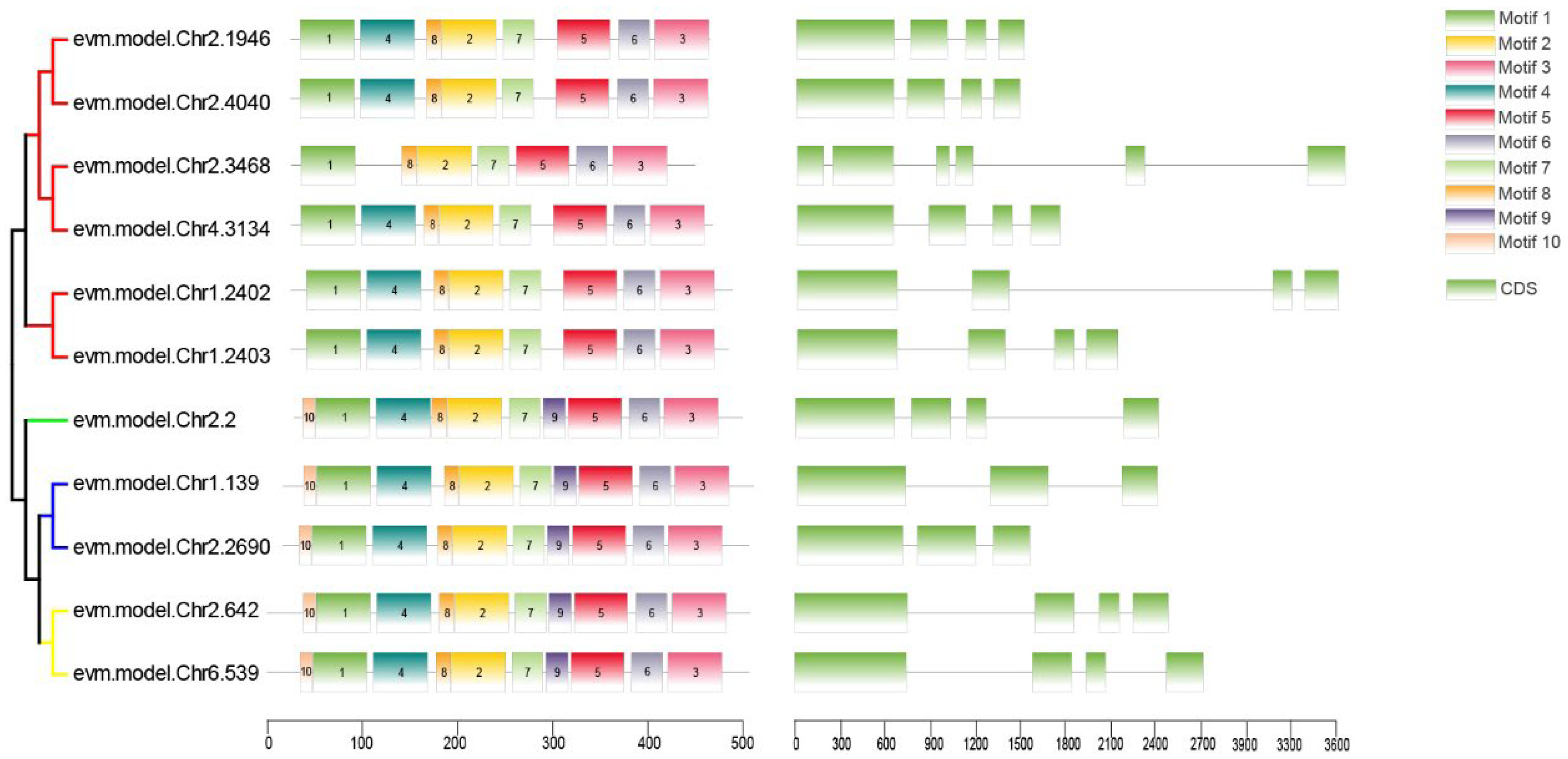
2.4. Chromosomal Location, Gene Structure, and Motif Analysis of RrYUCs Family
2.5. RNA Extraction and RT-qPCR
2.6. VIGS
2.7. Transgenic Assays Mediated by Agrobacterium Rhizogenes
2.8. Root Configuration Observation and Analysis
3. Results
3.1. Analysis of Hormone Types in Callus and Root
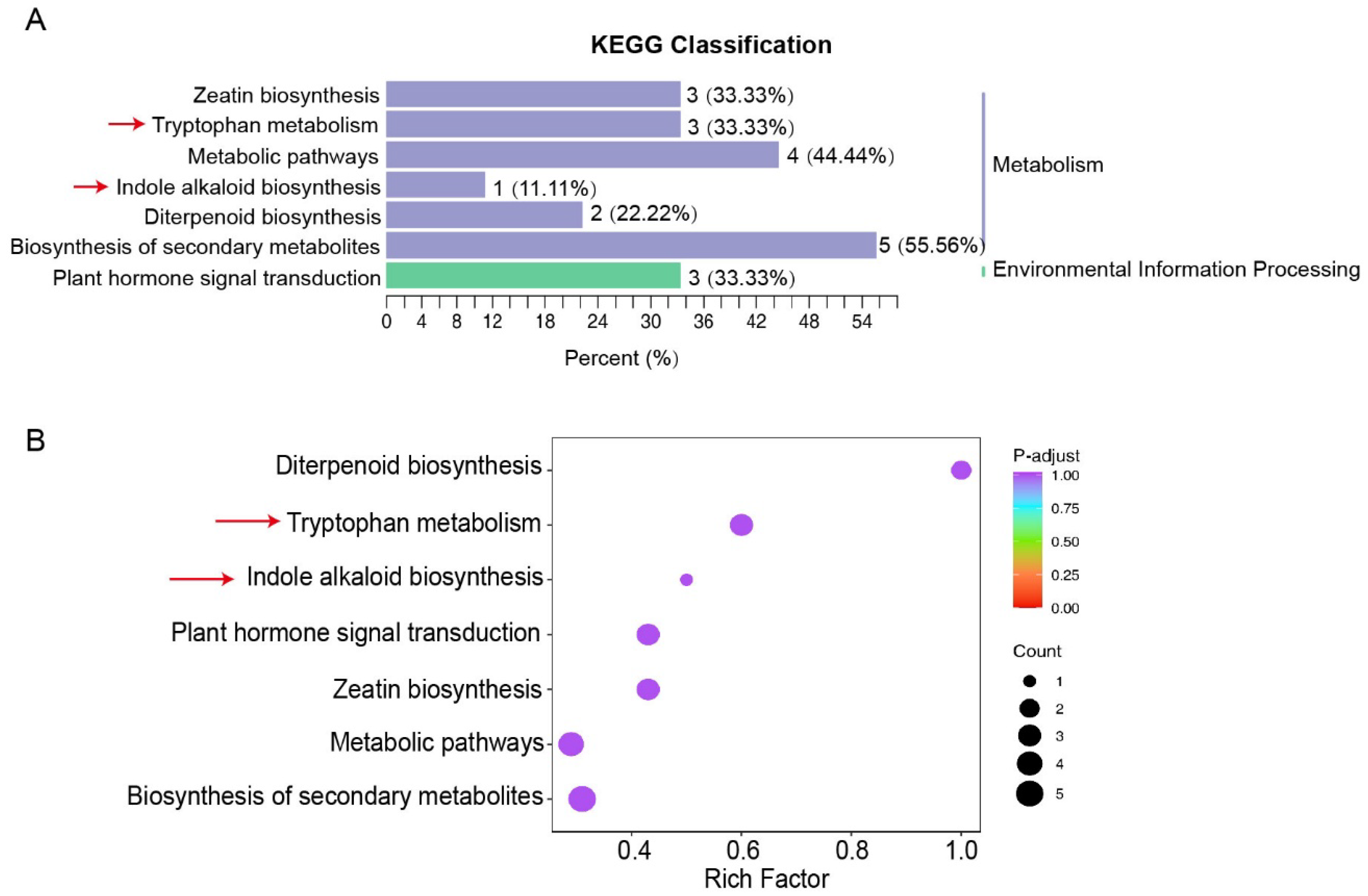
3.2. RrYUCs Family Identification in R. rugosa
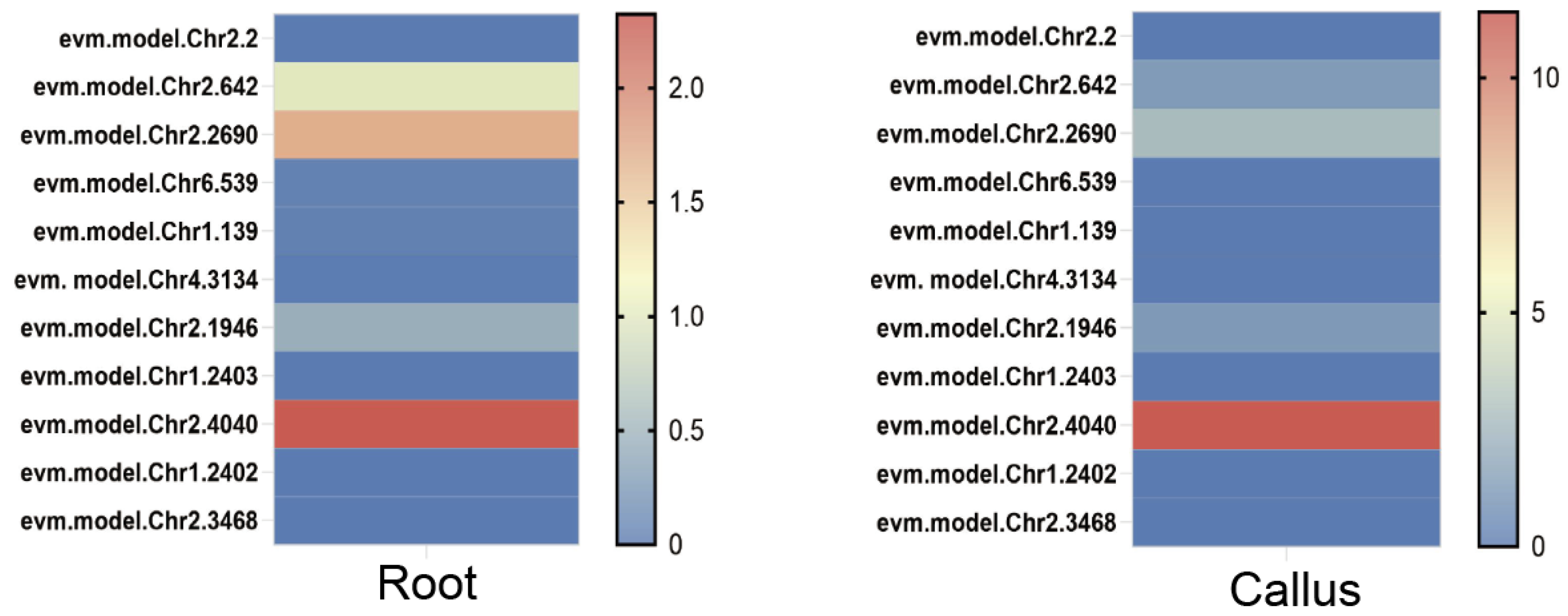
3.3. Expression Profiling of RrYUCs Genes in Roots and Calluses
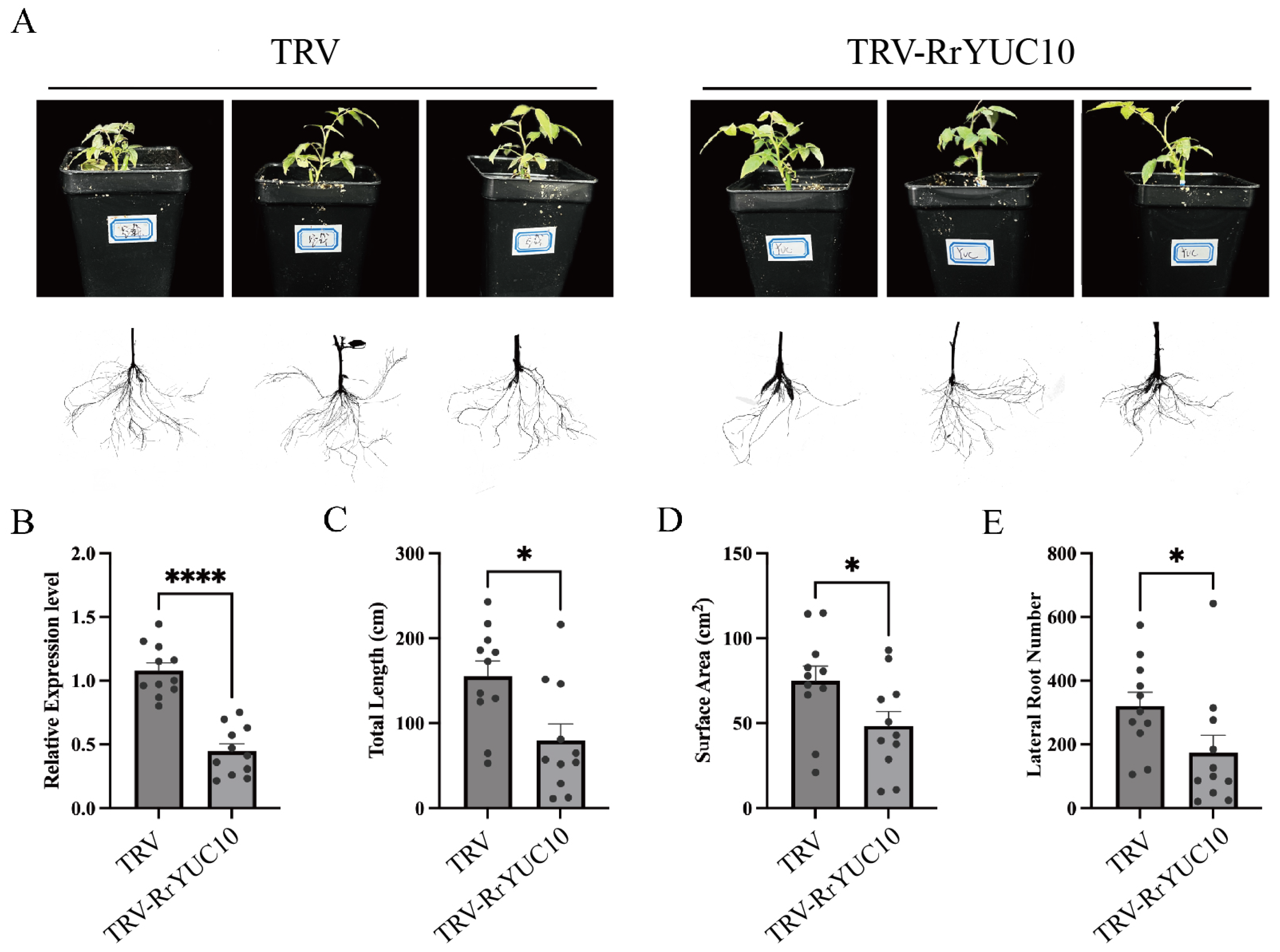
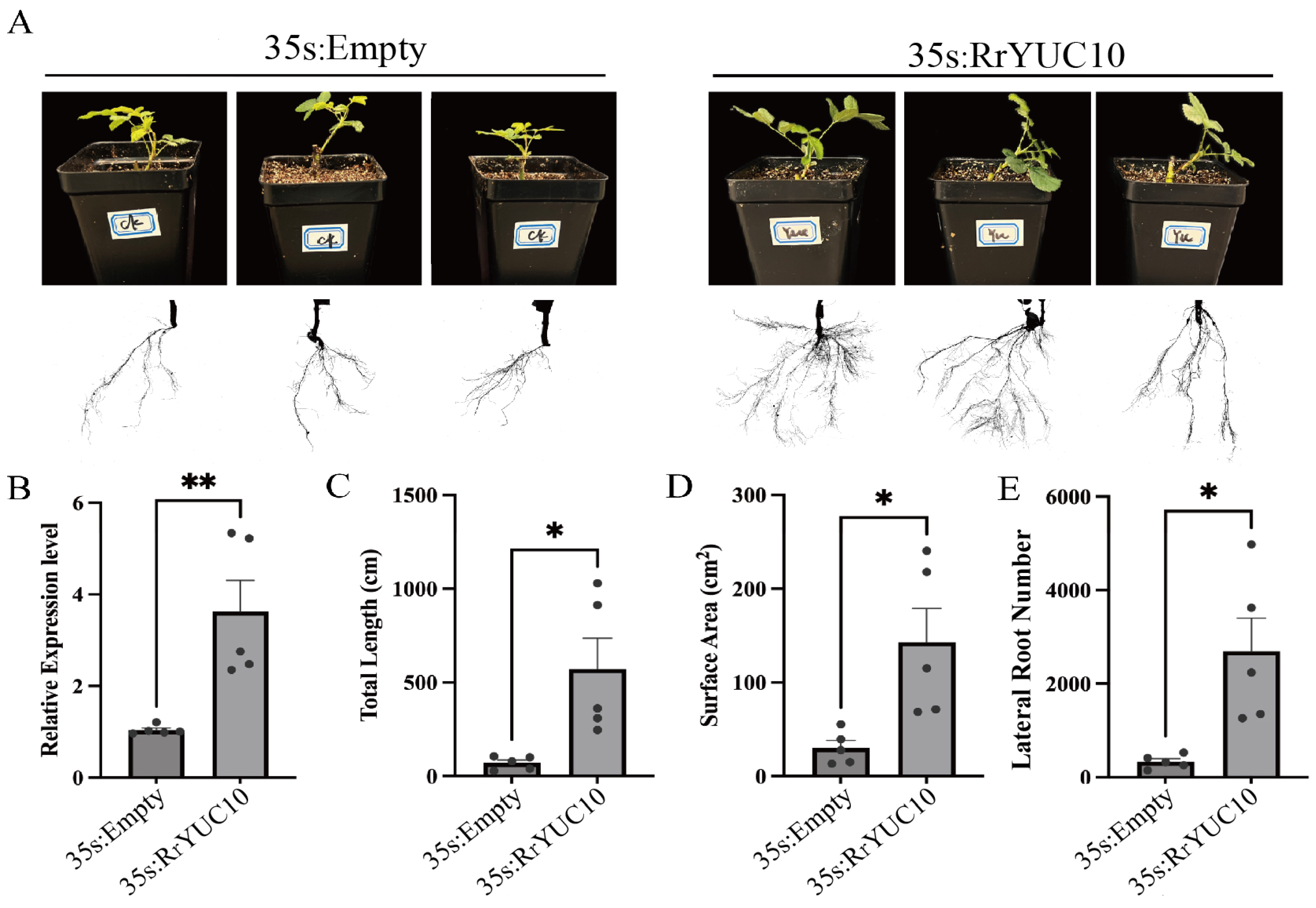
3.4. RrYUC10 Positively Regulates Adventitious Rooting in R. rugosa
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Li, S.W.; Xue, L.G.; Xu, S.J.; Feng, H.Y.; An, L.Z. Mediators, genes and signaling in adventitious rooting. Bot. Rev. 2009, 75, 230–247. [Google Scholar] [CrossRef]
- Steffens, B.; Rasmussen, A. The physiology of adventitious roots. Plant Physiol. 2016, 170, 603–617. [Google Scholar] [CrossRef] [PubMed]
- Ge, Y.C.; Fang, X.; Liu, W.; Sheng, L.H.; Xu, L. Adventitious lateral rooting: The plasticity of root system architecture. Physiol. Plant 2019, 165, 39–43. [Google Scholar] [CrossRef] [PubMed]
- da Costa, C.T.; de Almeida, M.R.; Ruedell, C.M.; Schwambach, J.; Maraschin, F.S.; Fett-Neto, A.G. When stress and development go hand in hand: Main hormonal controls of adventitious rooting in cuttings. Front. Plant Sci. 2013, 4, 133. [Google Scholar] [CrossRef] [PubMed]
- Fleck, J.D.; Schwambach, J.; Almeida, M.E.; Yendo, A.C.A.; De Costa, F.; Gosmann, G.; Fett-Neto, A.G. Immunoadjuvant saponin production in seedlings and micropropagated plants of Quillaja brasiliensis. In Vitro Cell Dev. Biol. Plant 2009, 45, 715–720. [Google Scholar] [CrossRef]
- Bellini, C.; Pacurar, D.I.; Perrone, I. Adventitious roots and lateral roots: Similarities and differences. Annu. Rev. Plant Biol. 2014, 65, 639–666. [Google Scholar] [CrossRef]
- Osterc, G.; Stefancic, M.; Stampar, F. Juvenile stockplant material enhances root development through higher endogenous auxin level. Acta Physiol. Plant 2009, 31, 899–903. [Google Scholar] [CrossRef]
- Druege, U. Overcoming physiological bottlenecks of leaf vitality and root development in cuttings: A systemic perspective. Front. Plant Sci. 2020, 11, 907. [Google Scholar] [CrossRef]
- Iglesias, M.J.; Sellaro, R.; Zurbriggen, M.D.; Casal, J.J. Multiple links between shade avoidance and auxin networks. J. Exp. Bot. 2018, 69, 213–228. [Google Scholar] [CrossRef]
- Lakehal, A.; Bellini, C. Control of adventitious root formation: Insights into synergistic and antagonistic hormonal interactions. Physiol. Plant 2019, 165, 90–100. [Google Scholar] [CrossRef]
- Pacurar, D.I.; Perrone, I.; Bellini, C. Auxin is a central player in the hormone cross-talks that control adventitious rooting. Physiol. Plant 2014, 151, 83–96. [Google Scholar] [CrossRef]
- Kevers, C.; Hausman, J.F.; Faivre-Rampant, O.; Evers, D.; Gaspar, T. Hormonal control of adventitious rooting: Progress and questions. Angew. Bot. 1997, 71, 71–79. [Google Scholar]
- Singh, Z.; Singh, H.; Garg, T.; Mushahary, K.K.K.; Yadav, S.R. Genetic and hormonal blueprint of shoot-borne adventitious root development in rice and maize. Plant Cell Physiol. 2023, 63, 1806–1813. [Google Scholar] [CrossRef] [PubMed]
- Oliveros-Valenzuela, M.D.; Reyes, D.; Sánchez-Bravo, J.; Acosta, M.; Nicolás, C. Isolation and characterization of a cDNA clone encoding an auxin influx carrier in carnation cuttings. Expression in different organs and cultivars and its relationship with cold storage. Plant Physiol. Biochem. 2008, 46, 1071–1076. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.H.; Zou, M.H.; Feng, B.H.; Huang, X.; Zhang, Z.; Sun, G.M. Molecular cloning and characterization of the genes encoding an auxin efflux carrier and the auxin influx carriers associated with the adventitious root formation in mango (Mangifera indica L.) cotyledon segments. Plant Physiol. Biochem. 2012, 55, 33–42. [Google Scholar] [CrossRef]
- Blakeslee, J.J.; Rossi, T.S.; Kriechbaumer, V. Auxin biosynthesis: Spatial regulation and adaptation to stress. J. Exp. Bot. 2019, 70, 5041–5049. [Google Scholar] [CrossRef]
- Zhao, Y.D. Auxin Biosynthesis: A simple two-step pathway converts tryptophan to indole-3-acetic acid in plants. Mol. Plant 2012, 5, 334–338. [Google Scholar] [CrossRef]
- Zhao, Y.D.; Christensen, S.K.; Fankhauser, C.; Cashman, J.R.; Cohen, J.D.; Weigel, D.; Chory, J. A role for flavin monooxygenase-like enzymes in auxin biosynthesis. Science 2001, 291, 306–309. [Google Scholar] [CrossRef]
- Zhao, Y. Auxin biosynthesis. Arab. Book. 2014, 12, e0173. [Google Scholar] [CrossRef]
- Chen, L.Q.; Tong, J.H.; Xiao, L.T.; Ruan, Y.; Liu, J.C.; Zeng, M.H.; Huang, H.; Wang, J.W.; Xu, L. YUCCA-mediated auxin biogenesis is required for cell fate transition occurring during de novo root organogenesis in Arabidopsis. J. Exp. Bot. 2016, 67, 4273–4284. [Google Scholar] [CrossRef]
- Hurst, C.C. Notes on the origin and evolution of our garden roses. J. R. Hortic. Soc. 1941, 66, 282–289. [Google Scholar]
- Martínez, M.C.; Santiago, J.L. Narcea-an unknown, ancient cultivated rose variety from northern Spain. Hortic. Res. 2020, 7, 44. [Google Scholar] [CrossRef]
- Lu, J.; Wang, C.Q. Medicinal components and pharmacological effects of Rosa rugosa. Rec. Nat. Prod. 2018, 12, 535–543. [Google Scholar] [CrossRef]
- Ng, T.B.; He, J.S.; Niu, S.M.; Zhao, L.; Pi, Z.F.; Shao, W.; Liu, F. A gallic acid derivative and polysaccharides with antioxidative activity from rose (Rosa rugosa) flowers. J. Pharm. Pharm. 2004, 56, 537–545. [Google Scholar] [CrossRef]
- Bai, M.J.; Liu, J.Y.; Fan, C.G.; Chen, Y.Q.; Chen, H.; Lu, J.; Sun, J.J.; Ning, G.G.; Wang, C.Q. KSN heterozygosity is associated with continuous flowering of Rosa rugosa Purple branch. Hortic. Res. 2021, 8, 26. [Google Scholar] [CrossRef]
- Otiende, M.A.; Fricke, K.; Nyabundi, J.O.; Ngamau, K.; Hajirezaei, M.R.; Druege, U. Involvement of the auxin-cytokinin homeostasis in adventitious root formation of rose cuttings as affected by their nodal position in the stock plant. Planta 2021, 254, 65. [Google Scholar] [CrossRef] [PubMed]
- Otiende, M.A.; Nyabundi, J.O.; Ngamau, K.; Opala, P. Effects of cutting position of rose rootstock cultivars on rooting and its relationship with mineral nutrient content and endogenous carbohydrates. Sci. Hortic. 2017, 225, 204–212. [Google Scholar] [CrossRef]
- Park, S.M.; Won, E.J.; Park, Y.G.; Jeong, B.R. Effects of node position, number of leaflets left, and light intensity during cutting propagation on rooting and subsequent growth of domestic roses. Hortic. Env. Biotechnol. 2011, 52, 339–343. [Google Scholar] [CrossRef]
- Floková, K.; Tarkowská, D.; Miersch, O.; Strnad, M.; Wasternack, C.; Novák, O. UHPLC-MS/MS based target profiling of stress-induced phytohormones. Phytochemistry 2014, 105, 147–157. [Google Scholar] [CrossRef]
- Chen, F.; Su, L.Y.; Hu, S.Y.; Xue, J.Y.; Liu, H.; Liu, G.H.; Jiang, Y.F.; Du, J.K.; Qiao, Y.S.; Fan, Y.N.; et al. A chromosome-level genome assembly of rugged rose (Rosa rugosa) provides insights into its evolution, ecology, and floral characteristics. Hortic. Res. 2021, 8, 141. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. Mega7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Chen, C.J.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.H.; Xia, R. Tbtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Luo, M.; Gao, Z.; Li, H.; Li, Q.; Zhang, C.X.; Xu, W.P.; Song, S.R.; Ma, C.; Wang, S.P. Selection of reference genes for miRNA qRT-PCR under abiotic stress in grapevine. Sci. Rep. 2018, 8, 4444. [Google Scholar] [CrossRef] [PubMed]
- Joshi, M.; Ginzberg, I. Adventitious root formation in crops-Potato as an example. Physiol. Plant 2021, 172, 124–133. [Google Scholar] [CrossRef]
- Ribeiro, C.L.; Silva, C.M.; Drost, D.R.; Novaes, E.; Novaes, C.R.; Dervinis, C.; Kirst, M. Integration of genetic, genomic and transcriptomic information identifies putative regulators of adventitious root formation in Populus. BMC Plant Biol. 2016, 16, 66. [Google Scholar] [CrossRef]
- Abarca, D.; Pizarro, A.; Hernández, I.; Sánchez, C.; Solana, S.P.; Del Amo, A.; Carneros, E.; Díaz-Sala, C. The GRAS gene family in pine: Transcript expression patterns associated with the maturation-related decline of competence to form adventitious roots. BMC Plant Biol. 2014, 14, 354. [Google Scholar] [CrossRef]
- Qiao, L.X.; Zhang, T.J.; Yang, H.Y.; Yang, S.H.; Wang, J.H. Overexpression of a SHORT-ROOT transcriptional factor enhances the auxin mediated formation of adventitious roots and lateral roots in poplar trees. Plant Sci. 2022, 323, 111408. [Google Scholar] [CrossRef]
- Meuwly, P.; Molders, W.; Buchala, A.; Metraux, J.P. Local and Systemic Biosynthesis of Salicylic Acid in Infected Cucumber Plants. Plant Physiol. 1995, 109, 1107–1114. [Google Scholar] [CrossRef]
- Pasternak, T.; Groot, E.P.; Kazantsev, F.V.; Teale, W.; Omelyanchuk, N.; Kovrizhnykh, V.; Palme, K.; Mironova, V.V. Salicylic Acid Affects Root Meristem Patterning via Auxin Distribution in a Concentration Dependent Manner. Plant Physiol. 2019, 180, 1725–1739. [Google Scholar] [CrossRef]
- Dong, C.J.; Liu, X.Y.; Xie, L.L.; Wang, L.L.; Shang, Q.M. Salicylic acid regulates adventitious root formation via competitive inhibition of the auxin conjugation enzyme CsGH3.5 in cucumber hypocotyls. Planta 2020, 252, 75. [Google Scholar] [CrossRef]
- Luo, W.G.; Xiao, N.; Wu, F.Y.; Mo, B.X.; Kong, W.W.; Yu, Y. Genome-Wide Identification and Characterization of YUCCA Gene Family in Mikania micrantha. Int. J. Mol. Sci. 2022, 23, 13037. [Google Scholar] [CrossRef]
- Uc-Chuc, M.A.; Kú-González, Á.F.; Jiménez-Ramírez, I.A.; Loyola-Vargas, V.M. Identification, analysis, and modeling of the YUCCA protein family genome-wide in Coffea canephora. Proteins 2022, 90, 1005–1024. [Google Scholar] [CrossRef] [PubMed]
- Song, C.H.; Zhang, D.; Zheng, L.W.; Shen, Y.W.; Zuo, X.Y.; Mao, J.P.; Meng, Y.A.; Wu, H.Q.; Zhang, Y.K.; Liu, X.Y.; et al. Genome-wide identification and expression profiling of the YUCCA gene family in Malus domestica. Sci. Rep. 2020, 10, 10866. [Google Scholar] [CrossRef] [PubMed]
- Wu, R.R.; Chen, J.B.; Lin, Y.; Jia, Q.Y.; Guo, Y.J.; Liu, J.Y.; Yan, Q.; Xue, C.C.; Chen, X.; Yuan, X.X. Genome-Wide Identification, Expression Analysis, and Potential Roles under Abiotic Stress of the YUCCA Gene Family in Mungbean (Vigna radiata L.). Int. J. Mol. Sci. 2023, 24, 1603. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.L.; Xu, T.; Wang, H.G.; Feng, D.S. Genome-wide identification and expression analysis of the TaYUCCA gene family in wheat. Mol. Biol. Rep. 2021, 48, 1269–1279. [Google Scholar] [CrossRef]
- Ye, X.L.; Sun, J.; Tian, Y.; Chen, J.W.; Yao, X.T.; Quan, X.H.; Huang, L. Identification of YUC genes associated with leaf wrinkling trait in Tacai variety of Chinese cabbage. PeerJ 2024, 12, e17337. [Google Scholar] [CrossRef]
- Naser, V.; Shani, E. Auxin response under osmotic stress. Plant Mol. Biol. 2016, 91, 661–672. [Google Scholar] [CrossRef]
- Cha, J.Y.; Kim, W.Y.; Kang, S.B.; Kim, J.I.; Baek, D.; Jung, I.J.; Kim, M.R.; Li, N.; Kim, H.J.; Nakajima, M.; et al. A novel thiol-reductase activity of Arabidopsis YUC6 confers drought tolerance independently of auxin biosynthesis. Nat. Commun. 2015, 6, 8041. [Google Scholar] [CrossRef]
- Ke, Q.B.; Wang, Z.; Ji, C.Y.; Jeong, J.C.; Lee, H.S.; Li, H.B.; Xu, B.C.; Deng, X.P.; Kwak, S.S. Transgenic poplar expressing Arabidopsis YUCCA6 exhibits auxin-overproduction phenotypes and increased tolerance to abiotic stress. Plant Physiol. Biochem. 2015, 94, 19–27. [Google Scholar] [CrossRef]
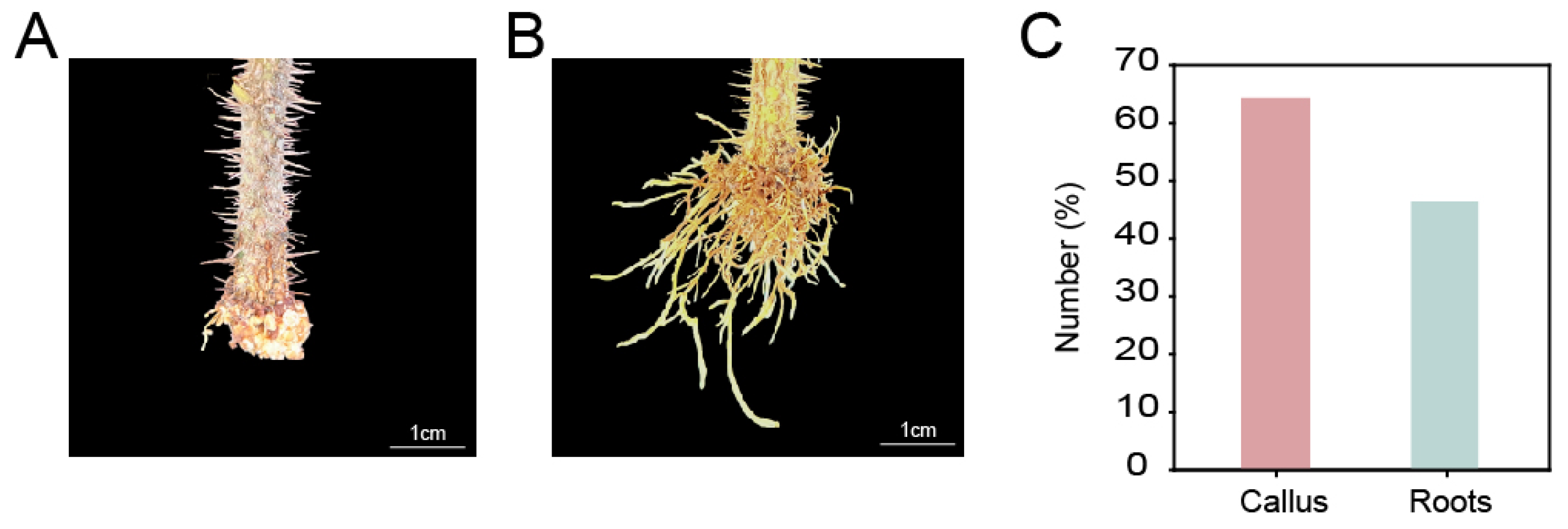
| Compounds | Callus (ng/g) | Root (ng/g) |
|---|---|---|
| TRP | 64,012.2649 ± 5782.02 | 48,337.16 ± 8157.33 |
| Indole | 9298.86 ± 1474.74 | 7988.50 ± 2832.12 |
| IAA | 3.56 ± 2.25 | 10.37 ± 0.71 |
| ABA | 108.14 ± 63.30 | 92.32 ± 22.28 |
| CK | 1.16 ± 0.84 | 2.25 ± 0.42 |
| Phe | 21,512.09 ± 2745.43 | 18,819.67 ± 2573.05 |
| SA | 353.11 ± 77.63 | 381.08 ± 35.09 |
| JA | 63.00 ± 11.41 | 81.94 ± 10.04 |
| GA | 244.15 ± 153.65 | 46.15 ± 7.94 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bai, M.; Xi, Y.; Xue, J.; Xu, X.; Xu, M.; Feng, L. RrYUC10 Positively Regulates Adventitious Root Formation in Rosa rugosa Stem Cuttings. Horticulturae 2025, 11, 1027. https://doi.org/10.3390/horticulturae11091027
Bai M, Xi Y, Xue J, Xu X, Xu M, Feng L. RrYUC10 Positively Regulates Adventitious Root Formation in Rosa rugosa Stem Cuttings. Horticulturae. 2025; 11(9):1027. https://doi.org/10.3390/horticulturae11091027
Chicago/Turabian StyleBai, Mengjuan, Yu Xi, Junqing Xue, Xiangfeng Xu, Mengmeng Xu, and Liguo Feng. 2025. "RrYUC10 Positively Regulates Adventitious Root Formation in Rosa rugosa Stem Cuttings" Horticulturae 11, no. 9: 1027. https://doi.org/10.3390/horticulturae11091027
APA StyleBai, M., Xi, Y., Xue, J., Xu, X., Xu, M., & Feng, L. (2025). RrYUC10 Positively Regulates Adventitious Root Formation in Rosa rugosa Stem Cuttings. Horticulturae, 11(9), 1027. https://doi.org/10.3390/horticulturae11091027





