1. Introduction
Transcription factors (TFs) are key regulators in plants that play crucial roles in various growth and developmental processes, as well as in responses to abiotic stresses [
1,
2,
3,
4]. The
GLABROUS1 Enhancer Binding Protein (GeBP) family is a family of transcription factors specific to plants whose members share a central DNA-binding domain and were initially discovered in
Arabidopsis thaliana. GeBP and its homologs share two conserved regions: an unknown motif in the central region and a C-terminal hypothesized leucine zipper motif [
5]. Both regions are crucial for downstream gene expression transactivation. At present, 16, 10, 10, 9, and 16
GeBP genes have been identified in
Arabidopsis [
5],
Solanum lycopersicum [
6],
Mangifera indica L [
7],
Glycine max [
8], and
Bam busoideae [
9], respectively. Previous studies highlight the importance of the GeBP gene family in plant growth and development. For instance, GeBP regulates trichome development through the expression control of the
GLABROUS1 (
GL1) gene [
10]. In
Arabidopsis, GeBP also influences trichome elongation by modulating gibberellins and cytokinins in vivo [
11].
Trichomes in the epidermis are hair-like structures and constitute the aerial part of most terrestrial plants [
12]. Trichomes are defined as unicellular or multicellular appendages, which are an extension of the above-ground epidermal cells in plants [
13]. These appendages play a key role in the development of plant sand, which occurs in a wide variety of species. Trichomes are a protective barrier against natural hazards, such as herbivores, ultraviolet (UV) irradiation, pathogen attacks, and excessive transpiration, and they aid in seed spread and seed protection [
14]. In
Arabidopsis, their initiation requires the activity of
GLABROUS1 (
GL1), which is expressed in the epidermis [
15]. Curaba et al. [
5] identified and isolated
GL1 enhancer-binding protein (GeBP), which specifically binds to the regulatory element of the promoter and regulates the expression of
GL1 [
5].
GeBP is predicted to play a role in various hormonal pathways [
16]. It is worth noting that the
GEBP/GPL genes represent a newly defined class of leucine zipper (
Leu zipper) transcription factors, and they play redundant roles in the regulation of the cytokinin hormone pathway [
10]. A recent study demonstrated that the
Arabidopsis GeBPLIKE 4 (GPL4) transcription factor, as an inhibitor of root growth, is induced rapidly in the root tips in response to cadmium (Cd) [
17]. These research outcomes suggest that the
GeBP family gene is not only involved in the developmental process of the plant but also protects against environmental stress. The constitutive expressor of pathogenesis-related gene-5 (
CPR5) in
Arabidopsis displays highly pleiotropic functions, particularly in pathogen responses, cell proliferation, cell expansion, and cell death. It was found that GeBP/GPLs are involved in the control of cell expansion in a CPR5-dependent manner but not in the control of cell proliferation by regulating a set of genes that represents a subset of the CPR5 pathway [
18].
Brassica oleracea is a diploid species (2n = 18) within the family Brassicaceae, encompassing a wide array of economically important vegetables, such as cabbage, broccoli, cauliflower, kale, brussels sprouts, and kohlrabi. These diverse morphotypes have arisen through centuries of selective breeding, leading to significant morphological and nutritional variation within the species [
19].
Native to the coastal regions of southern and western Europe,
B. oleracea has been cultivated since ancient times. Its wild ancestors are believed to have originated in the eastern Mediterranean, with early cultivation records dating back to Greek and Roman periods [
20].
The species has undergone whole-genome duplication events, contributing to its genetic complexity and adaptability. Recent advancements have led to the development of high-quality reference genomes for various
B. oleracea cultivars, including cabbage and broccoli, facilitating comparative genomic studies and providing insights into gene family evolution, structural variations, and metabolic regulation [
21,
22,
23]. These genomic resources have proven invaluable in understanding the domestication processes, morphological diversification, and stress response mechanisms in
B. oleracea, thereby supporting breeding programs aimed at improving crop yield, nutritional quality, and resilience to environmental stresses. In practice, the availability of high-resolution genomic data enables the identification of key loci associated with traits such as disease resistance, flavor enhancement, and texture improvement, thereby facilitating marker-assisted selection and accelerating the development of superior cabbage cultivars.
The genus
Brassica includes economically important horticultural crops [
24], with
Brassica napus being an allopolyploid derived from hybridization between
B. oleracea and
Brassica rapa [
25]. Understanding the evolutionary trajectories and functional diversification of gene families in these species can directly inform breeding strategies for yield stability, nutritional improvement, and stress resilience. Moreover,
Arabidopsis, as the model plant of the family Brassicaceae, offers a well-annotated genomic reference and serves as a valuable experimental control for in silico comparative analyses [
26].
Recently, GeBP genes have been characterized in many species. However, little is known about the evolutionary dynamics of the GeBP family in B. oleracea. In this study, we applied bioinformatic methods to predict and analyze 28 BoGeBP genes in B. oleracea, including phylogenetic analysis, chromosomal localization, homology analysis, tissue expression analysis, gene structure and protein structure analyses, and codon preference analysis. The general objective of this study is to generate fundamental genomic and functional insights into the GeBP transcription factor family in B. oleracea, with specific aims to (i) characterize its evolutionary expansion and duplication mechanisms in relation to B. rapa, B. napus, and Arabidopsis; (ii) predict functional diversification based on structural and regulatory features; and (iii) identify candidate genes with potential applications in molecular breeding for horticultural trait improvement. We provide a reference and theoretical basis for further functional studies of BoGeBP genes.
3. Results
3.1. Identification and Retrieval of GeBP Gene Family Members
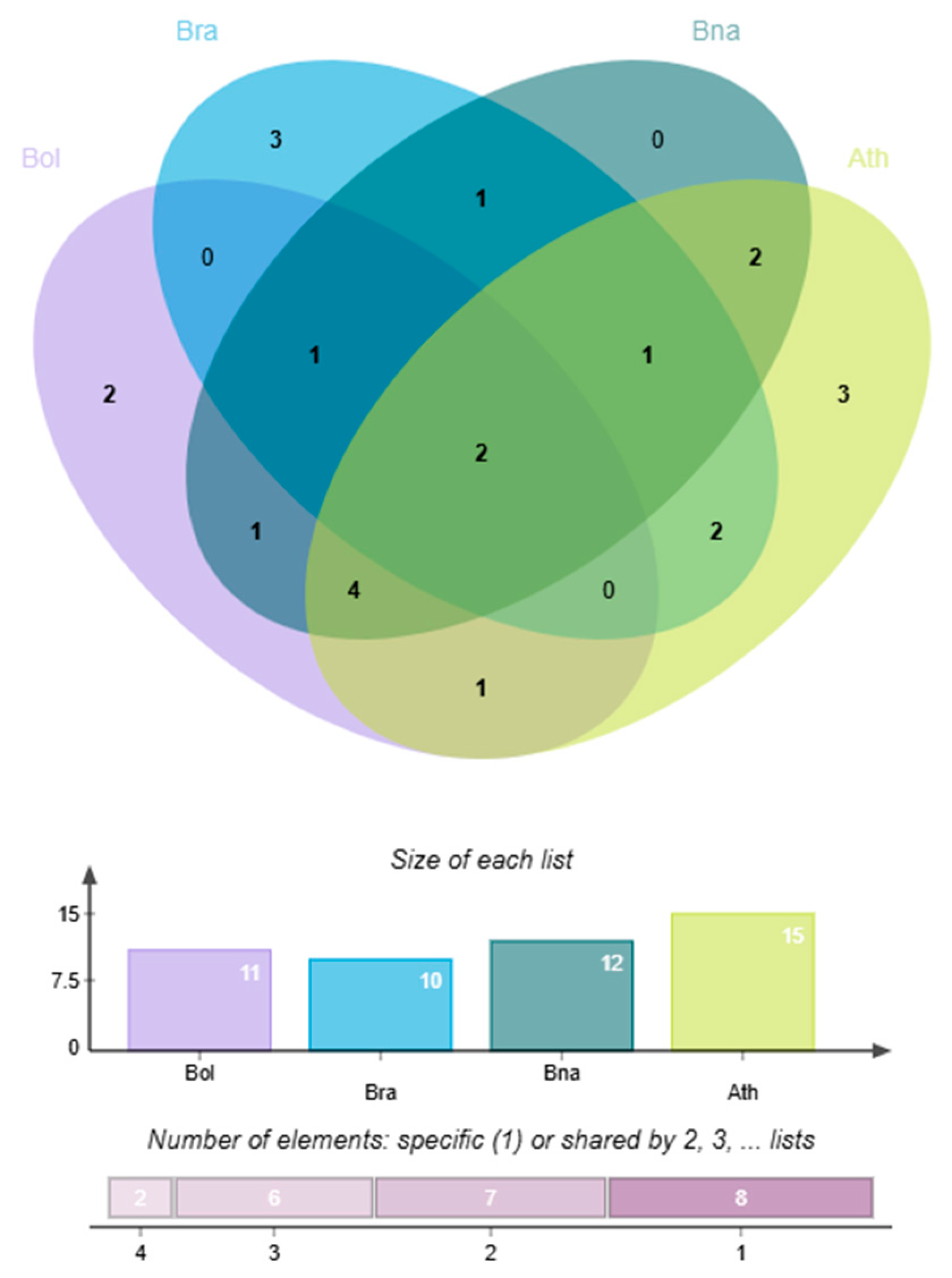
Using the downloaded Hidden Markov Model (HMM) profile of GeBP, HMMER searches were conducted against the B. oleracea genome database. Candidate genes were further validated through domain confirmation using InterPro, SMART, and CDD databases. The intersection of results from these three tools yielded 28 B. oleracea GeBP genes: BolC01g001690.2J, BolC01g019250.2J, BolC01g019630.2J, BolC01g037440.2J, BolC01g055250.2J, BolC03g069820.2J, BolC04g027520.2J, BolC04g041600.2J, BolC04g042880.2J, BolC04g042890.2J, BolC04g042900.2J, BolC04g042910.2J, BolC04g042920.2J, BolC04g042930.2J, BolC04g047580.2J, BolC04g061600.2J, BolC05g009330.2J, BolC05g060710.2J, BolC06g017320.2J, BolC06g018980.2J, BolC06g018990.2J, BolC07g015340.2J, BolC07g039050.2J, BolC07g051920.2J, BolC08g008250.2J, BolC08g055380.2J, BolC09g005140.2J, and BolC09g019010.2J.
Using the same approach, 20
GeBP gene family members were identified in
B. rapa, and 44 were identified in
B. napus (
Table 1).
Generally, most whole-genome replication events include whole-genome duplication (WGD) and whole-genome triplication (WGT), with substantial gene losses also associated with the replication process [
52,
53].
Brassica ancestor species’ genomes experienced WGD events, separating from the
Arabidopsis lineage and then undergoing WGT events specific to the
Brassicaceae lineage, then
B. rapa and
B. oleracea were hybridized to form allotetraploid
B. napus, which also included much genome reorganization and gene loss [
21,
25,
54]. There are 23
GeBP family genes in
Arabidopsis, so, in theory, there should be 69 (23 × 3), 69 (23 × 3), and 138 (23 × 3 × 2)
GeBP genes, respectively, in
B. rapa,
B. oleracea, and
B. napus, but their actual numbers are 20, 28, and 44. Hence, after the WGT event occurred in ancestral
Brassica,
B. rapa lost 49 genes,
B. oleracea lost 41 genes, and
B. napus lost 94 genes in the
GeBP family. And four (20 + 28 − 44) genes of
B. napus were lost after hybridization between
B. rapa and
B. oleracea.
3.2. Multiple Sequence Alignment and Phylogenetic Tree Construction of the GeBP Gene Family
Based on the conserved domain and MEME motif analyses of GeBP family members from
B. oleracea,
B. rapa,
B. napus, and
Arabidopsis, a maximum likelihood (ML) phylogenetic tree was constructed and classified into four groups: Group A (yellow), Group B (blue), Group C (green), and Group D (red) (
Figure 1). Among them,
B. oleracea contains 11 genes in Group A, 4 in Group B, 6 in Group C, and 7 in Group D.
The GeBP genes from the four species were largely clustered together, suggesting a relatively close evolutionary relationship. Notably, the three Brassica species (B. oleracea, B. rapa, and B. napus)—all belonging to the genus Brassica—demonstrated closer phylogenetic relationships compared to Arabidopsis, which exhibited fewer clustering events, reflecting greater divergence from the Brassica lineage.
3.3. Gene Structure and Conserved Motif Analyses of the GeBP Gene Family in B. oleracea
Among the 28
GeBP genes identified in
B. oleracea, none contained UTR regions (
Table 2). The gene with the greatest number of introns and CDSs was
BolC04g061600.2J in Group C. The domain positions within each group were generally consistent, with all genes containing at least motif 10 (blue), motif 2 (orange), and motif 3 (red). In addition to Group D, the other three groups also contained motif 5 (
Figure 2). Most members of Groups A and D included motif 6 (purple), except for
BolC01g001690.2J and
BolC03g069820.2J, which lacked this motif.
3.4. Physicochemical Properties of BoGeBP Gene Family Members
All 28 GeBP proteins in
B. oleracea were predicted to be hydrophilic (
Table 3). Most proteins were localized in the nucleus. Specifically, BolC01g019630.2J (Group A) was located in the plasma membrane, BolC09g005140.2J and BolC01g055250.2J (Group B) in both the chloroplast and nucleus, BolC06g018980.2J (Group C) in the chloroplast and Golgi apparatus, and BolC03g069820.2J (Group D) in both the plasma membrane and nucleus.
The number of amino acids varied greatly among the proteins, ranging from 126 (BolC04g042900.2J) to 881 (BolC01g019630.2J). Similarly, the molecular weight spanned a wide range from 1.7 kDa (BolC04g042890.2J) to 96.1 kDa (BolC01g019630.2J). The isoelectric point (pI) values ranged from 4.45 to 9.63, with most proteins falling within the acidic to neutral range (4.45–7.89), and a few exhibiting basic properties, such as BolC06g018990.2J (pI = 9.63).
Most proteins had instability indices greater than 40, indicating a tendency toward instability in vitro. For example, BolC04g042900.2J (Group D) had an instability index of 79.05, suggesting that it may require rapid degradation or depend on post-translational modifications for functional stability. By contrast, BolC04g041600.2J (Group A) showed a relatively stable profile with an index of 35.4, implying potential for prolonged existence in membrane structures.
The aliphatic index ranged from 48.58 to 92.59, suggesting that most BoGeBP proteins possess favorable thermostability. Notably, BolC03g069820.2J (Group D) had the highest aliphatic index (92.59), indicating potential adaptation to high-temperature environments. All proteins exhibited negative GRAVY (Grand Average of Hydropathy) values (ranging from –1.942 to –0.027), supporting their overall hydrophilic nature.
Additionally, two BoGeBP proteins were identified as transmembrane proteins: BolC01g019630.2J (Group A), with transmembrane regions at positions 413–431, 461–483, 490–512, 522–544, 557–579, 583–605, 657–679, 728–750, 757–779, 799–821, and 828–850; and BolC03g069820.2J (Group D), with transmembrane regions at positions 160–182 and 589–611.
3.5. Chromosomal Distribution of BoGeBP Gene Family Members
The chromosomal locations of
BoGeBP genes were mapped across the
B. oleracea genome (
Figure 3). Chromosome 1 contains five
BoGeBP genes, while chromosome 3 harbors one. Chromosome 4 carries the highest number, with ten
BoGeBP genes, indicating the presence of tandem gene duplications. Chromosomes 5, 8, and 9 each contain two
BoGeBP genes. Chromosomes 6 and 7 contain three
BoGeBP genes each.
3.6. Protein Structure Analysis of BoGeBP Family Members
The secondary structure analysis revealed distinct patterns among the four groups of BoGeBP proteins (
Table 4). In Group A, the average content of α-helices was 44.76%, while random coils accounted for 42.93%, indicating a relatively balanced structure. This balance suggests that these proteins possess both structural stability and potentially functional active regions. Group B proteins exhibited an average α-helix content of 34.15% and a significantly higher proportion of random coils at 57.05%, implying a more flexible and less ordered structure. Group C proteins displayed a notably high α-helix content, averaging 53.05%, with some proteins reaching nearly 60% (e.g., BolC06g018980.2J), suggesting a strong helical character. In Group D, the average α-helix content was 39.01%, with substantial variability ranging from 19.58% (BolC03g069820.2J) to 53.55% (BolC04g042890.2J). The average random coil proportion was 44.47%. Some proteins in this group also showed a relatively high proportion of extended chain structures, such as BolC03g069820.2J (27.08%), which may indicate specialized functions.
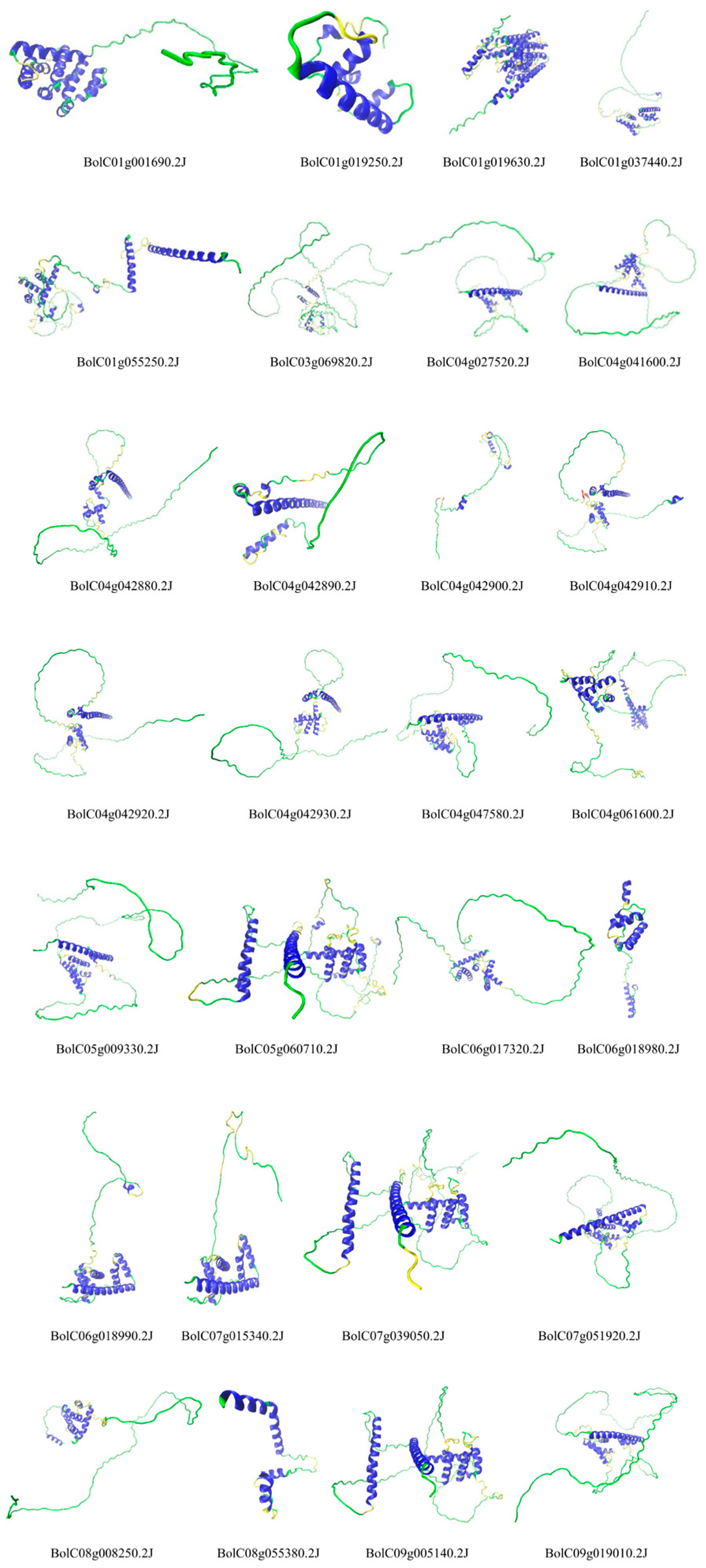
The tertiary structure analysis revealed substantial diversity across the entire gene family, while members within the same group exhibited relatively similar structures (
Figure 4). For example, Group A members, including BolC04g041600.2J, BolC07g051920.2J, BolC04g027520.2J, BolC09g019010.2J, BolC05g009330.2J, and BolC04g047580.2J, showed structural resemblance. Similar intra-group structural consistency was observed in Group B (e.g., BolC09g005140.2J, BolC07g039050.2J, BolC05g060710.2J), Group C (e.g., BolC06g018990.2J, and BolC07g015340.2J), and Group D (e.g., BolC04g042880.2J, BolC04g042930.2J, BolC04g042920.2J, and BolC04g042910.2J). Proteins with similar tertiary structures may possess analogous functions and possibly share closer evolutionary relationships.
3.7. Cis-Acting Regulatory Elements in the Promoter Regions of BoGeBP Genes
Among the
cis-acting regulatory elements identified in the promoter regions of the 28
BoGeBP genes, light-responsive elements accounted for the highest proportion (46%), followed by hormone-responsive elements (25%), stress-responsive elements (21%), and other (8%) (
Figure 5).
This result suggests that most BoGeBP genes are potentially regulated by light signals, indicating that GeBP genes may be involved not only in stress responses but also in photomorphogenesis or light signal transduction pathways.
The substantial presence of hormone-responsive elements—such as ABRE (involved in abscisic acid response) and CGTCA-motif (related to methyl jasmonate response)—suggests that BoGeBP genes may play important roles in hormone-mediated stress responses, highlighting their potential for improving plant stress resistance through molecular breeding.
Among all genes, the following three had the highest number of cis-elements: BolC06g017320.2J (24 elements), which exhibited an even distribution among light-, stress-, and hormone-responsive elements, indicating its diverse regulatory functions in plant development and stress response; BolC07g051920.2J (21 elements), which predominantly contained light-responsive elements (13 in total), including 4 G-box and 3 GT1 motifs, suggesting a strong role in light signaling; and BolC08g055380.2J (20 elements), which contained 8 light-responsive and 7 hormone-responsive elements, further supporting its involvement in light and hormonal regulatory pathways.
3.8. Protein–Protein Interaction Network Prediction of BoGeBP Family Members
Except for BolC03g069820.2J, all members of the BoGeBP family exhibit complex predicted protein–protein interaction networks (
Figure 6). Although most proteins, such as BolC01g001690.2J and BolC04g042900.2J, were annotated as “uncharacterized proteins,” the functions of their interacting partners provided crucial clues. For instance, BolC01g001690.2J interacts with members of the ATG9 family (e.g., A0A0D3BT90 and A0A0D3C3E1), suggesting its potential involvement in autophagosome formation or cytoplasm-to-vacuole targeting (Cvt) processes. This implies that this gene cluster may regulate the assembly of autophagy-related membrane structures in response to nutrient deprivation or pathogen invasion.
BolC05g060710.2J, annotated as a PALP domain-containing protein, interacts with numerous thioredoxin domain-containing proteins (e.g., A0A0D3A515 and A0A0D3BVL7), indicating a possible role in redox homeostasis or sulfur metabolism regulation.
BolC04g047580.2J and BolC04g042900.2J interact with A0A0D3C304, a phytocyanin domain-containing protein, suggesting that the BoGeBP family plays an important role in the function of this protein, which may act as a signal transduction component or a metal ion-binding protein involved in intercellular communication or copper/iron homeostasis. Proteins such as BolC04g042880.2J, BolC04g042890.2J, and BolC04g042900.2J interact with A0A0D3E8D6, which contains an ERCC4 domain frequently associated with DNA repair, indicating a potential core role for the family in maintaining genome stability. BolC07g039050.2J and BolC09g005140.2J interact with bifunctional dihydrofolate reductase-thymidylate synthase proteins (e.g., A0A0D3A1Z0 and A0A0D3B7L5), which are directly involved in dTMP synthesis and folate metabolism, possibly influencing DNA replication and repair efficiency. BolC01g019630.2J, annotated as an MFS transporter protein, interacts with pectinesterase and peroxidase proteins, potentially coordinating cell wall softening (via pectin degradation) and reactive oxygen species scavenging, thus contributing to pathogen defense or developmental regulation.
3.9. GO and KEGG Analyses of BoGeBP Family Members
Gene Ontology (GO) functional annotation was performed for the
BoGeBP family, and 28
BoGeBP genes were assigned a total of 133 GO terms (
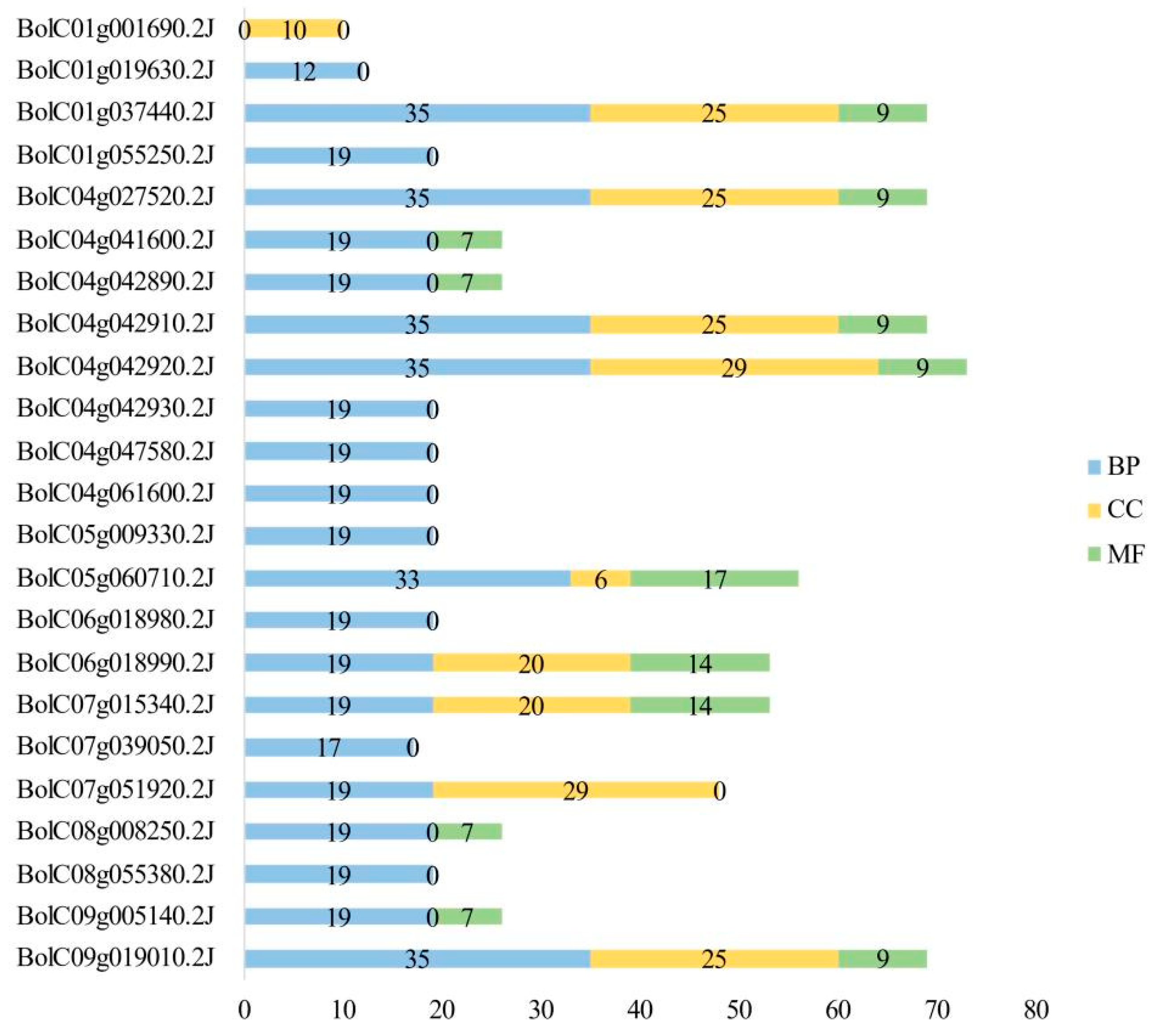
Table S1). These GO terms were classified into three main categories: molecular function (MF), cellular component (CC), and biological process (BP).
Among the 133 GO terms, 77 were associated with BP. Of these, 17 terms were enriched in 19 BoGeBP members, while 41 terms were enriched in only one member. For the CC category, 29 GO terms were identified, including 5 terms enriched in 10 members, 5 in 9 members, and 10 in 8 members. In the MF category, 27 GO terms were annotated, with 4 terms enriched in 12 members and 13 terms enriched in only one member.
A statistical analysis of the number of GO terms assigned to the 28
BoGeBP members (
Figure 7) showed that GO terms related to biological processes were the most abundant, with a total of 503 annotations. The number of BP-related terms per gene ranged from 0 to 35. GO terms under the cellular component category totaled 214, with most genes associated with approximately 25 terms. GO terms under molecular function were the least numerous, with only 118 annotations, and most genes were associated with around 9 terms.
These findings suggest that BoGeBP family members are extensively involved in various biological processes in B. oleracea, while also playing important roles in cellular structure and molecular function.
According to the analysis of
Table 5 and
Table 6,
BolC05g060710.2J was annotated with K01738, which encodes cysteine synthase (EC 2.5.1.47). This enzyme catalyzes a key step in cysteine biosynthesis by combining O-acetylserine and sulfide to form cysteine. It is directly involved in both cysteine and methionine metabolism (map00270) and sulfur metabolism (map00920)
, acting as a core enzyme in the incorporation of sulfur into amino acids. Moreover, it is also associated with carbon metabolism (map01200)
, biosynthesis of amino acids (map01230), and biosynthesis of secondary metabolites (map01110), indicating its multifunctional role in primary metabolism.
BolC07g039050.2J was enriched in K12951 [cobalt/nickel transporting P-type ATPase (EC 7.2.2.-)] and K15441 [tRNA-specific adenosine deaminase 2 (EC 3.5.4.-)], suggesting its potential roles in transmembrane transport of heavy metal ions and in tRNA editing, which may contribute to the regulation of translation fidelity.
BolC01g019630.2J was annotated with K13783, a member of the major facilitator superfamily (MFS), predicted to function as a glycerol-3-phosphate transporter. This implies its possible involvement in energy metabolism (e.g., the glycerol-3-phosphate shuttle) or lipid biosynthesis, thereby influencing cellular energy homeostasis.
3.10. Codon Usage Bias Analysis of the GeBP Gene Family in B. oleracea
The Nc (effective number of codons) values of the
BoGeBP genes ranged from 47.84 to 59.89, with an average of 53.6282. The CAI (codon adaptation index) values ranged from 0.166 to 0.301, with an average of 0.2318. The CBI (codon bias index) values ranged from −0.158 to 0.052, with a mean of −0.0432 (
Table 7). For
BrGeBP genes, the Nc values ranged from 47.74 to 57.67 (mean: 52.289), the CAI values ranged from 0.132 to 0.287 (mean: 0.2330), and the CBI values ranged from −0.188 to 0.053 (mean: −0.0450). For
BnGeBP genes, the Nc values ranged from 37.25 to 58.36 (mean: 52.5648), the CAI values ranged from 0.168 to 0.298 (mean: 0.2284), and the CBI values ranged from −0.249 to 0.067 (mean: −0.0440). For
AtGeBP, the Nc values ranged from 40.95 to 61.00, with an average of 50.2983; the CAI values ranged from 0.189 to 0.301 (mean: 0.2393), and the CBI values ranged from −0.229 to 0.081 (mean: −0.0641). These results indicate that
GeBP genes in all studied species exhibit relatively weak codon usage bias.
According to
Figure 8 and
Table 8,
BoGeBP genes possessed 11 optimal codons, of which 8 ended with A/U(T) and 3 with G/C. The high proportion of optimal codons ending in A/U suggests that members of the gene family may tend to be expressed under conditions of low translational efficiency or in stress-specific contexts.
BrGeBP genes had 10 optimal codons (6 ending with A/U and 4 with G/C), while
BnGeBP had 12 optimal codons (8 ending with A/U and 4 with G/C).
AtGeBP genes had the highest number of optimal codons, totaling 15, with 13 ending in A/U and only 2 in G/C. These findings suggest that the
GeBP gene family across all studied species preferentially uses codons ending in A or U. Two optimal codons, UUG and UCU, were found to be conserved across all four species, while UGU was conserved among species of the genus
Brassica.
3.11. Tissue-Specific Expression Analysis of BoGeBP Family Members
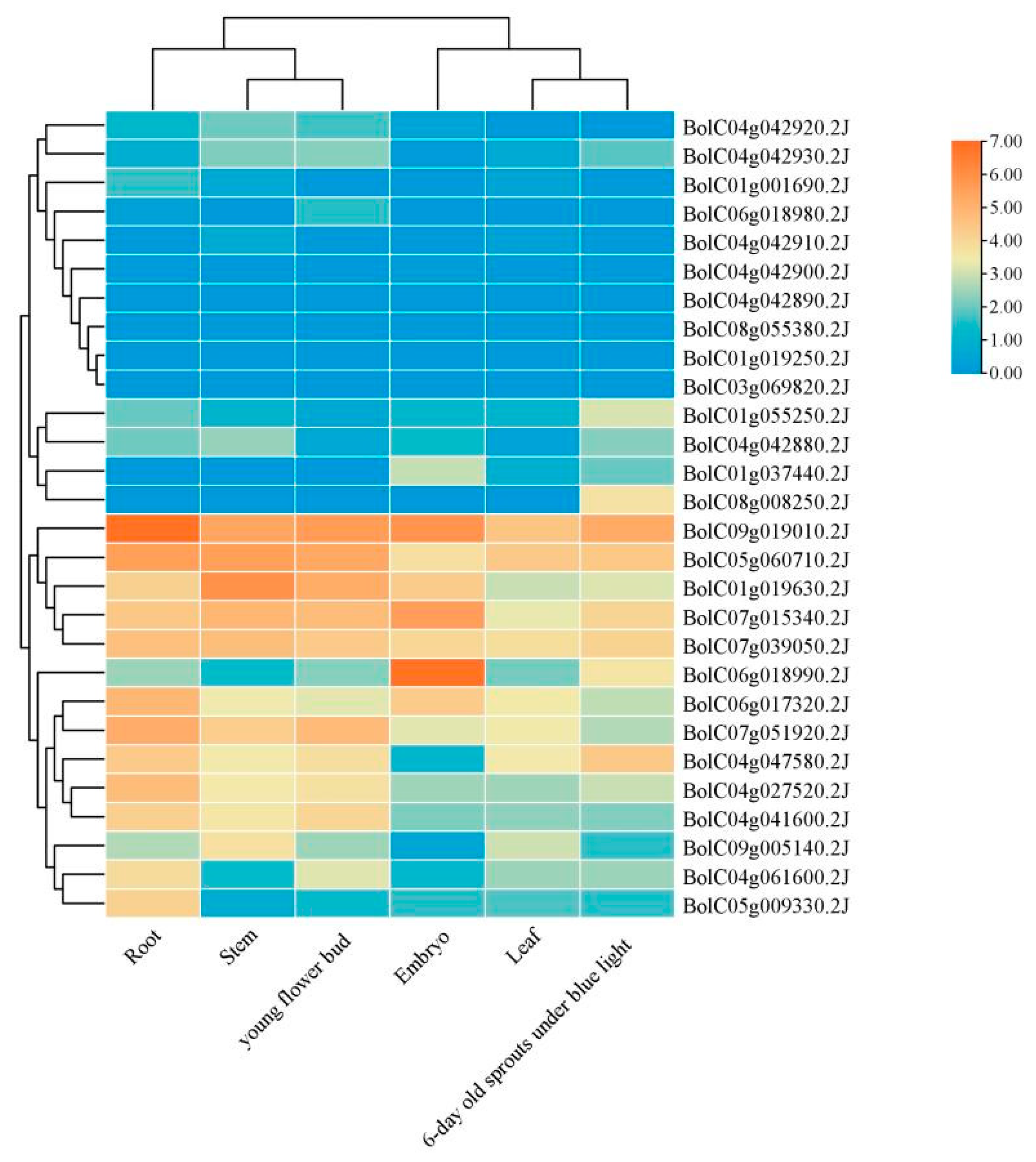
BoGeBP genes exhibited tissue-specific expression patterns (
Figure 9). The highest expression levels were observed in roots, followed by stems and floral buds. Among all members,
BolC09g019010.2J showed the highest overall expression, indicating its potential key role in specific tissue functions. Notably,
BolC08g055380.2J showed no detectable expression in any of the three tested tissues, suggesting it may be a pseudogene or only expressed under very specific circumstances not included in this study.
3.12. Synteny Analysis of GeBP Gene Family Among Cabbage and Other Species
To investigate the evolutionary relationships of the
GeBP gene family, a synteny analysis was conducted among
B. oleracea (
BoGeBP),
Arabidopsis (
AtGeBP),
B. rapa (
BrGeBP), and
B. napus (
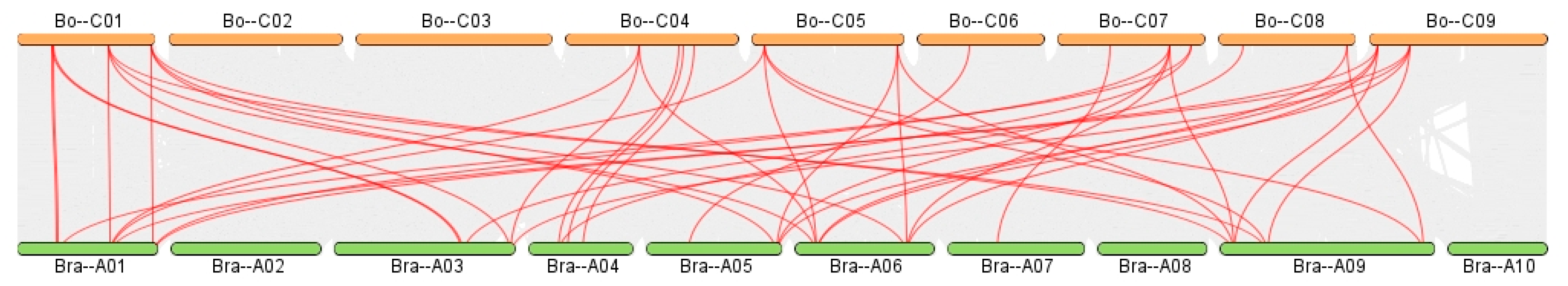
BnGeBP). A total of 19 syntenic gene pairs were identified between
BoGeBP and
AtGeBP (
Figure 10), 45 pairs between
BoGeBP and
BrGeBP (
Figure 11), and up to 72 pairs between
BoGeBP and
BnGeBP (
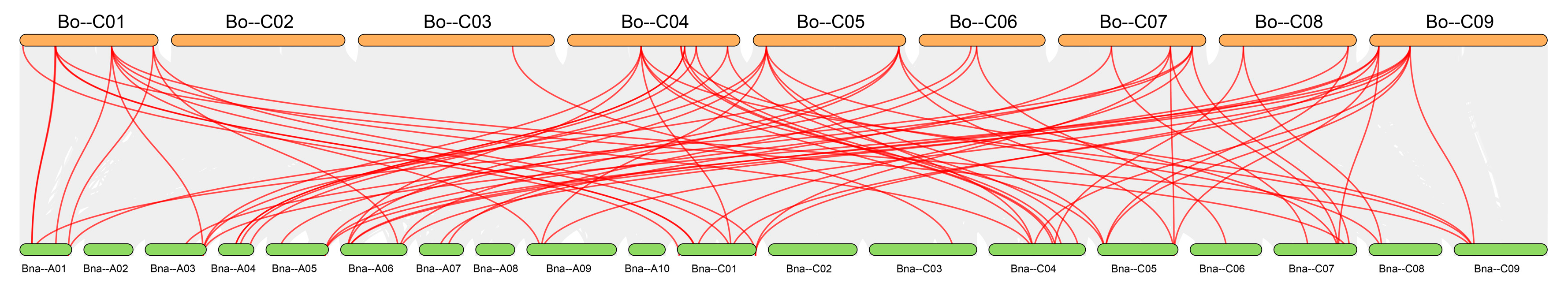
Figure 12), revealing distinct differences in the degree of genomic collinearity among species.
The degree of synteny was positively correlated with phylogenetic proximity: the closer the evolutionary relationship between species, the greater the number of conserved syntenic gene pairs. This pattern arises because more recently diverged species have experienced fewer chromosomal rearrangements, gene losses, and sequence divergences, so that ancestral genomic blocks remain more intact; furthermore, genes with essential or conserved functions are often subject to purifying selection, which helps to maintain their genomic context and reinforces synteny among close relatives. The highest number of collinear pairs was found between
BoGeBP and
BnGeBP (72 pairs), consistent with their close taxonomic relationship—
B. napus is an allotetraploid derived from hybridization between
B. oleracea and
B. rapa [
25]. This result not only reflects their shared evolutionary history but also suggests that a large number of conserved genome duplication blocks in the
GeBP gene family have been retained between these two species.
Although the number of syntenic pairs between BoGeBP and BrGeBP (45 pairs) is fewer than with BnGeBP, it is still considerably higher than that with AtGeBP (19 pairs), which aligns with the fact that both BoGeBP and BrGeB are diploid members of the genus Brassica. By contrast, the lowest number of syntenic gene pairs between BoGeBP and AtGeBP reflects their greater evolutionary divergence and lower conservation within the GeBP gene family.
3.13. Analysis of Gene Duplication Types in the GeBP Gene Family
The
BoGeBP genes predominantly originated from tandem duplications (32%) and whole-genome duplications (WGD; 57%), suggesting that local tandem duplication may have rapidly generated functionally redundant genes, which, in combination with WGD, contributed to enhanced adaptability. In
BrGeBP genes, dispersed duplication accounted for the majority (55%), along with a moderate proportion of WGD duplication (30%). This pattern implies that transposition or chromosomal rearrangement may play a role in dynamically modulating gene function. For
BnGeBP genes, WGD duplication contributed to 82% of the family members, consistent with the allotetraploid origin of
B. napus, indicating that large-scale genome duplication events, such as hybridization, are the primary drivers of gene family expansion (
Table 9). By contrast,
AtGeBP genes were primarily derived from dispersed duplication (57%) and lacked both tandem and proximal duplication types, suggesting that the flexibility of the
Arabidopsis genome may facilitate rapid adaptation through dispersed duplication events.
4. Discussion
4.1. Expansion Mechanisms and Structural Conservation of the GeBP Gene Family
In this study, 28
GeBP genes were identified in
B. oleracea, a number intermediate between the diploid
B. rapa (20 genes) and the allotetraploid
B. napus (44 genes), and significantly higher than that in the model plant
Arabidopsis (23 genes) [
21]. This pattern reflects the pivotal roles of polyploidization and gene duplication in the expansion of the
GeBP family, consistent with previous findings that whole-genome duplication events underpin transcription factor family proliferation in
Brassica and other plant lineages [
25,
55]. Phylogenetic analysis classified all GeBP proteins into four subgroups (A–D), with members from different
Brassica species clustering tightly within each subgroup, suggesting a conserved and lineage-shared expansion during species evolution. Conserved domain analysis further revealed that all
B. oleracea members retained motifs 2, 3, and 10 across subgroups, indicating strong structural conservation related to DNA binding and transcriptional regulation, likely maintained by purifying selection following duplication [
56,
57]. Chromosomal mapping showed that
BoGeBP genes are unevenly distributed across the
B. oleracea genome, with chromosome 4 harboring ten genes, many of which form tandem duplications. Duplication type analysis indicated that 57% of
BoGeBP genes originated from whole-genome or segmental duplication (WGD) and 32% from tandem duplication, implying that multiple duplication mechanisms have collectively driven the family expansion. The preferential retention of WGD-derived genes may preserve dosage-sensitive core functions in polyploid genomes, as predicted by the gene balance hypothesis and observed in other plant systems [
58]. By contrast, tandem duplications—frequently implicated in forming localized resistance and stress-responsive gene clusters—provide raw material for rapid environmental adaptation [
59].
By comparison,
GeBP genes in
Arabidopsis and
B. rapa are mainly derived from dispersed duplication, which is often associated with transposable element-mediated relocation and the acquisition of novel regulatory elements, thereby facilitating functional innovation and tissue-specific expression divergence [
60].
In summary, the expansion of the GeBP gene family in B. oleracea reflects both the conserved legacy of polyploidy and the adaptive contributions of tandem duplication. These divergent duplication patterns underscore distinct genome evolutionary strategies between the genera Brassica and Arabidopsis and provide a foundation for leveraging specific GeBP members in breeding programs to enhance stress resilience and agronomic performance in B. oleracea.
4.2. Structural Diversity and Cis-Regulatory Characteristics Reveal the Potential Functional Divergence and Applied Value of BoGeBP Genes
BoGeBP proteins are generally hydrophilic and predominantly localized in the nucleus, consistent with typical features of transcription factors [
61]. However, some members, such as BolC01g019630.2J (Group A) and BolC03g069820.2J (Group D), possess not only predicted transmembrane helical regions but also structural domains associated with transporters like MFS or P-ATPase, suggesting their potential dual role in signal sensing and transcriptional regulation. This type of “membrane–nucleus dual function” is rare among known GeBP proteins [
10] and holds significant research and application potential.
From a structural perspective, BoGeBP members exhibit notable differences in their secondary structure composition. Proteins in Clade C have the highest α-helix content (averaging 53%), suggesting strong structural stability and possible roles in sustained signal transduction or structural support [
62]. Clade B is characterized by a high proportion of random coils (average 57%), indicating greater flexibility and dynamic regulatory potential, possibly contributing to rapid responses to environmental stress [
63]. Clade D shows the highest structural variability, hinting at a trend toward functional diversification. Additionally, variations in protein instability index and aliphatic index across groups imply differing physiological roles in heat adaptation and protein stability.
Promoter cis-acting element analysis further supported these functional inferences. Among all identified cis-elements, 46% are associated with light response, 25% with plant hormone responses (such as ABA and MeJA), and 21% with abiotic stresses, indicating that BoGeBP genes are widely involved in crosstalk among light, hormonal, and stress signaling pathways. BolC06g017320.2J, enriched with all three types of regulatory elements and highly expressed across multiple tissues, is presumed to be a core integrator of multiple signals. BolC07g051920.2J is highly expressed in leaf tissue, its promoter contains abundant light-responsive elements, and it interacts with several photosynthesis-related proteins, suggesting a potential role in photosynthetic regulation and photomorphogenesis.
By integrating structural features, expression patterns, and functional annotations, this study identifies several key candidate genes with potential biological significance. BolC05g060710.2J is a well-characterized cysteine synthase gene involved in sulfur metabolism and amino acid biosynthesis. It is significantly upregulated under drought stress [
64] and may play a central role in antioxidant responses and the synthesis of sulfur-containing secondary metabolites (e.g., glucosinolates) [
65], making it a promising candidate for stress resistance regulation. BolC01g019630.2J is a large MFS-type transporter potentially associated with energy metabolism, cell wall remodeling, and signal transduction. It is well suited for functional validation under pathogen stress or salt/drought conditions (
Table 5) [
66]. BolC03g069820.2J is a protein enriched in β-sheet structures and has a high aliphatic index, suggesting strong thermostability and membrane localization. It is speculated to play a unique role in responses to heat or heavy metal stress.
In summary, the BoGeBP gene family exhibits a unique combination of structural conservation and diversity, and demonstrates high potential for functional differentiation in terms of cis-regulation, subcellular localization, and biological function. These insights lay the groundwork for exploring their roles in light response, hormone signaling, and stress adaptation and identifies key genes that may serve as valuable genetic resources for the breeding of stress-tolerant, nutrient-rich, or pest-resistant cabbage cultivars.
4.3. Future Perspectives
By integrating synteny and gene duplication pattern analyses, this study reveals that the expansion strategies of the
GeBP gene family differ significantly among
Brassica species. In future research, the following approaches are recommended to further elucidate the biological roles and regulatory mechanisms of
GeBP genes. CRISPR/Cas9-mediated functional validation: this genome-editing approach enables precise dissection of the roles of key
GeBP genes in growth, development, and stress responses, providing direct causal evidence for gene function. Its advantages include high specificity and the ability to generate targeted knockouts or allelic variants; however, challenges may arise from genetic redundancy within the family, potential off-target effects, and genotype-dependent transformation efficiency in
B. oleracea [
67]. Integration of protein interactomics and metabolomics: this combined strategy can reveal the protein partners and downstream metabolic pathways of GeBP proteins, particularly those with predicted dual functions in signaling and transcriptional regulation. The main advantage lies in capturing the multi-layered regulatory network; however, the challenge is that data integration is computationally intensive, and protein detection may be hindered by low abundance or transient interactions [
68]. Spatiotemporal transcriptome analysis under environmental gradients: this approach provides high-resolution insight into the dynamic expression patterns of
GeBP genes across tissues, developmental stages, and diverse abiotic stress or hormonal conditions. It offers a comprehensive view of regulatory plasticity, but the large-scale datasets generated require robust statistical models, and distinguishing primary from secondary stress responses remains a methodological hurdle [
69].
Collectively, these strategies will systematically clarify the synergistic roles of GeBP members in light, hormone, and stress signaling networks, while also highlighting their functional diversity. The anticipated advantages of these methods can accelerate functional characterization and breeding application, although the outlined challenges should be addressed to ensure reproducibility and translational impact. Ultimately, this integrated framework will provide valuable insights and genetic resources for molecular design breeding of cabbage and other Brassica vegetables.
4.4. Limitations and Significance of This Study
This study systematically analyzed the evolutionary characteristics, structural features, and expression patterns of the GeBP transcription factor family in B. oleracea, identifying several key candidate genes with potential roles in stress resistance and regulatory functions. However, there are some limitations to this study. Firstly, all conclusions were drawn based on public databases and bioinformatic analyses, lacking experimental validation such as confirmation of transcription factor binding sites or functional assays through gene knockout/overexpression. Secondly, the expression data were primarily derived from reference genomes and a limited number of samples, resulting in constraints regarding environmental and varietal diversity. Additionally, the accuracy of protein interaction predictions, functional annotations, and GO/KEGG enrichment analyses is influenced by algorithmic limitations and the completeness of current database annotations.
Despite these constraints, the results reveal that BoGeBP genes exhibit both structural conservation and regulatory divergence, suggesting their roles in coordinating light, hormone, and abiotic stress responses. From a horticultural perspective, the identification of lineage-specific expansion patterns and stress-responsive cis-regulatory modules offers practical avenues for crop improvement. WGD-derived genes may function as stable regulators of core metabolic processes, while tandem duplications enriched in stress-related elements could mediate rapid responses to environmental stimuli. Notably, key candidates such as BolC05g060710.2J, BolC01g019630.2J, and BolC06g017320.2J hold promise for molecular breeding through marker-assisted selection or gene editing, especially in the development of cabbage cultivars with enhanced resilience, nutritional quality, and environmental adaptability. This research therefore contributes valuable knowledge for advancing sustainable and resilient horticultural production systems and highlights the importance of integrating bioinformatics with applied breeding strategies in Brassica crops.
5. Conclusions
This study provides new insights into the evolutionary dynamics and regulatory complexity of the GeBP transcription factor family in B. oleracea. By resolving lineage-specific expansion patterns and uncovering structural divergence—particularly the unexpected identification of members (e.g., BolC01g019630.2J) that potentially bridge membrane perception and nuclear transcription—the work refines our understanding of how transcription factor families diversify and integrate environmental signals in polyploid crop genomes. The characterization of promoter architectures that coordinate light, hormone, and stress responsiveness further illuminates the multilayered regulatory roles of GeBP proteins and suggests how signal convergence is achieved at the cis-regulatory level.
Beyond descriptive cataloguing, the study advances the field by prioritizing concrete candidate genes (BolC05g060710.2J, BolC01g019630.2J, and BolC06g017320.2J) with putative central roles in metabolism, signaling, and stress integration, thus providing a rational basis for downstream functional genomics and metabolic engineering. The integrative framework—combining synteny, structural motif conservation, expression specificity, and cis-element profiling—serves as a transferable template for dissecting other transcription factor families in complex plant genomes.
Ultimately, these findings contribute both to basic evolutionary biology (by clarifying duplication-driven innovation and conservation in a recently diversified gene family) and applied crop improvement, offering prioritized molecular targets and mechanistic hypotheses to accelerate the breeding of cabbage and related Brassica vegetables with enhanced stress resilience and trait optimization.