Exploring Factors Influencing the Consumption of Grape Skins: A Review
Abstract
1. Introduction
2. Development and Skin Characteristics of Grapes
2.1. Growth and Development of Grapes
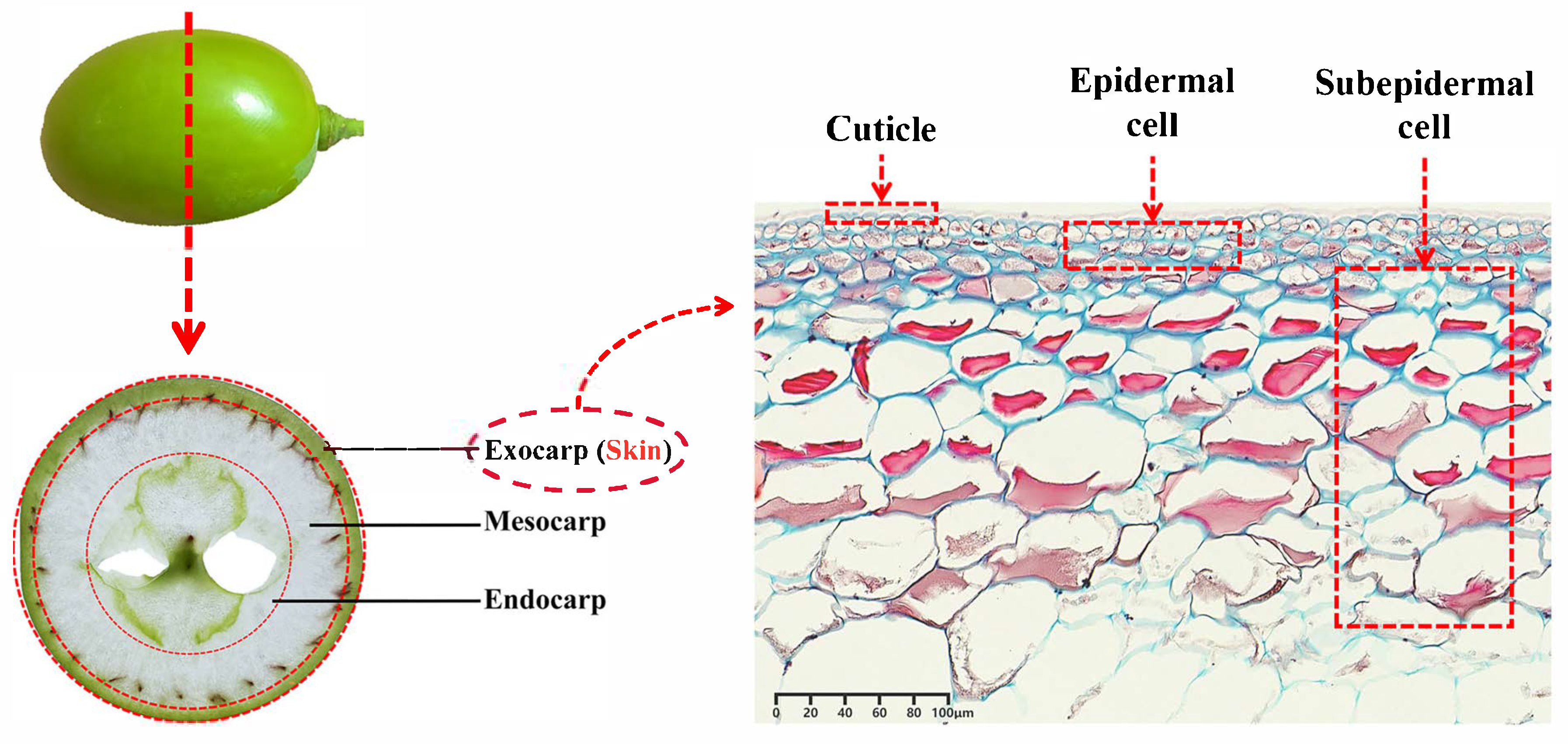
2.2. Structure of Grape Skin
2.3. Sensory Characteristics of Grape Skin
3. Influencing Factors and Molecular Regulation of Skin Sensory Characteristics
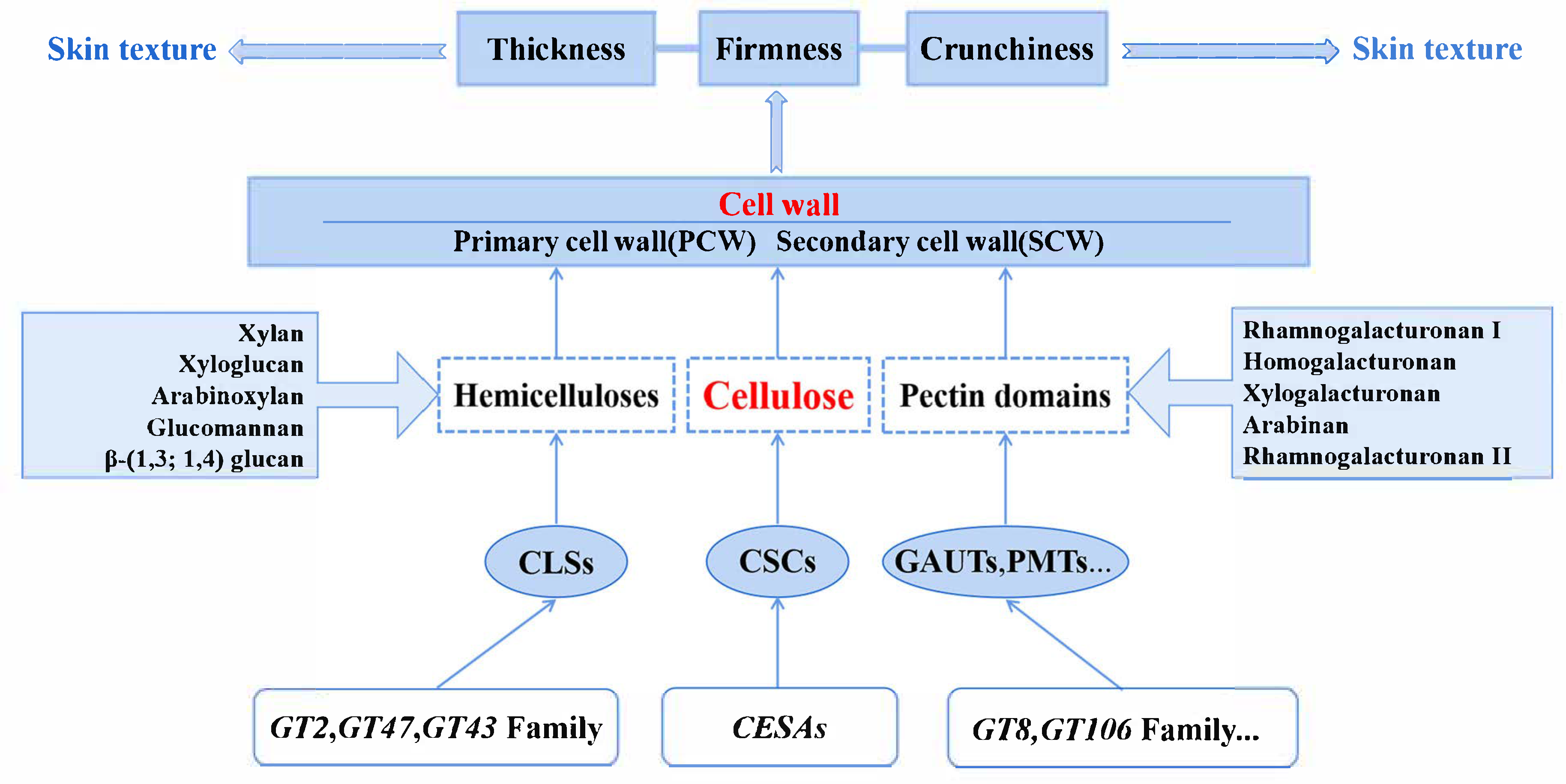
3.1. Grape Skin Texture
3.1.1. Composition of Grape Skin Cell Wall
3.1.2. Transcription Factors Involved in the Regulation of Cell Wall Material Content
3.1.3. Possible Ways to Change the Texture of Skin
3.2. Astringency of Grape Skin
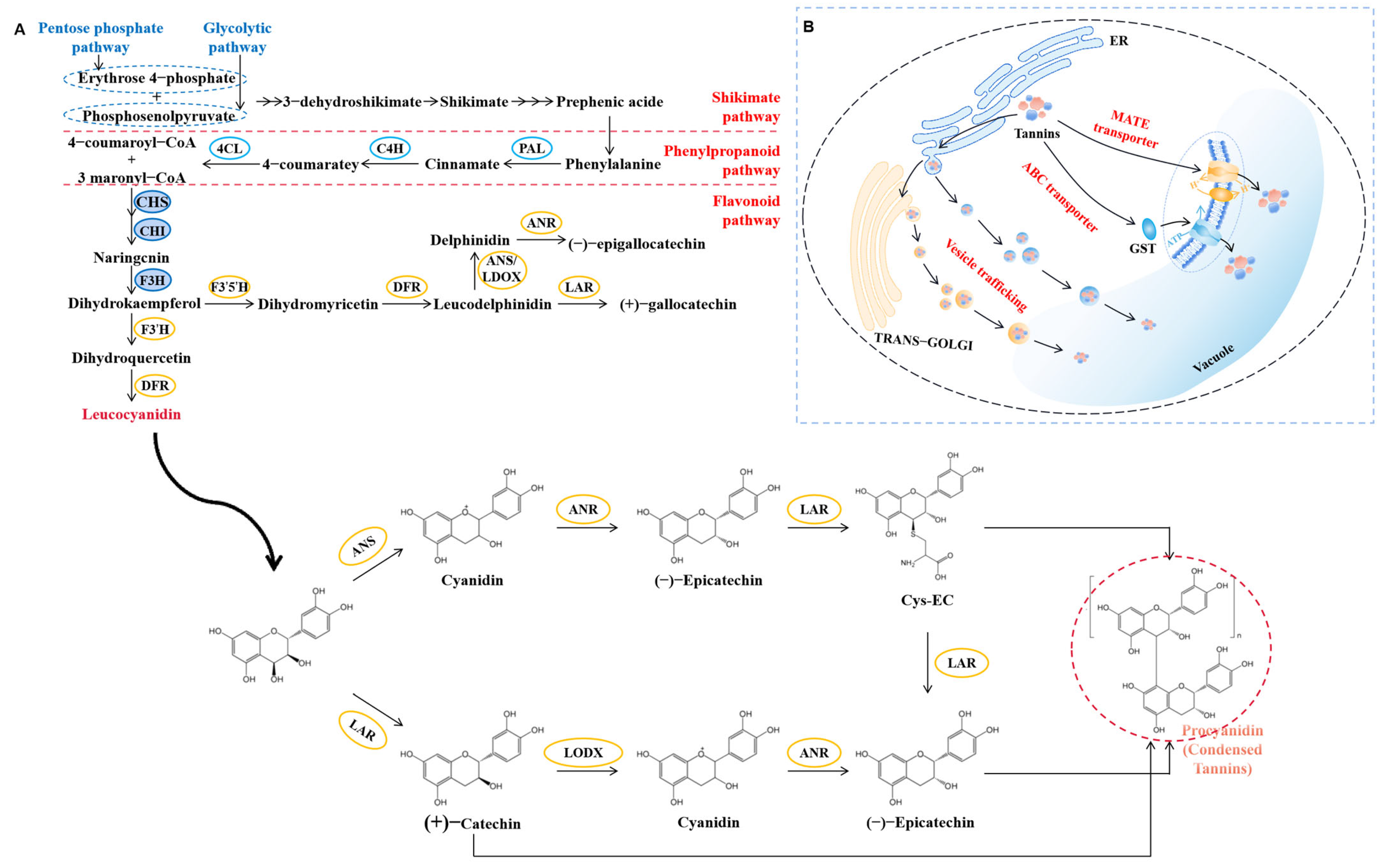
3.2.1. Composition of Tannins
3.2.2. The Biosynthetic Pathway of Tannins
3.2.3. Transportation Pathways of Tannins
3.2.4. Possible Ways to Remove Astringency
3.3. Other Factors
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Atak, A. Vitis species for stress tolerance/resistance. Genet. Resour. Crop Evol. 2025, 72, 2425–2444. [Google Scholar] [CrossRef]
- Wu, Y.; Duan, S.; Zhao, L.; Gao, Z.; Luo, M.; Song, S.; Xu, W.; Zhang, C.; Ma, C.; Wang, S. Aroma characterization based on aromatic series analysis in table grapes. Sci. Rep. 2016, 6, 31116. [Google Scholar] [CrossRef] [PubMed]
- Sabra, A.; Netticadan, T.; Wijekoon, C. Grape bioactive molecules, and the potential health benefits in reducing the risk of heart diseases. Food Chem. X 2021, 12, 100149. [Google Scholar] [CrossRef]
- Xia, L.; Xu, C.; Huang, K.; Lu, J.; Zhang, Y. Evaluation of phenolic compounds, antioxidant and antiproliferative activities of 31 grape cultivars with different genotypes. J. Food Biochem. 2019, 43, e12626. [Google Scholar] [CrossRef]
- Nassiri-Asl, M.; Hosseinzadeh, H. Review of the Pharmacological Effects of Vitis vinifera (Grape) and its Bioactive Constituents: An Update. Phytother. Res. 2016, 30, 1392–1403. [Google Scholar] [CrossRef] [PubMed]
- Tabeshpour, J.; Mehri, S.; Shaebani Behbahani, F.; Hosseinzadeh, H. Protective effects of Vitis vinifera (grapes) and one of its biologically active constituents, resveratrol, against natural and chemical toxicities: A comprehensive review. Phytother. Res. 2018, 32, 2164–2190. [Google Scholar] [CrossRef]
- Lluís, L.; Muñoz, M.; Nogués, M.R.; Sánchez-Martos, V.; Romeu, M.; Giralt, M.; Valls, J.; Solà, R. Toxicology evaluation of a procyanidin-rich extract from grape skins and seeds. Food Chem. Toxicol. 2011, 49, 1450–1454. [Google Scholar] [CrossRef] [PubMed]
- Chengolova, Z.; Ivanov, Y.; Godjevargova, T. Comparison of Identification and Quantification of Polyphenolic Compounds in Skins and Seeds of Four Grape Varieties. Molecules 2023, 28, 4061. [Google Scholar] [CrossRef]
- Atak, A.; Göksel, Z.; Yılmaz, Y. Changes in Major Phenolic Compounds of Seeds, Skins, and Pulps from Various Vitis spp. and the Effect of Powdery and Downy Mildew Diseases on Their Levels in Grape Leaves. Plants 2021, 10, 2554. [Google Scholar] [CrossRef]
- Yilmaz, Y.; Göksel, Z.; Erdoğan, S.S.; Öztürk, A.; Atak, A.; Özer, C. Antioxidant Activity and Phenolic Content of Seed, Skin and Pulp Parts of 22 Grape (Vitis vinifera L.) Cultivars (4 Common and 18 Registered or Candidate for Registration). J. Food Process. Preserv. 2015, 39, 1682–1691. [Google Scholar] [CrossRef]
- Mochida, K.; Maki, S.; Onishi, A.; Uchida, Y.; Kurahashi, T. Effects of CPPU Treatment Methods on the Fruit Quality of ‘Shine Muscat’ Grape. Hortic. Res. 2013, 12, 155–163. [Google Scholar] [CrossRef][Green Version]
- Wei, X.; Ju, Y.; Ma, T.; Zhang, J.; Fang, Y.; Sun, X. New perspectives on the biosynthesis, transportation, astringency perception and detection methods of grape proanthocyanidins. Crit. Rev. Food Sci. Nutr. 2021, 61, 2372–2398. [Google Scholar] [CrossRef]
- Cliff, M.A.; Dever, M.C.; Reynolds, A.G. Descriptive Profiling of New and Commercial British Columbia Table Grape Cultivars. Am. J. Enol. Vitic. 1996, 47, 301. [Google Scholar] [CrossRef]
- Yu, J.; Wang, R.; Ma, W.; Lei, S.; Zhu, M.; Yang, G. Pectate Lyase Gene VvPL1 Plays a Role in Fruit Cracking of Table Grapes. J. Agric. Food Chem. 2023, 71, 1643–1654. [Google Scholar] [CrossRef] [PubMed]
- Schlosser, J.; Olsson, N.; Weis, M.; Reid, K.; Peng, F.; Lund, S.; Bowen, P. Cellular expansion and gene expression in the developing grape (Vitis vinifera L.). Protoplasma 2008, 232, 255–265. [Google Scholar] [CrossRef]
- Malacarne, G.; Lagreze, J.; Rojas San Martin, B.; Malnoy, M.; Moretto, M.; Moser, C.; Dalla Costa, L. Insights into the cell-wall dynamics in grapevine berries during ripening and in response to biotic and abiotic stresses. Plant Mol. Biol. 2024, 114, 38. [Google Scholar] [CrossRef] [PubMed]
- Nakagawa, S.; Nanjo, Y. A Morphological Study of Delaware Grape Berries. J. Jpn. Soc. Hortic. Sci. 1965, 34, 85–95. [Google Scholar] [CrossRef]
- Agudelo-Romero, P.; Erban, A.; Rego, C.; Carbonell-Bejerano, P.; Nascimento, T.; Sousa, L.; Martínez-Zapater, J.M.; Kopka, J.; Fortes, A.M. Transcriptome and metabolome reprogramming in Vitis vinifera cv. Trincadeira berries upon infection with Botrytis cinerea. J. Exp. Bot. 2015, 66, 1769–1785. [Google Scholar] [CrossRef] [PubMed]
- Hardie, W.J.; O’Brien, T.P.; Jaudzems, V.G. Morphology, anatomy and development of the pericarp after anthesis in grape, Vitis vinifera L. Aust. J. Grape Wine Res. 1996, 2, 97–142. [Google Scholar] [CrossRef]
- Chang, B.-M.; Keller, M. Cuticle and skin cell walls have common and unique roles in grape berry splitting. Hortic. Res. 2021, 8, 168. [Google Scholar] [CrossRef]
- Ginzberg, I.; Stern, R.A. Strengthening fruit-skin resistance to growth strain by application of plant growth regulators. Sci. Hortic. 2016, 198, 150–153. [Google Scholar] [CrossRef]
- Cadot, Y.; Miñana-Castelló, M.T.; Chevalier, M. Anatomical, histological, and histochemical changes in grape seeds from Vitis vinifera L. cv Cabernet franc during fruit development. J. Agric. Food Chem. 2006, 54, 9206–9215. [Google Scholar] [CrossRef]
- Ollat, N.; Carde, J.-P.; Gaudillère, J.-P.; Barrieu, F.; Diakou-Verdin, P.; Moing, A. Grape berry development: A review. OENO One 2002, 36, 109–131. [Google Scholar] [CrossRef]
- Wang, H.; Chen, Y.; Lin, H.; Lin, M.; Chen, Y.; Lin, Y. 1-Methylcyclopropene containing-papers suppress the disassembly of cell wall polysaccharides in Anxi persimmon fruit during storage. Int. J. Biol. Macromol. 2020, 151, 723–729. [Google Scholar] [CrossRef]
- Lin, H.; Ma, L.; Guo, Q.; Liu, C.; Hou, Y.; Liu, Z.; Zhao, Y.; Jiang, C.; Guo, X.; Guo, Y. Berry texture QTL and candidate gene analysis in grape (Vitis vinifera L.). Hortic. Res. 2023, 10, uhad226. [Google Scholar] [CrossRef]
- Pedneault, K.; Provost, C. Fungus resistant grape varieties as a suitable alternative for organic wine production: Benefits, limits, and challenges. Sci. Hortic. 2016, 208, 57–77. [Google Scholar] [CrossRef]
- Atak, A. New Perspectives in Grapevine (Vitis spp.) Breeding. In Case Studies of Breeding Strategies in Major Plant Species; Wang, H., Ed.; IntechOpen: London, UK, 2022. [Google Scholar] [CrossRef]
- Zepeda, B.; Olmedo, P.; Ejsmentewicz, T.; Sepúlveda, P.; Balic, I.; Balladares, C.; Delgado-Rioseco, J.; Fuentealba, C.; Moreno, A.A.; Defilippi, B.G.; et al. Cell wall and metabolite composition of berries of Vitis vinifera (L.) cv. Thompson Seedless with different firmness. Food Chem. 2018, 268, 492–497. [Google Scholar] [CrossRef] [PubMed]
- Sato, A.; Yamada, M.; Iwanami, H. Estimation of the Proportion of Offspring Having Genetically Crispy Flesh in Grape Breeding. J. Am. Soc. Hortic. Sci. 2006, 131, 46–52. [Google Scholar] [CrossRef]
- Gombau, J.; Nadal, P.; Canela, N.; Gómez-Alonso, S.; García-Romero, E.; Smith, P.; Hermosín-Gutiérrez, I.; Canals, J.M.; Zamora, F. Measurement of the interaction between mucin and oenological tannins by Surface Plasmon Resonance (SPR); relationship with astringency. Food Chem. 2019, 275, 397–406. [Google Scholar] [CrossRef]
- Posé, S.; Paniagua, C.; Matas, A.J.; Gunning, A.P.; Morris, V.J.; Quesada, M.A.; Mercado, J.A. A nanostructural view of the cell wall disassembly process during fruit ripening and postharvest storage by atomic force microscopy. Trends Food Sci. Technol. 2019, 87, 47–58. [Google Scholar] [CrossRef]
- Rojas, B.; Suárez-Vega, F.; Saez-Aguayo, S.; Olmedo, P.; Zepeda, B.; Delgado-Rioseco, J.; Defilippi, B.G.; Pedreschi, R.; Meneses, C.; Pérez-Donoso, A.G.; et al. Pre-Anthesis Cytokinin Applications Increase Table Grape Berry Firmness by Modulating Cell Wall Polysaccharides. Plants 2021, 10, 2642. [Google Scholar] [CrossRef]
- Vicente, A.R.; Saladié, M.; Rose, J.K.C.; Labavitch, J.M. The linkage between cell wall metabolism and fruit softening: Looking to the future. J. Sci. Food Agric. 2007, 87, 1435–1448. [Google Scholar] [CrossRef]
- Nunan, K.J.; Davies, C.; Robinson, S.P.; Fincher, G.B. Expression patterns of cell wall-modifying enzymes during grape berry development. Planta 2001, 214, 257–264. [Google Scholar] [CrossRef]
- Iwatani, S.-I.; Yakushiji, H.; Mitani, N.; Sakurai, N. Evaluation of grape flesh texture by an acoustic vibration method. Postharvest Biol. Technol. 2011, 62, 305–309. [Google Scholar] [CrossRef]
- Szymanski, D.B.; Cosgrove, D.J. Dynamic coordination of cytoskeletal and cell wall systems during plant cell morphogenesis. Curr. Biol. 2009, 19, R800–R811. [Google Scholar] [CrossRef] [PubMed]
- Landrein, B.; Hamant, O. How mechanical stress controls microtubule behavior and morphogenesis in plants: History, experiments and revisited theories. Plant J. 2013, 75, 324–338. [Google Scholar] [CrossRef] [PubMed]
- Lionetti, V.; Raiola, A.; Mattei, B.; Bellincampi, D. The Grapevine VvPMEI1 Gene Encodes a Novel Functional Pectin Methylesterase Inhibitor Associated to Grape Berry Development. PLoS ONE 2015, 10, e0133810. [Google Scholar] [CrossRef] [PubMed]
- Rytioja, J.; Hildén, K.; Yuzon, J.; Hatakka, A.; de Vries, R.P.; Mäkelä, M.R. Plant-polysaccharide-degrading enzymes from Basidiomycetes. Microbiol. Mol. Biol. Rev. 2014, 78, 614–649. [Google Scholar] [CrossRef]
- Zhang, B.; Gao, Y.; Zhang, L.; Zhou, Y. The plant cell wall: Biosynthesis, construction, and functions. J. Integr. Plant Biol. 2021, 63, 251–272. [Google Scholar] [CrossRef]
- Cosgrove, D.J. Growth of the plant cell wall. Nat. Rev. Mol. Cell Biol. 2005, 6, 850–861. [Google Scholar] [CrossRef]
- Höfte, H.; Voxeur, A. Plant cell walls. Curr. Biol. 2017, 27, R865–R870. [Google Scholar] [CrossRef]
- Schuetz, M.; Smith, R.; Ellis, B. Xylem tissue specification, patterning, and differentiation mechanisms. J. Exp. Bot. 2013, 64, 11–31. [Google Scholar] [CrossRef]
- Somerville, C.; Bauer, S.; Brininstool, G.; Facette, M.; Hamann, T.; Milne, J.; Osborne, E.; Paredez, A.; Persson, S.; Raab, T.; et al. Toward a systems approach to understanding plant cell walls. Science 2004, 306, 2206–2211. [Google Scholar] [CrossRef]
- Liu, C.; Wang, X.; Shulaev, V.; Dixon, R.A. A role for leucoanthocyanidin reductase in the extension of proanthocyanidins. Nat. Plants 2016, 2, 16182. [Google Scholar] [CrossRef]
- Purushotham, P.; Ho, R.; Zimmer, J. Architecture of a catalytically active homotrimeric plant cellulose synthase complex. Science 2020, 369, 1089–1094. [Google Scholar] [CrossRef]
- Peng, L.; Kawagoe, Y.; Hogan, P.; Delmer, D. Sitosterol-beta-glucoside as primer for cellulose synthesis in plants. Science 2002, 295, 147–150. [Google Scholar] [CrossRef]
- Scheller, H.V.; Ulvskov, P. Hemicelluloses. Annu. Rev. Plant Biol. 2010, 61, 263–289. [Google Scholar] [CrossRef] [PubMed]
- Richmond, T.A.; Somerville, C.R. The cellulose synthase superfamily. Plant Physiol. 2000, 124, 495–498. [Google Scholar] [CrossRef] [PubMed]
- Atmodjo, M.A.; Hao, Z.; Mohnen, D. Evolving views of pectin biosynthesis. Annu. Rev. Plant Biol. 2013, 64, 747–779. [Google Scholar] [CrossRef] [PubMed]
- Mitsuda, N.; Seki, M.; Shinozaki, K.; Ohme-Takagi, M. The NAC transcription factors NST1 and NST2 of Arabidopsis regulate secondary wall thickenings and are required for anther dehiscence. Plant Cell 2005, 17, 2993–3006. [Google Scholar] [CrossRef]
- Vincken, J.-P.; Schols, H.A.; Oomen, R.J.F.J.; McCann, M.C.; Ulvskov, P.; Voragen, A.G.J.; Visser, R.G.F. If homogalacturonan were a side chain of rhamnogalacturonan I. Implications for cell wall architecture. Plant Physiol. 2003, 132, 1781–1789. [Google Scholar] [CrossRef]
- Jones, L.; Milne, J.L.; Ashford, D.; McQueen-Mason, S.J. Cell wall arabinan is essential for guard cell function. Proc. Natl. Acad. Sci. USA 2003, 100, 11783–11788. [Google Scholar] [CrossRef] [PubMed]
- Zykwinska, A.W.; Ralet, M.-C.J.; Garnier, C.D.; Thibault, J.-F.J. Evidence for in vitro binding of pectin side chains to cellulose. Plant Physiol. 2005, 139, 397–407. [Google Scholar] [CrossRef] [PubMed]
- Kimura, S.; Laosinchai, W.; Itoh, T.; Cui, X.; Linder, C.R.; Brown, R.M. Immunogold labeling of rosette terminal cellulose-synthesizing complexes in the vascular plant Vigna angularis. Plant Cell 1999, 11, 2075–2086. [Google Scholar] [CrossRef]
- Polko, J.K.; Kieber, J.J. The Regulation of Cellulose Biosynthesis in Plants. Plant Cell 2019, 31, 282–296. [Google Scholar] [CrossRef]
- Zhang, Y.; Nikolovski, N.; Sorieul, M.; Vellosillo, T.; McFarlane, H.E.; Dupree, R.; Kesten, C.; Schneider, R.; Driemeier, C.; Lathe, R.; et al. Golgi-localized STELLO proteins regulate the assembly and trafficking of cellulose synthase complexes in Arabidopsis. Nat. Commun. 2016, 7, 11656. [Google Scholar] [CrossRef]
- Arioli, T.; Peng, L.; Betzner, A.S.; Burn, J.; Wittke, W.; Herth, W.; Camilleri, C.; Höfte, H.; Plazinski, J.; Birch, R.; et al. Molecular analysis of cellulose biosynthesis in Arabidopsis. Science 1998, 279, 717–720. [Google Scholar] [CrossRef]
- Pear, J.R.; Kawagoe, Y.; Schreckengost, W.E.; Delmer, D.P.; Stalker, D.M. Higher plants contain homologs of the bacterial celA genes encoding the catalytic subunit of cellulose synthase. Proc. Natl. Acad. Sci. USA 1996, 93, 12637–12642. [Google Scholar] [CrossRef]
- Desprez, T.; Juraniec, M.; Crowell, E.F.; Jouy, H.; Pochylova, Z.; Parcy, F.; Höfte, H.; Gonneau, M.; Vernhettes, S. Organization of cellulose synthase complexes involved in primary cell wall synthesis in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2007, 104, 15572–15577. [Google Scholar] [CrossRef]
- Malinovsky, F.G.; Fangel, J.U.; Willats, W.G.T. The role of the cell wall in plant immunity. Front. Plant Sci. 2014, 5, 178. [Google Scholar] [CrossRef] [PubMed]
- Persson, S.; Paredez, A.; Carroll, A.; Palsdottir, H.; Doblin, M.; Poindexter, P.; Khitrov, N.; Auer, M.; Somerville, C.R. Genetic evidence for three unique components in primary cell-wall cellulose synthase complexes in Arabidopsis. Proc. Natl. Acad. Sci. USA 2007, 104, 15566–15571. [Google Scholar] [CrossRef] [PubMed]
- Taylor, N.G.; Howells, R.M.; Huttly, A.K.; Vickers, K.; Turner, S.R. Interactions among three distinct CesA proteins essential for cellulose synthesis. Proc. Natl. Acad. Sci. USA 2003, 100, 1450–1455. [Google Scholar] [CrossRef]
- Brown, D.M.; Zeef, L.A.H.; Ellis, J.; Goodacre, R.; Turner, S.R. Identification of novel genes in Arabidopsis involved in secondary cell wall formation using expression profiling and reverse genetics. Plant Cell 2005, 17, 2281–2295. [Google Scholar] [CrossRef]
- Brown, D.M.; Zhang, Z.; Stephens, E.; Dupree, P.; Turner, S.R. Characterization of IRX10 and IRX10-like reveals an essential role in glucuronoxylan biosynthesis in Arabidopsis. Plant J. 2009, 57, 732–746. [Google Scholar] [CrossRef]
- Wolf, S.; van der Does, D.; Ladwig, F.; Sticht, C.; Kolbeck, A.; Schürholz, A.-K.; Augustin, S.; Keinath, N.; Rausch, T.; Greiner, S.; et al. A receptor-like protein mediates the response to pectin modification by activating brassinosteroid signaling. Proc. Natl. Acad. Sci. USA 2014, 111, 15261–15266. [Google Scholar] [CrossRef]
- Du, J.; Anderson, C.T.; Xiao, C. Dynamics of pectic homogalacturonan in cellular morphogenesis and adhesion, wall integrity sensing and plant development. Nat. Plants 2022, 8, 332–340. [Google Scholar] [CrossRef]
- Amos, R.A.; Pattathil, S.; Yang, J.-Y.; Atmodjo, M.A.; Urbanowicz, B.R.; Moremen, K.W.; Mohnen, D. A two-phase model for the non-processive biosynthesis of homogalacturonan polysaccharides by the GAUT1:GAUT7 complex. J. Biol. Chem. 2018, 293, 19047–19063. [Google Scholar] [CrossRef]
- Sterling, J.D.; Atmodjo, M.A.; Inwood, S.E.; Kumar Kolli, V.S.; Quigley, H.F.; Hahn, M.G.; Mohnen, D. Functional identification of an Arabidopsis pectin biosynthetic homogalacturonan galacturonosyltransferase. Proc. Natl. Acad. Sci. USA 2006, 103, 5236–5241. [Google Scholar] [CrossRef]
- Kim, S.-J.; Held, M.A.; Zemelis, S.; Wilkerson, C.; Brandizzi, F. CGR2 and CGR3 have critical overlapping roles in pectin methylesterification and plant growth in Arabidopsis thaliana. Plant J. 2015, 82, 208–220. [Google Scholar] [CrossRef] [PubMed]
- Krupková, E.; Immerzeel, P.; Pauly, M.; Schmülling, T. The TUMOROUS SHOOT DEVELOPMENT2 gene of Arabidopsis encoding a putative methyltransferase is required for cell adhesion and co-ordinated plant development. Plant J. 2007, 50, 735–750. [Google Scholar] [CrossRef] [PubMed]
- Miao, Y.; Li, H.-Y.; Shen, J.; Wang, J.; Jiang, L. QUASIMODO 3 (QUA3) is a putative homogalacturonan methyltransferase regulating cell wall biosynthesis in Arabidopsis suspension-cultured cells. J. Exp. Bot. 2011, 62, 5063–5078. [Google Scholar] [CrossRef] [PubMed]
- Mouille, G.; Ralet, M.-C.; Cavelier, C.; Eland, C.; Effroy, D.; Hématy, K.; McCartney, L.; Truong, H.N.; Gaudon, V.; Thibault, J.-F.; et al. Homogalacturonan synthesis in Arabidopsis thaliana requires a Golgi-localized protein with a putative methyltransferase domain. Plant J. 2007, 50, 605–614. [Google Scholar] [CrossRef] [PubMed]
- Takenaka, Y.; Kato, K.; Ogawa-Ohnishi, M.; Tsuruhama, K.; Kajiura, H.; Yagyu, K.; Takeda, A.; Takeda, Y.; Kunieda, T.; Hara-Nishimura, I.; et al. Pectin RG-I rhamnosyltransferases represent a novel plant-specific glycosyltransferase family. Nat. Plants 2018, 4, 669–676. [Google Scholar] [CrossRef] [PubMed]
- Dumont, M.; Lehner, A.; Bouton, S.; Kiefer-Meyer, M.C.; Voxeur, A.; Pelloux, J.; Lerouge, P.; Mollet, J.-C. The cell wall pectic polymer rhamnogalacturonan-II is required for proper pollen tube elongation: Implications of a putative sialyltransferase-like protein. Ann. Bot. 2014, 114, 1177–1188. [Google Scholar] [CrossRef]
- Liu, L.; Shang-Guan, K.; Zhang, B.; Liu, X.; Yan, M.; Zhang, L.; Shi, Y.; Zhang, M.; Qian, Q.; Li, J.; et al. Brittle Culm1, a COBRA-like protein, functions in cellulose assembly through binding cellulose microfibrils. PLoS Genet. 2013, 9, e1003704. [Google Scholar] [CrossRef]
- Nakashima, K.; Takasaki, H.; Mizoi, J.; Shinozaki, K.; Yamaguchi-Shinozaki, K. NAC transcription factors in plant abiotic stress responses. Biochim. Et Biophys. Acta (BBA)-Gene Regul. Mech. 2012, 1819, 97–103. [Google Scholar] [CrossRef]
- Yin, Q.; Qin, W.; Zhou, Z.; Wu, A.-M.; Deng, W.; Li, Z.; Shan, W.; Chen, J.-Y.; Kuang, J.-F.; Lu, W.-J. Banana MaNAC1 activates secondary cell wall cellulose biosynthesis to enhance chilling resistance in fruit. Plant Biotechnol. J. 2024, 22, 413–426. [Google Scholar] [CrossRef]
- Zhong, R.; Demura, T.; Ye, Z.-H. SND1, a NAC domain transcription factor, is a key regulator of secondary wall synthesis in fibers of Arabidopsis. Plant Cell 2006, 18, 3158–3170. [Google Scholar] [CrossRef]
- Zhong, R.; Richardson, E.A.; Ye, Z.-H. Two NAC domain transcription factors, SND1 and NST1, function redundantly in regulation of secondary wall synthesis in fibers of Arabidopsis. Planta 2007, 225, 1603–1611. [Google Scholar] [CrossRef]
- Forlani, S.; Mizzotti, C.; Masiero, S. The NAC side of the fruit: Tuning of fruit development and maturation. BMC Plant Biol. 2021, 21, 238. [Google Scholar] [CrossRef]
- Zhong, R.; Lee, C.; Zhou, J.; McCarthy, R.L.; Ye, Z.-H. A battery of transcription factors involved in the regulation of secondary cell wall biosynthesis in Arabidopsis. Plant Cell 2008, 20, 2763–2782. [Google Scholar] [CrossRef]
- Ko, J.-H.; Kim, W.-C.; Han, K.-H. Ectopic expression of MYB46 identifies transcriptional regulatory genes involved in secondary wall biosynthesis in Arabidopsis. Plant J. 2009, 60, 649–665. [Google Scholar] [CrossRef] [PubMed]
- McCarthy, R.L.; Zhong, R.; Ye, Z.-H. MYB83 is a direct target of SND1 and acts redundantly with MYB46 in the regulation of secondary cell wall biosynthesis in Arabidopsis. Plant Cell Physiol. 2009, 50, 1950–1964. [Google Scholar] [CrossRef]
- Zhong, R.; Richardson, E.A.; Ye, Z.-H. The MYB46 transcription factor is a direct target of SND1 and regulates secondary wall biosynthesis in Arabidopsis. Plant Cell 2007, 19, 2776–2792. [Google Scholar] [CrossRef] [PubMed]
- Ko, J.H.; Jeon, H.W.; Kim, W.C.; Kim, J.Y.; Han, K.H. The MYB46/MYB83-mediated transcriptional regulatory programme is a gatekeeper of secondary wall biosynthesis. Ann. Bot. 2014, 114, 1099–1107. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Dai, J.; Liang, L.; Zhang, Y.; He, Y.; Xing, L.; Ma, J.; Zhang, D.; Zhao, C. Chitinase-Like Protein PpCTL1 Contributes to Maintaining Fruit Firmness by Affecting Cellulose Biosynthesis during Peach Development. Foods 2023, 12, 2503. [Google Scholar] [CrossRef]
- Li, G.; Liu, X.; Liang, Y.; Zhang, Y.; Cheng, X.; Cai, Y. Genome-wide characterization of the cellulose synthase gene superfamily in Pyrus bretschneideri and reveal its potential role in stone cell formation. Funct. Integr. Genom. 2020, 20, 723–738. [Google Scholar] [CrossRef]
- Yi, Z.; Sharif, R.; Gulzar, S.; Huang, Y.; Ning, T.; Zhan, H.; Meng, Y.; Xu, C. Changes in hemicellulose metabolism in banana peel during fruit development and ripening. Plant Physiol. Biochem. 2024, 215, 109025. [Google Scholar] [CrossRef]
- Lahaye, M.; Tabi, W.; Le Bot, L.; Delaire, M.; Orsel, M.; Campoy, J.A.; Quero Garcia, J.; Le Gall, S. Comparison of cell wall chemical evolution during the development of fruits of two contrasting quality from two members of the Rosaceae family: Apple and sweet cherry. Plant Physiol. Biochem. 2021, 168, 93–104. [Google Scholar] [CrossRef]
- Sanhueza, D.; Balic-Norambuena, I.; Sepúlveda-Orellana, P.; Siña-López, S.; Moreno, A.A.; Moya-León, M.A.; Saez-Aguayo, S. Unraveling cell wall polysaccharides during blueberry ripening: Insights into the roles of rhamnogalacturonan-I and arabinogalactan proteins in fruit firmness. Front. Plant Sci. 2024, 15, 1422917. [Google Scholar] [CrossRef]
- Huang, W.; Chen, M.; Zhao, T.; Han, F.; Zhang, Q.; Liu, X.; Jiang, C.; Zhong, C. Genome-Wide Identification and Expression Analysis of Polygalacturonase Gene Family in Kiwifruit (Actinidia chinensis) during Fruit Softening. Plants 2020, 9, 327. [Google Scholar] [CrossRef]
- Qian, M.; Xu, Z.; Zhang, Z.; Li, Q.; Yan, X.; Liu, H.; Han, M.; Li, F.; Zheng, J.; Zhang, D.; et al. The downregulation of PpPG21 and PpPG22 influences peach fruit texture and softening. Planta 2021, 254, 22. [Google Scholar] [CrossRef]
- Al Hinai, T.Z.S.; Vreeburg, R.A.M.; Mackay, C.L.; Murray, L.; Sadler, I.H.; Fry, S.C. Fruit softening: Evidence for pectate lyase action in vivo in date (Phoenix dactylifera) and rosaceous fruit cell walls. Ann. Bot. 2021, 128, 511–525. [Google Scholar] [CrossRef]
- Rossetti, D.; Yakubov, G.E.; Stokes, J.R.; Williamson, A.M.; Fuller, G.G. Interaction of human whole saliva and astringent dietary compounds investigated by interfacial shear rheology. Food Hydrocoll. 2008, 22, 1068–1078. [Google Scholar] [CrossRef]
- Pires, M.A.; Pastrana, L.M.; Fuciños, P.; Abreu, C.S.; Oliveira, S.M. Sensorial Perception of Astringency: Oral Mechanisms and Current Analysis Methods. Foods 2020, 9, 1124. [Google Scholar] [CrossRef] [PubMed]
- Ma, W.; Guo, A.; Zhang, Y.; Wang, H.; Liu, Y.; Li, H. A review on astringency and bitterness perception of tannins in wine. Trends Food Sci. Technol. 2014, 40, 6–19. [Google Scholar] [CrossRef]
- Rossetti, D.; Bongaerts, J.H.H.; Wantling, E.; Stokes, J.R.; Williamson, A.M. Astringency of tea catechins: More than an oral lubrication tactile percept. Food Hydrocoll. 2009, 23, 1984–1992. [Google Scholar] [CrossRef]
- Halsam, E.; Lilley, T.H. Natural astringency in foodstuffs—A molecular interpretation. Crit. Rev. Food Sci. Nutr. 1988, 27, 1–40. [Google Scholar] [CrossRef]
- Koleckar, V.; Kubikova, K.; Rehakova, Z.; Kuca, K.; Jun, D.; Jahodar, L.; Opletal, L. Condensed and hydrolysable tannins as antioxidants influencing the health. Mini Rev. Med. Chem. 2008, 8, 436–447. [Google Scholar] [CrossRef]
- Yonemori, K.; Matsushima, J. Property of Development of the Tannin Cells in Non-Astringent Type Fruits of Japanese Persimmon (Diospyros kaki) and Its Relationship to Natural Deastringency. J. Jpn. Soc. Hortic. Sci. 1985, 54, 201–208. [Google Scholar] [CrossRef]
- Basalekou, M.; Kyraleou, M.; Pappas, C.; Tarantilis, P.; Kotseridis, Y.; Kallithraka, S. Proanthocyanidin content as an astringency estimation tool and maturation index in red and white winemaking technology. Food Chem. 2019, 299, 125135. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, D.; Slade, D. Oligomeric proanthocyanidins: Naturally occurring O-heterocycles. Nat. Prod. Rep. 2002, 19, 517–541. [Google Scholar] [CrossRef]
- Mané, C.; Souquet, J.M.; Ollé, D.; Verriés, C.; Véran, F.; Mazerolles, G.; Cheynier, V.; Fulcrand, H. Optimization of simultaneous flavanol, phenolic acid, and anthocyanin extraction from grapes using an experimental design: Application to the characterization of champagne grape varieties. J. Agric. Food Chem. 2007, 55, 7224–7233. [Google Scholar] [CrossRef]
- Verries, C.; Guiraud, J.-L.; Souquet, J.-M.; Vialet, S.; Terrier, N.; Ollé, D. Validation of an extraction method on whole pericarp of grape berry (Vitis vinifera L. cv. Shiraz) to study biochemical and molecular aspects of flavan-3-ol synthesis during berry development. J. Agric. Food Chem. 2008, 56, 5896–5904. [Google Scholar] [CrossRef] [PubMed]
- Soares, S.; Brandão, E.; Guerreiro, C.; Soares, S.; Mateus, N.; de Freitas, V. Tannins in Food: Insights into the Molecular Perception of Astringency and Bitter Taste. Molecules 2020, 25, 2590. [Google Scholar] [CrossRef] [PubMed]
- Haslam, E. Vegetable tannins-lessons of a phytochemical lifetime. Phytochemistry 2007, 68, 2713–2721. [Google Scholar] [CrossRef]
- Okuda, T.; Ito, H. Tannins of Constant Structure in Medicinal and Food Plants—Hydrolyzable Tannins and Polyphenols Related to Tannins. Molecules 2011, 16, 2191–2217. [Google Scholar] [CrossRef]
- Akagi, T.; Katayama-Ikegami, A.; Kobayashi, S.; Sato, A.; Kono, A.; Yonemori, K. Seasonal abscisic acid signal and a basic leucine zipper transcription factor, DkbZIP5, regulate proanthocyanidin biosynthesis in persimmon fruit. Plant Physiol. 2012, 158, 1089–1102. [Google Scholar] [CrossRef]
- Mir, R.; Jallu, S.; Singh, T.P. The shikimate pathway: Review of amino acid sequence, function and three-dimensional structures of the enzymes. Crit. Rev. Microbiol. 2015, 41, 172–189. [Google Scholar] [CrossRef]
- Hichri, I.; Barrieu, F.; Bogs, J.; Kappel, C.; Delrot, S.; Lauvergeat, V. Recent advances in the transcriptional regulation of the flavonoid biosynthetic pathway. J. Exp. Bot. 2011, 62, 2465–2483. [Google Scholar] [CrossRef]
- Petit, P.; Granier, T.; d’Estaintot, B.L.; Manigand, C.; Bathany, K.; Schmitter, J.-M.; Lauvergeat, V.; Hamdi, S.; Gallois, B. Crystal structure of grape dihydroflavonol 4-reductase, a key enzyme in flavonoid biosynthesis. J. Mol. Biol. 2007, 368, 1345–1357. [Google Scholar] [CrossRef]
- Tanner, G.J.; Francki, K.T.; Abrahams, S.; Watson, J.M.; Larkin, P.J.; Ashton, A.R. Proanthocyanidin biosynthesis in plants. Purification of legume leucoanthocyanidin reductase and molecular cloning of its cDNA. J. Biol. Chem. 2003, 278, 31647–31656. [Google Scholar] [CrossRef] [PubMed]
- Yu, K.; Jun, J.H.; Duan, C.; Dixon, R.A. VvLAR1 and VvLAR2 Are Bifunctional Enzymes for Proanthocyanidin Biosynthesis in Grapevine. Plant Physiol. 2019, 180, 1362–1374. [Google Scholar] [CrossRef]
- Jun, J.H.; Lu, N.; Docampo-Palacios, M.; Wang, X.; Dixon, R.A. Dual activity of anthocyanidin reductase supports the dominant plant proanthocyanidin extension unit pathway. Sci. Adv. 2021, 7, eabg4682. [Google Scholar] [CrossRef] [PubMed]
- Jun, J.H.; Xiao, X.; Rao, X.; Dixon, R.A. Proanthocyanidin subunit composition determined by functionally diverged dioxygenases. Nat. Plants 2018, 4, 1034–1043. [Google Scholar] [CrossRef]
- Yu, K.; Dixon, R.A.; Duan, C. A role for ascorbate conjugates of (+)-catechin in proanthocyanidin polymerization. Nat. Commun. 2022, 13, 3425. [Google Scholar] [CrossRef] [PubMed]
- Goodman, C.D.; Casati, P.; Walbot, V. A multidrug resistance-associated protein involved in anthocyanin transport in Zea mays. Plant Cell 2004, 16, 1812–1826. [Google Scholar] [CrossRef]
- Wu, W.; Zhu, Q.-G.; Wang, W.-Q.; Grierson, D.; Yin, X.-R. Molecular basis of the formation and removal of fruit astringency. Food Chem. 2022, 372, 131234. [Google Scholar] [CrossRef]
- Pérez-Díaz, R.; Madrid-Espinoza, J.; Salinas-Cornejo, J.; González-Villanueva, E.; Ruiz-Lara, S. Differential Roles for VviGST1, VviGST3, and VviGST4 in Proanthocyanidin and Anthocyanin Transport in Vitis vinífera. Front. Plant Sci. 2016, 7, 1166. [Google Scholar] [CrossRef]
- Chen, S.-Y.; Tang, Y.-M.; Hu, Y.-Y.; Wang, Y.; Sun, B.; Wang, X.-R.; Tang, H.-R.; Chen, Q. FaTT12-1, a multidrug and toxin extrusion (MATE) member involved in proanthocyanidin transport in strawberry fruits. Sci. Hortic. 2018, 231, 158–165. [Google Scholar] [CrossRef]
- Sun, L.; Huo, J.; Liu, J.; Yu, J.; Zhou, J.; Sun, C.; Wang, Y.; Leng, F. Anthocyanins distribution, transcriptional regulation, epigenetic and post-translational modification in fruits. Food Chem. 2023, 411, 135540. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Pang, Y.; Dixon, R.A. The mysteries of proanthocyanidin transport and polymerization. Plant Physiol. 2010, 153, 437–443. [Google Scholar] [CrossRef] [PubMed]
- Bogs, J.; Downey, M.O.; Harvey, J.S.; Ashton, A.R.; Tanner, G.J.; Robinson, S.P. Proanthocyanidin synthesis and expression of genes encoding leucoanthocyanidin reductase and anthocyanidin reductase in developing grape berries and grapevine leaves. Plant Physiol. 2005, 139, 652–663. [Google Scholar] [CrossRef]
- Hichri, I.; Heppel, S.C.; Pillet, J.; Léon, C.; Czemmel, S.; Delrot, S.; Lauvergeat, V.; Bogs, J. The Basic Helix-Loop-Helix Transcription Factor MYC1 Is Involved in the Regulation of the Flavonoid Biosynthesis Pathway in Grapevine. Mol. Plant 2010, 3, 509–523. [Google Scholar] [CrossRef]
- Amato, A.; Cavallini, E.; Walker, A.R.; Pezzotti, M.; Bliek, M.; Quattrocchio, F.; Koes, R.; Ruperti, B.; Bertini, E.; Zenoni, S.; et al. The MYB5-driven MBW complex recruits a WRKY factor to enhance the expression of targets involved in vacuolar hyper-acidification and trafficking in grapevine. Plant J. 2019, 99, 1220–1241. [Google Scholar] [CrossRef]
- Feng, J.; Zhang, W.; Wang, W.; Nieuwenhuizen, N.J.; Atkinson, R.G.; Gao, L.; Hu, H.; Zhao, W.; Ma, R.; Zheng, H.; et al. Integrated Transcriptomic and Proteomic Analysis Identifies Novel Regulatory Genes Associated with Plant Growth Regulator-Induced Astringency in Grape Berries. J. Agric. Food Chem. 2024, 72, 4433–4447. [Google Scholar] [CrossRef]
- An, J.-P.; Li, R.; Qu, F.-J.; You, C.-X.; Wang, X.-F.; Hao, Y.-J. R2R3-MYB transcription factor MdMYB23 is involved in the cold tolerance and proanthocyanidin accumulation in apple. Plant J. 2018, 96, 562–577. [Google Scholar] [CrossRef]
- Kyraleou, M.; Kotseridis, Y.; Koundouras, S.; Chira, K.; Teissedre, P.-L.; Kallithraka, S. Effect of irrigation regime on perceived astringency and proanthocyanidin composition of skins and seeds of Vitis vinifera L. cv. Syrah grapes under semiarid conditions. Food Chem. 2016, 203, 292–300. [Google Scholar] [CrossRef]
- Xu, F.; Gao, X.; Xi, Z.-M.; Zhang, H.; Peng, X.-Q.; Wang, Z.-Z.; Wang, T.-M.; Meng, Y. Application of exogenous 24-epibrassinolide enhances proanthocyanidin biosynthesis in Vitis vinifera ‘Cabernet Sauvignon’ berry skin. Plant Growth Regul. 2015, 75, 741–750. [Google Scholar] [CrossRef]
- Tessmer, M.A.; Kluge, R.A.; Appezzato-da-Glória, B. The accumulation of tannins during the development of ‘Giombo’ and ‘Fuyu’ persimmon fruits. Sci. Hortic. 2014, 172, 292–299. [Google Scholar] [CrossRef]
- Henry-Kirk, R.A.; McGhie, T.K.; Andre, C.M.; Hellens, R.P.; Allan, A.C. Transcriptional analysis of apple fruit proanthocyanidin biosynthesis. J. Exp. Bot. 2012, 63, 5437–5450. [Google Scholar] [CrossRef] [PubMed]
- Deluc, L.; Barrieu, F.; Marchive, C.; Lauvergeat, V.; Decendit, A.; Richard, T.; Carde, J.-P.; Mérillon, J.-M.; Hamdi, S. Characterization of a grapevine R2R3-MYB transcription factor that regulates the phenylpropanoid pathway. Plant Physiol. 2006, 140, 499–511. [Google Scholar] [CrossRef]
- Wang, N.; Qu, C.; Jiang, S.; Chen, Z.; Xu, H.; Fang, H.; Su, M.; Zhang, J.; Wang, Y.; Liu, W.; et al. The proanthocyanidin-specific transcription factor MdMYBPA1 initiates anthocyanin synthesis under low-temperature conditions in red-fleshed apples. Plant J. 2018, 96, 39–55. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.-F.; Vialet, S.; Guiraud, J.-L.; Torregrosa, L.; Bertrand, Y.; Cheynier, V.; This, P.; Terrier, N. A negative MYB regulator of proanthocyanidin accumulation, identified through expression quantitative locus mapping in the grape berry. New. Phytol. 2014, 201, 795–809. [Google Scholar] [CrossRef]
- Huang, Y.; Liang, D.; Xia, H.; Lin, L.-J.; Wang, J.; Lv, X.-L. Lignin and Quercetin Synthesis Underlies Berry Russeting in ‘Sunshine Muscat’ Grape. Biomolecules 2020, 10, 690. [Google Scholar] [CrossRef]
- Falginella, L.; Cipriani, G.; Monte, C.; Gregori, R.; Testolin, R.; Velasco, R.; Troggio, M.; Tartarini, S. A major QTL controlling apple skin russeting maps on the linkage group 12 of ‘Renetta Grigia di Torriana’. BMC Plant Biol. 2015, 15, 150. [Google Scholar] [CrossRef]
- Knoche, M.; Khanal, B.P.; Stopar, M. Russeting and Microcracking of ‘Golden Delicious’ Apple Fruit Concomitantly Decline Due to Gibberellin A4+7 Application. J. Amer. Soc. Hort. Sci. 2011, 136, 159–164. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wan, S.-Y.; Li, Y.-M.; Xie, Z.-S. Exploring Factors Influencing the Consumption of Grape Skins: A Review. Horticulturae 2025, 11, 962. https://doi.org/10.3390/horticulturae11080962
Wan S-Y, Li Y-M, Xie Z-S. Exploring Factors Influencing the Consumption of Grape Skins: A Review. Horticulturae. 2025; 11(8):962. https://doi.org/10.3390/horticulturae11080962
Chicago/Turabian StyleWan, Si-Yuan, You-Mei Li, and Zhao-Sen Xie. 2025. "Exploring Factors Influencing the Consumption of Grape Skins: A Review" Horticulturae 11, no. 8: 962. https://doi.org/10.3390/horticulturae11080962
APA StyleWan, S.-Y., Li, Y.-M., & Xie, Z.-S. (2025). Exploring Factors Influencing the Consumption of Grape Skins: A Review. Horticulturae, 11(8), 962. https://doi.org/10.3390/horticulturae11080962





