Abstract
Lycium barbarum is a traditional medicinal and edible plant species in China, exhibiting notable salt tolerance that enables cultivation in salt-affected soils. However, intensifying soil salinization has rendered severe salt stress a critical limiting factor for its fruit yield and quality. Universal stress proteins (USPs) serve as crucial regulators for plant abiotic stress responses through developmental process modulation. Nevertheless, the characteristics and functional divergence of USP gene family members remain unexplored in L. barbarum. Here, we performed genome-wide identification and characterization of the USP gene family in L. barbarum, revealing 52 members unevenly distributed across all 12 chromosomes. Phylogenetic analysis classified these LbUSP members into four distinct groups, demonstrating the integration of the conserved USP domain and diverse motifs within each group. Collinearity analysis indicated a stronger synteny of LbUSPs with orthologs in Solanum lycopersicum than with other species (Arabidopsis thaliana, Vitis vinifera, and Oryza sativa), demonstrating that gene duplication coupled with functional conservation represented the primary mechanism underlying USP family expansion in L. barbarum. In silico promoter screening detected abundant cis-acting elements associated with abiotic/biotic stress responses (MYB and MYC binding sites), phytohormone regulation (ABRE motif), and growth/development processes (Box-4 and G-box). Transcriptome sequencing and RT-qPCR validation revealed tissue-specific differential expression patterns of LbUSP8, LbUSP11, LbUSP12, LbUSP23, and LbUSP25 in roots and stems under salt stress, identifying them as prime candidates for mediating salt resistance in L. barbarum. Our findings establish a foundation for the functional characterization of LbUSPs and molecular breeding of salt-tolerant L. barbarum cultivars.
1. Introduction
Terrestrial plants, unlike motile animals, cannot evade environmental stresses and are consequently confronted with multifarious abiotic stressors throughout their life cycle. These stressors, including salinity, drought, temperature extremes, chemical toxicity, and oxidative stress, inflict significant damage and profoundly influence growth, development, agronomic yield, and qualitative traits [1,2,3,4]. To alleviate such pressures, plants have evolved complex regulatory networks through natural selection [5]. Stressors directly induce physicochemical alterations in cellular biomolecules, triggering stress responses that involve cascading events, including stress sensing, signal transduction, transcription, transcript processing, translation, and post-translational modifications. These processes are initiated across diverse compartments (e.g., cell wall, plasma membrane, cytoplasm, nucleus, chloroplasts, mitochondria, endoplasmic reticulum, peroxisomes), with key regulators identified through genetic, biochemical, and molecular studies.
As crucial regulators of stress adaptation in plants, universal stress proteins (USPs) were initially identified in bacteria during studies of stress responses [6]. When bacteria were exposed to a diverse range of environmental stressors, including high temperatures, oxidative stress, and nutrient deficiencies, the expression levels of a specific set of proteins were significantly elevated, and these were later defined as the Usps [6,7,8]. In bacteria, Usps are characterized by a conserved Usp domain (140–160 amino acids in length), which serves as a molecular scaffold for coordinating interactions with diverse cellular ligands through its solvent-accessible binding pocket [9,10]. Six Usp members have been identified in Escherichia coli, namely UspA, UspC, UspD, UspE, UspF, and UspG, which can be phylogenetically categorized into three distinct classes [11,12]. Class I consists of Usps that lack ATP-binding motifs, including UspA, UspC, and UspD. Class II, represented by UspF and UspG, is evolutionarily distinct from Class I and is defined by conserved ATP-binding motifs [G-2X-G-9X-(S/T)] that enable direct ATP interaction. Class III is exclusively represented by UspE, which is distinguished by a bifunctional domain architecture comprising two tandem-repeated Usp domains (designated E1 and E2 domains). Structural and phylogenetic analyses reveal that E1 shows high sequence homology with Class I Usp domains, while E2 exhibits convergent structural motifs and functional parallels with the ATP-binding Usp clade from Class II [12,13]. Each class of Usps from E. coli demonstrates specialized functional adaptations for countering specific environmental stressors [14,15]. The first structural entries of Usp deposited in the Protein Data Bank (PDB) originated from Methanocaldococcus jannaschii hypothetical protein MJ0577 (PDB ID: 1MJH) [16]. This archetypal UspA protein was structurally resolved in a nucleotide-bound state and crystallized in a complex with ATP, thereby establishing its functional association with ATP binding. Subsequent determination of the Haemophilus influenzae Usp structure (PDB ID: 1JMV) confirmed its lack of detectable ATP-binding capability in contrast to the M. jannaschii ortholog [10]. These contrasting biochemical properties prompted the early categorization of USPs into nucleotide-binding and non-nucleotide-binding subclasses, a classification framework rooted in their differential ligand interactions [17].
In plants, proteins possessing UspA domains that show similarity to those found in bacteria are designated as USPs [17,18]. The first reported plant USP was identified in Oryza sativa, where OsUSP1 functions as an ethylene-responsive modulator that enhanced hypoxia adaptation under submergence stress through the precise regulation of intracellular ethylene concentrations [19]. To date, numerous USP genes have been meticulously characterized across many plant species, including Arabidopsis thaliana [20], Gossypium hirsutum [21], Solanum tuberosum [22], Vitis vinifera [23], etc. Plant USPs are intricately involved in counteracting a diverse spectrum of abiotic stressors, including extreme temperatures, drought, salinity, and nutrient deficiencies, which pose significant challenges to plant growth and development [15]. In A. thaliana, AtUSP17 exerted a negative modulation on salt tolerance via the regulation of ethylene, abscisic acid (ABA), reactive oxygen species (ROS), and G-protein-mediated signaling cascades and associated physiological responses [24]. Conversely, overexpression of SbUSP from Salicornia brachiata enhanced the tolerance of tobacco to salt and osmotic stress [25]. Homologous overexpression of MsUSPA reduced the accumulation of ROS and elevated the drought tolerance of Malus sieversii [26]. Studies on the Triticum aestivum USP gene family revealed that overexpression of TaUSP_5D-1 improved the drought tolerance of transgenic lines via an improvement in the lateral root network in A. thaliana [27]. Among the 71 members of the USP gene family in Cajanus cajan, 49 members were identified to be drought-responsive proteins [28]. Homologous overexpression of USP1 in Solanum lycopersicum reduced stomatal density on leaf surfaces, directly enhancing water retention capacity and thereby improving thermotolerance and drought resistance in transgenic lines [29]. In addition, the ATP-binding homodimer SlRd2 (USP) was phosphorylated by SlCIPK6 in the cytoplasm, and transient SlCIPK6 expression in SlRd2-overexpressing plants reduced ROS accumulation versus wild-type, indicating that SlRd2 acts as both an interactor and phosphotarget of SlCIPK6 to regulate stress responses [7]. Furthermore, the blueberry VcUSP1 protein could bind to the promoter region of VcMYB4a to negatively regulate ultraviolet-B-induced anthocyanin biosynthesis [30]. Beyond abiotic stress responses, USPs contribute to plant immunity by countering pathogen attacks. Studies on A. thaliana demonstrated that AtPHOS32/34 were phosphorylated upon microbial challenge. Among these, AtPHOS32 was biochemically validated as a direct phosphorylation target of MAPK3/MAPK6 kinases, establishing its role in antipathogen signaling cascades [31,32]. In Astragalus sinicus, the USP gene ASD243 was transcriptionally upregulated during symbiotic interactions with the nitrogen-fixing bacterium Mesorhizobium huakuii [33]. These studies indicate that USP genes play a crucial role in plants’ adaptive responses, enabling them to maintain cellular homeostasis and enhance their resilience under adverse environmental conditions. However, the exact molecular function and regulatory response processes of USPs in many plants remain to be elucidated.
Lycium barbarum, a perennial deciduous shrub belonging to the Solanaceae family, is a functionally significant medicinal plant species in China [34]. Modern medical studies have demonstrated that the fruits of L. barbarum contain abundant bioactive constituents, including polysaccharides, multiple amino acids, and betaines, which exhibit significant pharmacological properties. These compounds collectively enhance metabolic processes, inhibit cellular senescence, and demonstrate anti-allergic, antioxidant, and anticarcinogenic effects [34,35]. In L. barbarum, these bioactive compounds originate from abundant secondary metabolites synthesized during adaptation to the unique saline–alkali soils and climatic conditions of northwestern China, which ultimately establish it as an authentic medicinal material [36]. As a salt-tolerant pioneer species for saline–alkali soil reclamation, L. barbarum delivers significant ecological and economic benefits in northwestern China, with these advantages further amplified by a well-established agro-industrial chain focused on value-added derivatives. According to the existing literature, L. barbarum exhibits optimal growth in saline soils with salt concentrations of 0.3% to 0.5%, whereas its development is significantly inhibited in highly saline soils with salt content exceeding 0.5% [37]. However, China ranks among the countries with extensive saline–alkali land coverage, possessing a total area of approximately 100 million hectares and accounting for ~10% of its territorial land area, with the highest concentration distributed in Northwestern China [38]. Even L. barbarum demonstrates adaptive growth in moderately saline soils, where escalating soil salinization has rendered severe salt stress a critical limiting factor for its productivity and yield. High-salinity soils induce reduced yield and compromised fruit quality, severely constraining the sustainable development of the L. barbarum industry and impeding agricultural efficiency enhancement. Consequently, developing effective strategies to improve the salt tolerance of L. barbarum represents an urgent priority for advancing industrial development. USP genes have been identified as playing critical regulatory roles in plant abiotic stress responses, but the abiotic stress processes in which their family members participate show significant differentiation among different plants. Therefore, it holds great significance to explore and precisely identify the key members of the USP gene family in L. barbarum that are responsible for salt stress.
In this study, a genome-wide identification of the L. barbarum USP gene family was conducted based on completed genome sequencing data. We then performed comprehensive analyses of USP gene family members, including protein physicochemical properties, phylogenetic relationships, and conserved motifs, as well as gene structures, collinearity, and promoter cis-acting elements. Subsequently, transcriptome sequencing and real-time quantitative PCR (RT-qPCR) were employed to quantify expression patterns of candidate differentially expressed genes (DEGs) in the roots and stems of L. barbarum under varying salt stress exposures, enabling a systematic identification of key USP family members participating in salt tolerance. The results establish a theoretical foundation for enhancing the stress resilience of L. barbarum while providing a basis for genetic breeding.
2. Materials and Methods
2.1. Identification and Analysis of Physical and Chemical Properties of LbUSP Genes
To identify putative LbUSP members from Lycium barbarum L., a dedicated protein database was constructed using the L. barbarum genome data previously obtained to facilitate systematically screening USP family members in L. barbarum. The hidden Markov model (HMM) file corresponding to the USP structure domain (PF00582) was obtained from the PFAM database (https://www.ebi.ac.uk/interpro/entry/pfam/PF00582/logo/, accessed on 9 July 2025). The obtained HMM profile was then used as a query to search for all possible USP sequences from the protein databases of L. barbarum using the Simple Hmm Search Program in TBtools-II software (v2.136) with an E-value threshold of <1.0 × 10−5 [39]. Candidate USPs subjected to rigorous one-to-one validation against PFAM and CDD databases were used to exclude proteins lacking conserved USP domains. The USP genes in S. lycopersicum and V. vinifera were identified by same method, while the USP sequences in A. thaliana and O. sativa were gained from the previous literature [20,40]. The chromosomal locations (start and end positions) of all LbUSP genes on the twelve chromosomes were obtained from the L. barbarum genome. The physicochemical properties of LbUSP, including the amino acid number, molecular weight (MW), theoretical isoelectric point (pI), instability index, aliphatic index, and grand average of hydropathicity (GRAVY) were analyzed using ExPASy (https://web.expasy.org/protparam/, accessed on 9 July 2025). WoLF PSORT (https://wolfpsort.hgc.jp, accessed on 9 July 2025) was employed to predict the subcellular localization of LbUSP.
2.2. Multiple Sequence Alignment and Phylogenetic Analysis
To explore the evolutionary relationship within the USP gene family, a multiple sequence alignment of USPs from L. barbarum, S. lycopersicum, A. thaliana, V. vinifera, and O. sativa was performed using the ClustalW program in MEGA 12 software. The phylogenetic tree was constructed using the maximum likelihood (ML) method with adaptive bootstrap (five threshold) replications.
2.3. Analysis of Gene Structure, Protein Conserved Motif, and Promoter Cis-Acting Elements
The exon–intron structure of LbUSP gene was defined by Tbtools-II software (v2.136), and the conserved motif of the LbUSP was obtained from the online MEME program (https://meme-suite.org/, accessed on 9 July 2025) with the following parameters: minimum width set to 6 bp, maximum width set to 50 bp, and maximum number of motifs set to ten. In order to ensure reliability, the LbUSP sequences were submitted to the MEME retrieval conserved motif, and the maximum number of motifs was set to ten. The structure and conserved domain of the LbUSPs were visualized by TBtools-II. The upstream 2000 bp sequence of the start codon of each LbUSP was obtained as the promoter from the L. barbarum genome for cis-acting element analysis by the PlantCARE database for online retrieval [41].
2.4. Collinearity and Ka/Ks Analysis of USP Genes
A collinearity analysis among the members of the LbUSP gene family was performed using MCScanX within Tbtools-II. Subsequently, the non-synonymous (Ka) and synonymous (Ks) substitution rates for each duplicated LbUSP gene were calculated via Tbtools-II. The dual synteny plot function was utilized to analyze the collinearity between LbUSPs and their orthologous genes in S. lycopersicum, A. thaliana, V. vinifera, and O. sativa, and the resulting data were visualized using Tbtools-II.
2.5. Preparation of Plant Materials and Application of Salt Treatment for Transcriptome Sequencing and RT-qPCR
Tissue culture plantlets of the L. barbarum cultivar ‘Ningqi 1’, derived from a single individual and maintained in our laboratory, were utilized as model material for salt treatment in transcriptome sequencing and RT-qPCR analyses. Briefly, stem segments (2.5–3.0 cm) excised from ‘Ningqi 1’ tissue culture plantlets were transferred onto solidified MS medium (containing MS salts and organic supplements, 15 g/L sucrose, and 7.4 g/L agar) supplemented with either 100 or 200 mmol/L NaCl. Stems maintained on standard MS medium without additional NaCl served as the control. Following 35 days of salt stress exposure, stems and regenerated roots were thoroughly rinsed with sterile water, flash-frozen in liquid nitrogen, and stored at –80 °C until RNA extraction and commercial sequencing (Metware Biotechnology Co., Ltd., Wuhan, China).
For RT-qPCR analysis, total RNA was isolated from stem and root samples using the RNA Easy Fast Plant Tissue Kit (Tiangen, Beijing, China), followed by DNase digestion to eliminate genomic DNA contamination. First-strand cDNA synthesis was performed using 1.0 μg of total RNA with the PrimeScript™ RT Reagent Kit (TaKaRa, Beijing, China). The resulting cDNA served as the template for RT-qPCR to quantify the transcriptional levels of target LbUSPs. To validate expression changes in candidate LbUSP genes involved in salt stress, transcripts exhibiting both relatively high expression levels (FPKM > 1.00) and significant salt stress responses in roots or stems (FDR < 0.05), as identified via transcriptome sequencing, were subjected to RT-qPCR analysis. The L. barbarum Actin gene was used as the reference gene to calculate the relative expression levels of LbUSPs using the 2−ΔCt method [42,43], with the expression level of Actin in each sample normalized to 1.00. Gene-specific primers (Table S1) were designed using Primer Premier 5.0. All RT-qPCR experiments, along with the previously described RNA-seq analyses, were performed with three independent biological replicates.
2.6. Data Analysis
Statistical analysis of the relative gene expression data from RT-qPCR was performed using GraphPad Prism v10.5. Data are presented as the mean ± standard deviation (SD) of three biological replicates. Significant differences among treatment groups were determined by a one-way analysis of variance (ANOVA) followed by Fisher’s LSD post hoc test for multiple comparisons. Differences were considered statistically significant at p < 0.05.
3. Results
3.1. Genome-Wide Identification of USP Members in L. barbarum
To investigate the functional roles of USPs in salt stress response in L. barbarum, we performed a genome-wide identification of USP gene family members using a conserved domain-based search strategy. The HMM profile of the USP domain (PF00582) was obtained from the PFAM database and employed to screen putative USP genes from the L. barbarum genome, leading to the identification of 52 USP gene family members. Based on the presence of functional domains, these proteins were categorized into two distinct categories, those containing solely the typical USP domain (38 members) and those exhibiting both USP and tyrosine kinase domains (14 members), designated as LbUSP and LbUtyK, respectively (Table S2).
3.2. Chromosome Distribution and Molecular Characterization of USPs in L. barbarum
The identified 52 USP gene family members were systematically designated as LbUSP1-LbUSP38 and LbUtyK1-LbUtyK14 according to their sequential chromosomal locations (Figure 1). Genomic distribution analysis revealed their uneven allocation across all 12 chromosomes of L. barbarum, with gene densities varying substantially from 3.8% to 13.2% per chromosome. Chromosomes 1 and 5 harbored the highest abundance (seven members each), while chromosomes 4 and 12 contained the fewest (two members each). Notably, USP genes were predominantly localized in terminal chromosomal regions with relative scarcity in central segments, suggesting their evolutionary origin could be attributed to non-uniform duplication events of chromosome fragments.
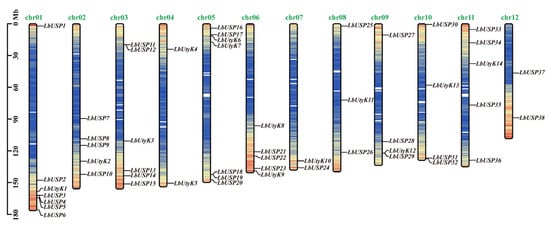
Figure 1.
Chromosomal distribution of LbUSP family members in L. barbarum. Distribution of the 52 identified LbUSP genes across the 12 chromosomes. Gene positions are mapped to exact chromosomal locations using the scale (left vertical bar, units in Megabases). Chromosome numbers appear at the top of each chromosome bar. Red and blue represent a high and low gene density of the chromosomal region, respectively.
A comprehensive physicochemical characterization of LbUSP family members is presented in Table 1. Amino acid lengths of LbUSPs varied from 142 aa (LbUSP7) to 880 aa (LbUtyK6), with an average length of 396 amino acids, and corresponding MWs spanned from 15.50 kDa (LbUSP7) to 99.02 kDa (LbUtyK6), averaging 43.97 kDa. Theoretical pI values ranged from 4.70 (LbUSP33) to 10.08 (LbUSP14), and aliphatic indices varied moderately between 74.33 (LbUSP36) and 114.08 (LbUSP37). Hydropathy profiles displayed pronounced divergence, with GRAVY scores extending from −0.639 (LbUSP14) to 0.432 (LbUSP37). The instability indices (spanning 8.34–57.31) of LbUSPs indicated that 23 proteins were classified as stable (instability index < 40), whereas 29 proteins exceeded the instability threshold (>40). Subcellular localization prediction analysis revealed distinct compartmentalization patterns among LbUSPs, with cytoplasmic localization representing the predominant distribution (24 members, 46.15% of total), followed by nuclear targeting (17/52, 32.69%), chloroplast localization (7/52, 13.46%), and plasma membrane association (3/52, 5.77%). Notably, only LbUSP20 was predicted to localize in the mitochondrion. These findings indicate that the variation in amino acid length and physicochemical properties among LbUSPs may enable functional diversification in response to different functional requirements.

Table 1.
Physicochemical properties of USPs from L. barbarum.
3.3. Phylogenetic Analysis of USP Gene Family
To elucidate the evolutionary patterns of USP family members across plant species, we performed a phylogenetic analysis using all 52 USP members from L. barbarum along with 180 USPs from four plants (Table S3), namely S. lycopersicum (50), A. thaliana (41), V. vinifera (46), and O. sativa (44). Based on evolutionary relationships, the USPs in the phylogenetic tree were classified into four groups (I–IV), containing 22, 12, 15, and 3 LbUSP members, respectively (Figure 2). Among them, groups I–III comprised USP members from all five tested species. Group I was the largest, encompassing 112 USPs (48.3% of the total) distributed across L. barbarum (22), S. lycopersicum (22), A. thaliana (17), V. vinifera (29), and O. sativa (22). In contrast, group IV was the smallest, containing only LbUSP17/28/37, VvUSP21, and SlUSP18. Phylogenetically, L. barbarum USPs exhibited a closer evolutionary distance to those of S. lycopersicum than to the other two eudicots (A. thaliana and V. vinifera) and the monocot O. sativa. Within finer phylogenetic clades, most LbUSPs clustered with their homologs from S. lycopersicum, indicating shared ancestral genes between these species. However, some specific LbUSPs (e.g., LbUSP21/30/31, LbUSP18/19, and LbUtyK3/6) formed paralogous clades unique to L. barbarum, implying species-specific gene duplication and possible functional divergence within L. barbarum.
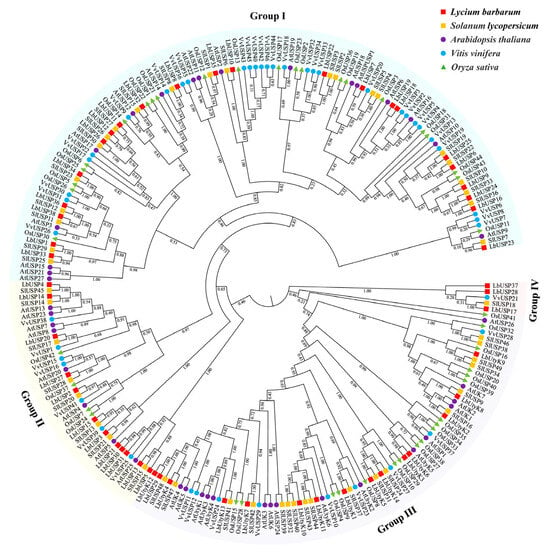
Figure 2.
Phylogenetic tree construction of USPs across plant species. The 52 LbUSPs and 180 homologous USPs from S. lycopersicum, A. thaliana, V. vinifera, and O. sativa were analyzed using the maximum likelihood (ML) method. USP sequences are listed in Tables S3 and S4. Colored arcs indicate group classifications. Species-specific symbols: red squares (L. barbarum), orange squares (S. lycopersicum), purple circles (A. thaliana), blue circles (V. vinifera), and green triangles (O. sativa).
3.4. Protein and Gene Structure Analyses of USPs in L. barbarum
Based on phylogenetic analysis, protein and gene structures of L. barbarum USP members were further characterized. Ten motifs in LbUSPs were predicted using the online MEME program, demonstrating conserved composition and spatial distribution within each group (Figure 3A and Table S4). Notably, motif 5 consistently localized to the N-terminus of LbUSPs in groups I–III, motif 8 occupied the C-terminus in group I and partial members of groups II–IV, and all LbUSP members except LbUSP17 shared motifs 2 or 9. These findings reveal that motifs 2, 5, 8, and 9 constitute the core USP domain (Figure 3B). Furthermore, detection of the conserved ATP-binding site in motif 2 prompted a sequence alignment analysis of motif 2-containing LbUSP members (Figure S1A). Thirteen of these twenty-three members were identified as harboring ATP-binding sites, indicating their ATP-binding activity (Figure S1B).
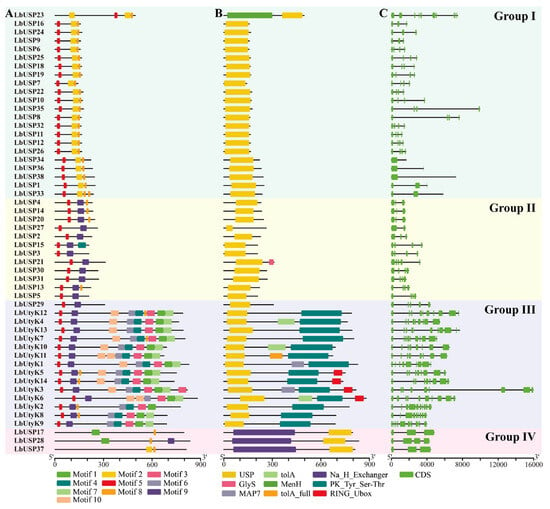
Figure 3.
In silico analyses of conserved motifs, domains, and gene structure of LbUSP members. (A) Schematic representation of conserved motifs in LbUSPs. Distinct colors correspond to unique motif identities. (B) Architecture of conserved domains in LbUSPs. Color-coded rectangles denote specific functional domains. (C) Exon–intron distribution of LbUSPs. Green boxes and gray lines indicate exon regions and introns, respectively.
As expected, the domain analysis confirmed that all LbUSPs contained one USP domain, with the USP domain spanning nearly the complete sequence of 34 LbUSP members (Figure 3B). However, unique domain architectures were found in specific LbUSPs. The LbUSP17/28/37 contained a Na+/H+ exchanger domain, while LbUSP23 featured a MenH superfamily domain, indicating potential functional specialization of LbUSP17/28/37 and LbUSP23 in sodium–proton homeostasis and menaquinone biosynthesis pathways, respectively. Additionally, a motif architecture (motifs 1, 3, 4, 6, 7) was observed in 14 LbUtyK proteins of group III (Figure 3A), all harboring PK_Tyr_Ser-Thr domains (tyrosine/serine/threonine kinases; Figure 3B), implying the probable involvement of LbUtyKs in kinase-mediated signaling cascade networks. The diverse motif patterns within each group support the hypothesis that these structural features may contribute to the functional diversification of LbUSPs.
Gene structure analysis revealed considerable variation in intron–exon organization among LbUSP family members (Figure 3C). The number of exons ranged from 2 to 11 across the LbUSP genes, with distinct patterns observed among phylogenetic groups. Most genes in groups I, II, and IV exhibited relatively simple structures containing 2–4 exons, whereas those in group III displayed more complex architectures with 5–11 exons. These structural differences likely reflected evolutionary modifications through exon deletion or acquisition events during long-term evolution. Importantly, the observed gene structure patterns correlated well with the identified motifs and domains, suggesting a coordinated evolution of these genomic features in LbUSP genes.
3.5. Analyses of Collinearity and Ka/Ks of USPs
To elucidate the expansion mechanisms of the USP gene family in L. barbarum, we characterized tandem and segmental duplications of all 38 LbUSPs and 14 LbUtyKs. Herein, 19 LbUSPs and 2 LbUtyKs were found to constitute 15 collinear gene pairs (Table 2). Genomic localization studies demonstrated that all these gene pairs resulted from the segmental type of duplication (Figure 4). To understand the evolutionary selection pressures acting on these duplicated genes, we calculated Ka/Ks ratios for all collinear gene pairs, following established methods [44]. Notably, all 15 homologous gene pairs exhibited Ka/Ks ratios ranging from 0.0809 to 0.4079 (Table 2), indicating strong purifying selection following duplication events. This finding shows that these genes have been evolutionarily constrained to maintain their function.

Table 2.
Ka and Ks substitution rates of collinear gene pairs in L. barbarum.
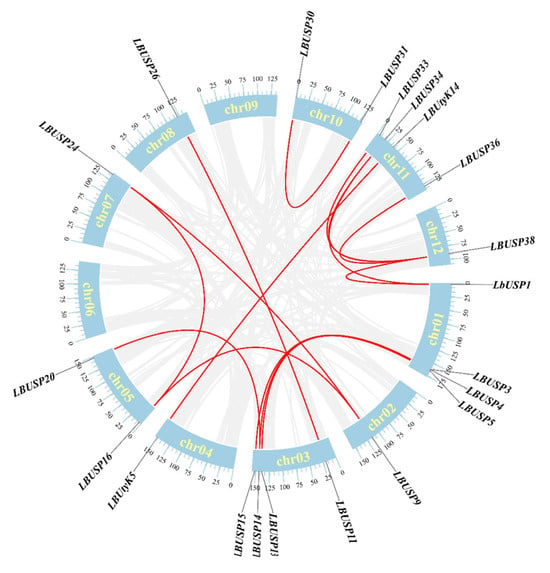
Figure 4.
Synteny analysis of the LbUSP family members. Gray lines indicate all synteny blocks in the L. barbarum genome; red lines denote LbUSP gene pairs. Chromosome numbers are positioned at the midpoint of each chromosome.
Further evolutionary relationships and conservation patterns of USP genes were obtained from a synteny analysis between L. barbarum and S. lycopersicum, A. thaliana, V. vinifera, and O. sativa (Figure 5). Analysis results revealed significant differences in gene synteny between dicot (S. lycopersicum, A. thaliana, V. vinifera) and monocot (O. sativa) species. Specifically, we identified 80 orthologous pairs between L. barbarum and S. lycopersicum, involving 46 LbUSPs and 39 StUSPs (Figure 5A and Table S5). Fewer collinear relationships were observed with V. vinifera (51 pairs; 35 LbUSPs and 22 VvUSPs), A. thaliana (43 pairs; 31 LbUSPs and 21 AtUSPs), and O. sativa (21 pairs; 14 LbUSPs and 12 OsUSPs) (Figure 5B–D and Tables S6–S8). This pattern reflects the closer phylogenetic relationship between L. barbarum and S. lycopersicum, both belonging to the Solanaceae family, compared to the more distantly related species.
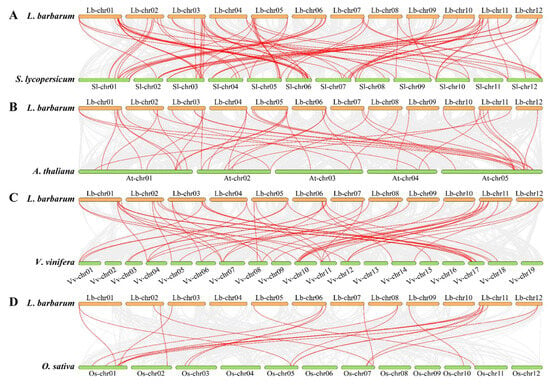
Figure 5.
Collinearity relationships of USP genes between L. barbarum and four other plants. (A) Synteny analysis of L. barbarum genome with S. lycopersicum genome. (B) Synteny analysis of L. barbarum genome with A. thaliana genome. (C) Synteny analysis of L. barbarum genome with V. vinifera genome. (D) Synteny analysis of L. barbarum genome with O. sativa genome. Gray lines represent homologous genomic blocks between L. barbarum and target species. Collinear gene pairs are highlighted with red lines.
3.6. Analysis of Cis-Acting Elements in Promoter of LbUSP Genes
To characterize potential regulatory elements governing LbUSP gene expression in response to salt stress, the 2000 bp upstream sequences of all 52 predicted LbUSP coding regions were extracted from the L. barbarum genome and subjected to comprehensive in silico analysis using PlantCARE to identify putative cis-acting elements (Figure 6 and Table S9). Based on the functional annotation [45], the identified cis-acting elements were classified into three major categories, which were abiotic and biotic stress, phytohormone-responsive, and plant growth and development.
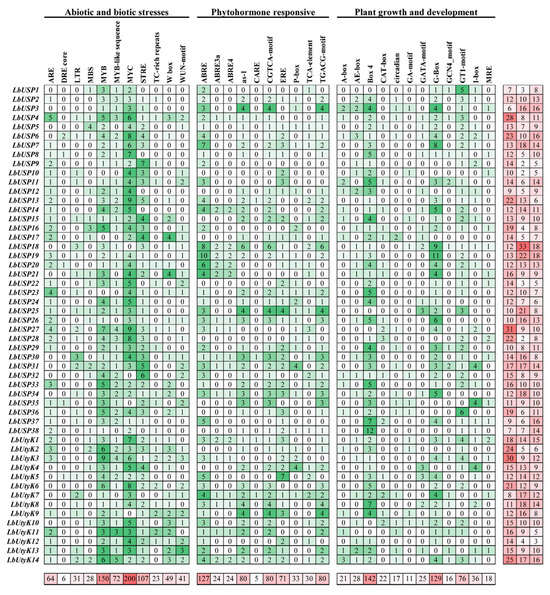
Figure 6.
Analysis of cis-acting elements in LbUSP promoter regions. Binding sites within promoter regions are represented by color-graded boxes indicating motif enrichment strength, and the graph shows the number of binding sites.
An analysis of stress-responsive cis-acting elements in LbUSP promoters identified diverse regulatory motifs associated with oxidation, defense, drought, wounding, heat, and low-temperature responses. The most abundant elements were the general stress-responsive MYB and MYC binding sites, accounting for 19% (154/791) and 26% (206/791) of the total cis-acting elements in the abiotic and biotic stress category, respectively. Additionally, several specialized stress-specific motifs were detected, such as WRKY-box (W-box), TC-rich repeat, and wound-responsive motif (WUN-motif) for wounding and pathogen response; stress-responsive element (STRE) and low-temperature responsive (LTR) motifs for temperature stress; the MYB-binding site (MBS) and the dehydration-responsive element (DRE) core for drought response; and the anaerobic response element (ARE) for anaerobic conditions.
An analysis of phytohormone-responsive cis-acting elements revealed abundant ABRE motifs (ABA responsiveness), CGTCA/TGACG motifs (MeJA responsiveness), and as-1 elements (SA/oxidative stress responsiveness) in LbUSP promoters, collectively accounting for >72% of all hormone-related regulatory elements. Additional motifs included TCA elements for SA response (30/568, 5%), P-boxes for GA signaling (34/568, 6%), and EREs for ethylene regulation (73/568, 13%). In the growth and development-associated category, light-responsive Box-4 (148/553, 27%) and G-box (132/553, 24%) predominated, highlighting the importance of light in regulating USP gene expression in L. barbarum. Some organs specifically expressed related motifs included endosperm-specific GCN4 motifs, meristem-associated CAT-boxes, and seed-specific RY-elements, suggesting potential roles of LbUSPs in modulating the developmental processes of L. barbarum.
3.7. Expression Profiling Analysis of LbUSPs in Response to Salt Stress
To assess the role of LbUSP genes in salt stress response, we analyzed the expression levels of all identified LbUSP members under varying NaCl concentrations (Figure 7). RNA-seq revealed a salt-responsive differential expression of multiple LbUSP genes in the roots (Figure 7A). Specifically, LbUSP7/20/32 were significantly upregulated after salt treatments. LbUSP19/21 were downregulated at 100 mmol/L NaCl and LbUSP12 was significantly suppressed at 200 mmol/L NaCl. The salt-responsive genes in stems differed from those in roots (Figure 7B). The transcript abundance of LbUSP8/34 and LbUtyK9 increased obviously, whereas LbUSP12/15/19 decreased post-treatment. The transcription of LbUSP13/20/25/36 and LbUSP2/5/22 was induced and suppressed significantly only at 200 mmol/L NaCl, respectively. Finally, 25 and 15 LbUSP genes showed no significant transcriptional changes under salt stress in roots and stems, respectively, while 19 and 21 members exhibited negligible expression (FPKM < 1.00) in roots and stems. This suggests that these genes likely do not function as core regulators in the salt stress adaptation of L. barbarum.
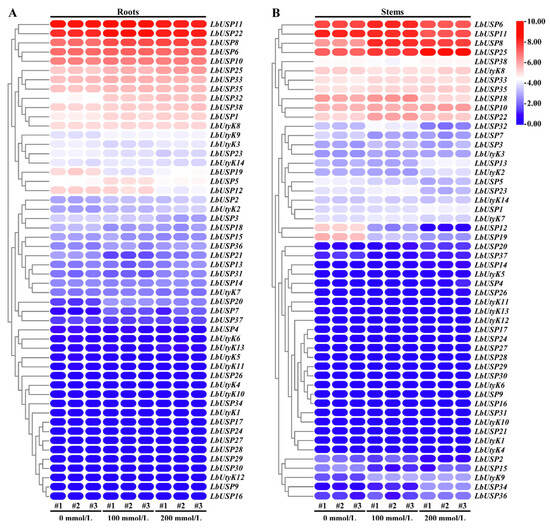
Figure 7.
Expression profiling of LbUSP genes in L. barbarum roots and stems under salt treatments. (A) Expression profiling of LbUSP genes in roots. (B) Expression profiling of LbUSP genes in stems. Samples treated with 0, 100, and 200 mmol/L NaCl, with three biological replicates per group. Heatmaps display log2-transformed expression values; red and blue indicate high and low relative expression levels of target genes, respectively.
To further validate the expression profiling of key LbUSP genes under salt stress, nine candidates (LbUSP3/5/8/11/12/19/23/25/34) showing relatively high expression levels and significant salt stress responses in roots or stems, as identified by transcriptome sequencing, were subjected to RT-qPCR assays (Figure 8). The RT-qPCR results broadly corroborated the transcriptome data. Specifically, salt stress induced the upregulation of LbUSP3/8/25/34 in both roots and stems, whereas LbUSP11/12/19 were downregulated. In contrast, LbUSP5/23 displayed opposing expression patterns between roots and stems, suggesting tissue-specific regulatory mechanisms and potentially divergent functions. These findings demonstrate that these LbUSP genes are most likely participants in the regulation of salt stress in L. barbarum.
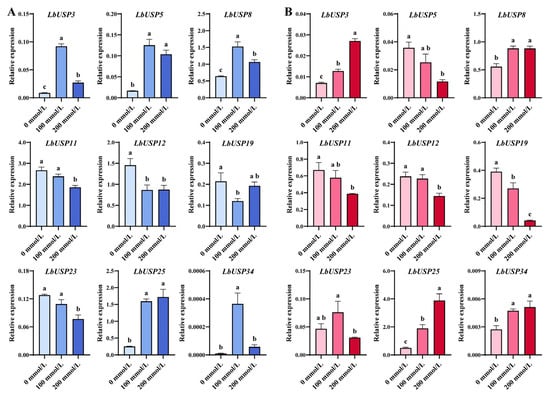
Figure 8.
RT-qPCR validation of LbUSP expression under salt stress. (A) Relative expression levels of LbUSPs in roots. (B) Relative expression levels of LbUSPs in stems. Data represent means ± SD (n = 3). Statistical significance determined by one-way ANOVA with Fisher’s LSD post hoc test (p < 0.05). Distinct letters indicate significant differences between treatment groups.
4. Discussion
L. barbarum, a medicinally and economically significant shrub, exhibits strong ecological adaptability to saline soils in northwestern China. However, intensifying soil salinization threatens its growth and economic value [34,46]. As essential regulators of cellular defense systems against multiple stressors, USPs modulate physiological processes and stress responses in plants [47,48,49]. Thus, we comprehensively characterize the physicochemical properties, phylogenetic relationships, and expression patterns of LbUSP gene family members, providing valuable guidance for elucidating the USP gene family expansion and salt tolerance mechanisms in L. barbarum and for breeding stress-resistant cultivars.
4.1. Molecular Characterization Revealing Diverse Functional Differentiation in L. barbarum USP Family
In this study, a substantial variation in physicochemical properties was observed in LbUSP family members identified by genome-wide analysis (Table 1). The composition of LbUSPs’ exons and introns varies greatly between the groups, but their genetic structures within the same group are similar (Figure 3C), as observed in other plant species [8,22,40,50]. All LbUSP family members retained the conserved USP domain, yet only 13 members possessed motif 2 harboring the ATP-binding sequence [G-2X-G-9X-(S/T)] (Figure S1). These findings demonstrate the structural conservation of LbUSP members within phylogenetic groups, alongside functional diversification across groups in L. barbarum.
The evolutionary integration of the conserved USP domain with diverse catalytic motifs is the primary factor that drives functional diversification in USPs, which is a molecular adaptation critical for enhanced stress tolerance in plants [15]. Notably, 27% of LbUSP family members (i.e., the 14 LbUtyKs) contained an additional PK_Tyr_Ser-Thr kinase domain alongside the conserved USP domain (Figure 3B). Previous research has indicated that PK_Tyr_Ser-Thr functions as constituent domains of high-affinity cell surface receptors, exemplified by receptor-like kinases (RLKs) in wheat, mediating phosphoryl transfer involved in stress resistance pathways [51,52,53]. However, subcellular localization results showing that 11 of 14 LbUtyKs localized to the nucleus (Table 1) imply their primary involvement in nuclear phosphorylation. This suggests that LbUtyKs operate stress responses through mechanisms distinct from those of cell surface receptors. Furthermore, six USPs (LbUSP17/28/37, VvUSP21 and SlUSP18) were found to contain a Na+/H+ exchanger domain (Figure 3B), forming a distinct phylogenetical clade absent in A. thaliana, O. sativa (Figure 2), Populus trichocarpa, Vaccinium corymbosum, G. hirsutum, and T. aestivum [21,27,54,55]. Previous studies have indicated that the integration of the cation transport motif confers upon USPs the capacity to utilize proton gradients for intracellular sodium export in bacteria [11]. The integration of the Na+/H+ exchanger domain enables LbUSP17/28/37 to mediate ion homeostasis and prevent cytotoxic accumulation, which is also consistent with their predicted plasma membrane localization (Table 1). Intriguingly, the identification of a unique MenH superfamily domain in LbUSP23 (Figure 3B) suggested its potential participation in the biosynthesis of phylloquinone, a key electron carrier in photosystem I essential for plant growth [56,57,58], implying that LbUSP23 likely facilitates the modulation of photosynthetic capacity in L. barbarum under stress conditions. Collectively, these findings reveal that the integration of diverse motifs in LbUSP members represents a significant contributor to the high-stress resistance of L. barbarum.
4.2. Gene Duplication-Driven Evolution and Expansion of USP Gene Family in L. barbarum
Phylogenetic analysis revealed a distinct evolutionary divergence between monocot and dicot USP family members (Figure 2). Notably, USP homologs from L. barbarum and S. lycopersicum predominantly clustered on terminal clades, whereas O. sativa USPs exhibited the greatest phylogenetic divergence from both Solanaceous species. Additionally, synteny analysis demonstrated a significantly stronger collinearity of LbUSP genes with dicots than with monocots (Figure 5). All these results illustrate a functional diversification of USP genes in angiosperm lineages [40].
Phylogenetic and collinear analyses further provide insights into USP gene family expansion mechanisms in L. barbarum. Despite possessing a genome nearly twice the size of S. lycopersicum [46,59], L. barbarum harbored comparable USP gene numbers (Figure 2). Additionally, pervasive purifying selection acting on duplicated USP genes was confirmed in L. barbarum (Table 2), aligning with previous reports that USP family diversity is driven primarily by ecological adaptation and evolutionary pressures rather than genome size [27]. Crucially, two specific LbUSP subsets (LbUSP1/33/34/36/38 and LbUSP9/16/24) exhibited different chromosomal distributions, yet they clustered closely in independent clades of a phylogenetic tree with prevalent collinearity and purifying selection (Figure 1, Figure 2 and Figure 4, Table 2). On the other hand, the Ks for LbUSP1/33/34/36/38 paralog pairs ranged from 1.27 to 2.79, significantly exceeding those of LbUSP9/16/24 paralogs (0.53 to 1.05), indicating an ancient duplication event in the former subset versus a recent duplication in the latter [60]. In summary, these findings collectively demonstrate that gene duplication coupled with functional conservation represents the primary mechanism underlying USP family expansion in L. barbarum.
4.3. Mining Key LbUSP Members’ Response to Salt Stress via Expression Profiling in Root and Stem of L. barbarum
Given that high temperatures, drought, salinity, and oxidative stress frequently co-occur and elicit analogous cellular damage in plants [61,62], USP genes generally exhibit functional versatility in adapting to multiple abiotic stressors [8,15]. Even the precise biological roles of LbUSP genes remain undefined, and functional evidence obtained from homologous USPs across diverse plant species shows the potentiality of LbUSPs in salt, drought, and thermal stress responses. For example, AtUSP10 (At2g47710) demonstrated inducted expression under salt and osmotic stresses in A. thaliana [20], SbUSP overexpression conferred enhanced salt and osmotic tolerance in transgenic tobacco [25], and OsUSP1 (designated OsUSP31 herein) augmented rice osmotic tolerance via ethylene modulation [19]. Here, these genes showed the highest homology with LbUSP25 (Figure 2), whose expression was induced significantly in the roots and stems of L. barbarum under salt stress (Figure 7), supporting the evolutionary preservation of USP-mediated stress adaptation mechanisms. On the other hand, functional diversification also exists among USP orthologs. AtUSP17 (At3g53990) negatively regulated salt tolerance in A. thaliana while enhancing cold and oxidative stress resilience [24,63,64,65], but GUSP1/2 and EuUSP1 were drought-responsive genes in Gossypium arboreum and Eucommia ulmoides, respectively [66,67,68]. Although these genes shared ancestry with LbUSP11/12, the expression levels of LbUSP11/12 were suppressed significantly by salt treatment in the roots and stems of L. barbarum, suggesting a negative functionality of LbUSP11/12 in salt stress responses, as is the case with AtUSP17. Crucially, LbUSP11 and LbUSP12 exhibited antagonistic expression dynamics following salt treatment (Figure 7 and Figure 8), implying their divergent regulatory roles in salinity adaptation.
Members of the LbUSP family may additionally contribute to salt stress adaptation through alternative signaling pathways. This mechanism was demonstrated by LbUSP23, a homolog of the oxidative stress-responsive USP domain protein SlRd2 in S. lycopersicum [7]. Under salt stress, LbUSP23 exhibited tissue-specific transcription, being suppressed in roots yet upregulated in stems, suggesting potential involvement in oxidative stress-mediated salt tolerance. Complementary evidence from MsUSPA indicated that its high-homology counterpart LbUSP8 may function in salt resistance via drought-response pathways in L. barbarum [26]. Notably, most kinase domain-containing LbUtyK proteins (excluding LbUtyk9) showed minimal expression changes under salt stress (Figure 7), implying their stress responses may occur primarily through the post-translational modulation of enzyme activity rather than transcriptional regulation.
The divergent expression profiles among LbUSP members likely reflect variations in cis-acting elements within their promoter regions [69]. Our in silico analysis identified multiple salt stress-associated cis-acting elements with notable prevalence across LbUSP promoters, including DRE-core, MYB, TC-rich repeats, W-box, and ABRE motifs [70,71,72,73,74]. Significantly, MYB and MYC motifs showed pronounced enrichment in LbUSP promoters (Figure 6), revealing functional coordination between LbUSP expression and salt stress adaptation. This finding aligns with the established roles of MYB/MYC transcription factors in mediating abiotic stress responses [71,75,76]. Notably, the promoters of five critical genes (LbUSP8/11/12/23/25) exhibited exceptional MYC motif density (≥2.75-fold enrichment over other cis-elements). This abundant MYC binding sites potentially drives enhanced salt tolerance functionality in candidate LbUSPs, highlighting that conserved regulatory modules govern LbUSP transcription under saline conditions.
In summary, the integration of transcriptome sequencing, RT-qPCR validation, and in silico promoter analysis identifies LbUSP8, LbUSP11, LbUSP12, LbUSP23, and LbUSP25 as putative key mediators of salt stress adaptation in L. barbarum. Following functional validation in future studies, these genes could enable the precise development of new salt-tolerant L. barbarum varieties through targeted gene editing, genetic transformation, and/or molecular marker-assisted breeding.
5. Conclusions
In this study, fifty-two USP gene family members were identified within the L. barbarum genome. A phylogenetic classification of 208 USPs resulted in four distinct groups, each characterized by conserved USP domains integrated with divergent functional motifs. Collinear and phylogenetic analyses revealed that gene duplication coupled with functional conservation represents the primary mechanism driving LbUSP family expansion. Integrated RNA-seq, RT-qPCR validation, and in silico promoter analyses indicated LbUSP8, LbUSP11, LbUSP12, LbUSP23, and LbUSP25 as putative key regulators in mediating salt stress adaptation in L. barbarum. A functional validation of these candidate genes through transgenic and gene-editing approaches in L. barbarum must be prioritized in future research. Nevertheless, this study identified critical LbUSP genes responding to salt stress in L. barbarum and established a molecular foundation for future targeted breeding strategies to enhance cultivation efficiency, fruit yield, and medicinal quality.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/horticulturae11080960/s1, Figure S1: Conserved motif signatures of LbUSPs and sequence alignment of LbUSP orthologs containing motif 2. (A) Sequence logos of conserved motifs in LbUSPs. Total stack height reflects sequence conservation at each position; single-letter amino acid abbreviations denote residue identity. (B) Multiple sequence alignment of LbUSP orthologs containing motif 2. Asterisks denote the ATP-binding site G-2X-G-9X-G-(S/T), and the conserved amino acids (G and S/T) are highlighted in red. Background shading indicates conservation levels: pink (100%), blue (≥75%), and green (≥50%); Table S1: The primers used for RT-qPCR in this study; Table S2: Accession numbers and characteristics of 52 LbUSP gene family members in L. barbarum; Table S3: List of USPs in S. lycopersicum, A. thaliana, V. vinifera, and O. sativa; Table S4: Conserved sequences of motifs in LbUSPs; Table S5: The collinear gene pairs between L. barbarum and S. lycopersicum; Table S6: The collinear gene pairs between L. barbarum and A. thaliana; Table S7: The collinear gene pairs between L. barbarum and V. vinifera; Table S8: The collinear gene pairs between L. barbarum and O. sativa; Table S9: Information on cis-acting elements predicted from 2000 bp upstream regions of 52 LbUSP gene family members.
Author Contributions
Conceptualization, J.L. and M.B.; methodology, J.L. and M.B.; software, J.L., M.B. and Y.X.; validation, Y.X.; formal analysis, J.L. and Y.X.; investigation, J.L. and M.B.; resources, J.Z. and Y.C.; data curation, J.L. and J.Z.; writing—original draft preparation, J.L. and M.B.; writing—review and editing, Y.X. and D.M.; visualization, J.L. and Y.X.; supervision, Y.X. and Y.C.; funding acquisition, Y.X., Y.C. and S.L. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Ningxia Natural Science Foundation (grant number 2024AAC02055) and the Key Project at Central Government Level: The Ability Establishment of Sustainable Use for Valuable Chinese Medicine Resources (grant number 2060302).
Data Availability Statement
The original contributions presented in the study are included in the article; further inquiries can be directed to the corresponding author.
Conflicts of Interest
The authors declare no conflicts of interest.
Abbreviations
The following abbreviations are used in this manuscript:
| aa | Amino acid |
| ABA | Abscisic acid |
| ARE | Anaerobic response element |
| DEG | Differentially expressed gene |
| DRE | Dehydration-responsive element |
| GRAVY | Grand average of hydropathicity |
| HMM | Hidden Markov model |
| Ka | Non-synonymous |
| Ks | Synonymous |
| LTR | Low-temperature responsive |
| MBS | MYB-binding site |
| ML | Maximum likelihood |
| MW | Molecular weight |
| PDB | Protein Data Bank |
| pI | Theoretical isoelectric point |
| RLK | Receptor-like kinase |
| ROS | Reactive oxygen species |
| STRE | Stress-responsive element |
| USP | Universal stress protein |
| W-box | WRKY-box |
| WUN-motif | Wound-responsive motif |
References
- Mittler, R.; Zandalinas, S.; Fichman, Y.; Van Breusegem, F. Reactive oxygen species signalling in plant stress responses. Nat. Rev. Mol. Cell Biol. 2022, 23, 663–679. [Google Scholar] [CrossRef]
- Suzuki, N.; Rivero, R.; Shulaev, V.; Blumwald, E.; Mittler, R. Abiotic and biotic stress combinations. New Phytol. 2014, 203, 32–43. [Google Scholar] [CrossRef]
- Zhang, H.; Zhu, J.; Gong, Z.; Zhu, J. Abiotic stress responses in plants. Nat. Rev. Genet. 2022, 23, 104–119. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J. Abiotic stress signaling and responses in plants. Cell 2016, 167, 313–324. [Google Scholar] [CrossRef] [PubMed]
- Osakabe, Y.; Kawaoka, A.; Nishikubo, N.; Osakabe, K. Responses to environmental stresses in woody plants: Key to survive and longevity. J. Plant Res. 2012, 125, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Nyström, T.; Neidhardt, F. Cloning, mapping and nucleotide sequencing of a gene encoding a universal stress protein in Escherichia coli. Mol. Microbiol. 1992, 6, 3187–3198. [Google Scholar] [CrossRef]
- Gutiérrez-Beltrán, E.; Personat, J.; de la Torre, F.; Del Pozo, O. A universal stress protein involved in oxidative stress is a phosphorylation target for protein kinase CIPK6. Plant Physiol. 2017, 173, 836–852. [Google Scholar] [CrossRef]
- Yan, T.; Li, M.; Wang, Q.; Wang, M.; Liu, L.; Ma, C.; Xiang, X.; Zhou, Q.; Liu, Z.; Gong, Z. Structures, functions, and regulatory networks of universal stress proteins in clinically relevant pathogenic bacteria. Cell. Signal. 2024, 116, 111032. [Google Scholar] [CrossRef]
- Aravind, L.; Anantharaman, V.; Koonin, E. Monophyly of class I aminoacyl tRNA synthetase, USPA, ETFP, photolyase, and PP-ATPase nucleotide-binding domains: Implications for protein evolution in the RNA. Proteins 2002, 48, 1–14. [Google Scholar] [CrossRef]
- Sousa, M.; McKay, D. Structure of the universal stress protein of Haemophilus influenzae. Structure 2001, 9, 1135–1141. [Google Scholar] [CrossRef]
- Chi, Y.; Koo, S.; Oh, H.; Lee, E.; Park, J.; Phan, K.; Wi, S.; Bae, S.; Paeng, S.; Chae, H.; et al. The physiological functions of universal stress proteins and their molecular mechanism to protect plants prom environmental stresses. Front. Plant Sci. 2019, 10, 750. [Google Scholar] [CrossRef] [PubMed]
- Gustavsson, N.; Diez, A.; Nyström, T. The universal stress protein paralogues of Escherichia coli are co-ordinately regulated and co-operate in the defence against DNA damage. Mol. Microbiol. 2002, 43, 107–117. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Guo, J.; Jin, X.; Kim, J.; Ji, Y.; Fan, S.; Ha, N.; Quan, C. Crystal structure and functional implications of the tandem-type universal stress protein UspE from Escherichia coli. BMC Struct. Biol. 2016, 16, 3. [Google Scholar] [CrossRef] [PubMed]
- Nachin, L.; Nannmark, U.; Nyström, T. Differential roles of the universal stress proteins of Escherichia coli in oxidative stress resistance, adhesion, and motility. J. Bacteriol. 2005, 187, 6265–6272. [Google Scholar] [CrossRef]
- Nabi, B.; Kumawat, M.; Ahlawat, N.; Ahlawat, S. Molecular, structural, and functional diversity of universal stress proteins (USPs) in bacteria, plants, and their biotechnological applications. Protein J. 2024, 43, 437–446. [Google Scholar] [CrossRef]
- Zarembinski, T.; Hung, L.; Mueller-Dieckmann, H.; Kim, K.; Yokota, H.; Kim, R.; Kim, S. Structure-based assignment of the biochemical function of a hypothetical protein: A test case of structural genomics. Proc. Natl. Acad. Sci. USA 1998, 95, 15189–15193. [Google Scholar] [CrossRef]
- Kvint, K.; Nachin, L.; Diez, A.; Nyström, T. The bacterial universal stress protein: Function and regulation. Curr. Opin. Microbiol. 2003, 6, 140–145. [Google Scholar] [CrossRef]
- Forêt, S.; Seneca, F.; de Jong, D.; Bieller, A.; Hemmrich, G.; Augustin, R.; Hayward, D.; Ball, E.; Bosch, T.; Agata, K.; et al. Phylogenomics reveals an anomalous distribution of USP genes in metazoans. Mol. Biol. Evol. 2011, 28, 153–161. [Google Scholar] [CrossRef]
- Sauter, M.; Rzewuski, G.; Marwedel, T.; Lorbiecke, R. The novel ethylene-regulated gene OsUsp1 from rice encodes a member of a plant protein family related to prokaryotic universal stress proteins. J. Exp. Bot. 2002, 53, 2325–2331. [Google Scholar] [CrossRef]
- Bhuria, M.; Goel, P.; Kumar, S.; Singh, A. Genome-wide identification and expression profiling of genes encoding universal stress proteins (USP) identify multi-stress responsive USP genes in Arabidopsis thaliana. Plant Physiol. Rep. 2019, 24, 434–445. [Google Scholar] [CrossRef]
- Li, Y.; Zheng, A.; Li, Z.; Wang, H.; Wang, J.; Dong, Z.; Yao, L.; Han, X.; Wei, F. Characterization and gene expression analysis reveal universal stress proteins respond to abiotic stress in Gossypium hirsutum. BMC Genom. 2024, 25, 98. [Google Scholar] [CrossRef]
- Qi, T.; He, F.; Zhang, X.; Wang, J.; Zhang, Z.; Jiang, H.; Zhao, B.; Du, C.; Che, Y.; Feng, X.; et al. Genome-wide identification and expression profiling of potato (Solanum tuberosum L.) universal stress proteins reveal essential roles in mechanical damage and deoxynivalenol stress. Int. J. Mol. Sci. 2024, 25, 1341. [Google Scholar] [CrossRef]
- Cui, X.; Zhang, P.; Hu, Y.; Chen, C.; Liu, Q.; Guan, P.; Zhang, J. Genome-wide analysis of the Universal stress protein A gene family in Vitis and expression in response to abiotic stress. Plant Physiol. Biochem. 2021, 165, 57–70. [Google Scholar] [CrossRef] [PubMed]
- Bhuria, M.; Goel, P.; Kumar, S.; Singh, A. AtUSP17 negatively regulates salt stress tolerance through modulation of multiple signaling pathways in Arabidopsis. Physiol. Plant. 2022, 174, e13635. [Google Scholar] [CrossRef] [PubMed]
- Udawat, P.; Jha, R.; Sinha, D.; Mishra, A.; Jha, B. Overexpression of a cytosolic abiotic stress responsive universal stress protein (SbUSP) mitigates salt and osmotic stress in transgenic tobacco plants. Front. Plant Sci. 2016, 7, 518. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Che, S.; Zhang, Y.; Wang, H.; Wei, T.; Yan, G.; Song, W.; Yu, W. Universal stress protein in Malus sieversii confers enhanced drought tolerance. J. Plant Res. 2019, 132, 825–837. [Google Scholar] [CrossRef]
- Singh, A.; Singhal, C.; Sharma, A.; Khurana, P. Identification of universal stress proteins in wheat and functional characterization during abiotic stress. Plant Cell Rep. 2023, 42, 1487–1501. [Google Scholar] [CrossRef]
- Varshney, R.; Chen, W.; Li, Y.; Bharti, A.; Saxena, R.; Schlueter, J.; Donoghue, M.; Azam, S.; Fan, G.; Whaley, A.; et al. Draft genome sequence of pigeonpea (Cajanus cajan), an orphan legume crop of resource-poor farmers. Nat. Biotechnol. 2012, 30, 83–89. [Google Scholar] [CrossRef]
- Zong, N.; Zhao, H.; Liu, K.; Lu, R.; Fan, Y.; Miao, M. Study on tomato USP1 in response to drought and high temperature stress. J. Anhui Agric. Univ. 2021, 48, 916–922. [Google Scholar] [CrossRef]
- Song, Y.; Ma, B.; Guo, Q.; Zhou, L.; Zhou, X.; Ming, Z.; You, H.; Zhang, C. MYB pathways that regulate UV-B-induced anthocyanin biosynthesis in blueberry (Vaccinium corymbosum). Front. Plant Sci. 2023, 14, 1125382. [Google Scholar] [CrossRef]
- Lenman, M.; Sörensson, C.; Andreasson, E. Enrichment of phosphoproteins and phosphopeptide derivatization identify universal stress proteins in elicitor-treated Arabidopsis. Mol. Plant Microbe Interact. 2008, 21, 1275–1284. [Google Scholar] [CrossRef]
- Merkouropoulos, G.; Andreasson, E.; Hess, D.; Boller, T.; Peck, S.C. An Arabidopsis protein phosphorylated in response to microbial elicitation, AtPHOS32, is a substrate of MAP kinases 3 and 6. J. Biol. Chem. 2008, 283, 10493–10499. [Google Scholar] [CrossRef] [PubMed]
- Chou, M.; Wei, X.; Chen, D.; Zhou, J. A novel nodule-enhanced gene encoding a putative universal stress protein from Astragalus sinicus. J. Plant Physiol. 2007, 164, 764–772. [Google Scholar] [CrossRef]
- Ma, R.; Zhang, X.; Ni, Z.; Thakur, K.; Wang, W.; Yan, Y.; Cao, Y.; Zhang, J.; Rengasamy, K.; Wei, Z. Lycium barbarum (Goji) as functional food: A review of its nutrition, phytochemical structure, biological features, and food industry prospects. Crit. Rev. Food Sci. Nutr. 2023, 63, 10621–10635. [Google Scholar] [CrossRef] [PubMed]
- Ni, Y.; Hu, Y.; Zhu, L.; Jiang, X.; Zhang, H.; Liu, J.; Zhao, Y. Lycium barbarum polysaccharide-derived nanoparticles dprotect visual function by inhibiting RGC ferroptosis and microglial activation in retinal ischemia-reperfusion mice. Adv. Healthc. Mater. 2024, 13, e2304285. [Google Scholar] [CrossRef]
- He, C.; Shi, X.; Lin, H.; Li, Q.; Xia, F.; Shen, G.; Feng, J. The combination of HSI and NMR techniques with deep learning for identification of geographical origin and GI markers of Lycium barbarum L. Food Chem. 2024, 461, 140903. [Google Scholar] [CrossRef]
- Zhang, T.; Dong, Q.; Zhan, X.; He, J.; Feng, H. Moving salts in an impermeable saline-sodic soil with drip irrigation to permit wolfberry production. Agric. Water Manag. 2019, 213, 636–645. [Google Scholar] [CrossRef]
- Sun, R.; Zhang, F.; Song, J.; Luo, C.; Li, K.; Wang, Y.; Chen, L. Research progress on the role of microorganisms in the remediation of saline-alkali land. Guangdong Agric. Sci. 2025, 52, 14–30. [Google Scholar]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y.; et al. TBtools-II: A “one for all, all for one” bioinformatics platform for biological big-data mining. Mol. Plant 2023, 16, 1733–1742. [Google Scholar] [CrossRef]
- Arabia, S.; Sami, A.; Akhter, S.; Sarker, R.; Islam, T. Comprehensive in silico characterization of universal stress proteins in rice (Oryza sativa L.) with insight into their stress-specific transcriptional modulation. Front. Plant Sci. 2021, 12, 712607. [Google Scholar] [CrossRef]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Li, H.; Huang, W.; Liu, X.; Zhu, X.; Ren, X.; An, W.; Zhou, J.; Zhao, J. Screening and validation of reference genes for real-time quantitative PCR in Lycium. Jiangsu Agric. Sci. 2023, 51, 41–51. [Google Scholar] [CrossRef]
- Livak, K.; Schmittgen, T. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Hurst, L. The Ka/Ks ratio: Diagnosing the form of sequence evolution. Trends Genet. 2002, 18, 486–487. [Google Scholar] [CrossRef]
- Mengarelli, D.; Zanor, M. Genome-wide characterization and analysis of the CCT motif family genes in soybean (Glycine max). Planta 2021, 253, 15. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Xu, Y.; Li, H.; An, W.; Yin, Y.; Wang, B.; Wang, L.; Wang, B.; Duan, L.; Ren, X.; et al. Metabolite-based genome-wide association studies enable the dissection of the genetic bases of flavonoids, betaine and spermidine in wolfberry (Lycium). Plant Biotechnol. J. 2024, 22, 1435–1452. [Google Scholar] [CrossRef] [PubMed]
- Sen, S.; Rai, R.; Chatterjee, A.; Rai, S.; Yadav, S.; Agrawal, C.; Rai, L. Molecular characterization of two novel proteins All1122 and Alr0750 of Anabaena PCC 7120 conferring tolerance to multiple abiotic stresses in Escherichia coli. Gene 2019, 685, 230–241. [Google Scholar] [CrossRef]
- Phan, K.; Paeng, S.; Chae, H.; Park, J.; Lee, E.; Wi, S.; Bae, S.; Kim, M.; Yun, D.; Kim, W.; et al. Universal stress protein regulates the circadian rhythm of central oscillator genes in Arabidopsis. FEBS Lett. 2022, 596, 1871–1880. [Google Scholar] [CrossRef]
- Cui, X.; Zhang, P.; Chen, C.; Zhang, J. VyUSPA3, a universal stress protein from the Chinese wild grape Vitis yeshanensis, confers drought tolerance to transgenic V. vinifera. Plant Cell Rep. 2023, 42, 181–196. [Google Scholar] [CrossRef]
- Fan, M.; Gao, S.; Yang, Y.; Yang, S.; Wang, H.; Shi, L. Genome-wide identification and expression analysis of the universal stress protein (USP) gene family in Arabidopsis thaliana, Zea mays, and Oryza sativa. Genetica 2024, 152, 119–132. [Google Scholar] [CrossRef]
- Cao, H.; Tian, Q.; Ju, M.; Duan, Y.; Li, G.; Ma, Q.; Zhang, H.; Zhang, X.; Miao, H. Genome-wide analysis of the U-box E3 ubiquitin ligase family role in drought tolerance in sesame (Sesamum indicum L.). Front. Plant Sci. 2023, 14, 1261238. [Google Scholar] [CrossRef] [PubMed]
- Lu, C.; Jiang, J.; Chen, Q.; Liu, H.; Ju, X.; Wang, H. Analysis and prediction of interactions between transmembrane and non-transmembrane proteins. BMC Genom. 2024, 25, 401. [Google Scholar] [CrossRef] [PubMed]
- Yan, J.; Su, P.; Meng, X.; Liu, P. Phylogeny of the plant receptor-like kinase (RLK) gene family and expression analysis of wheat RLK genes in response to biotic and abiotic stresses. BMC Genom. 2023, 24, 224. [Google Scholar] [CrossRef] [PubMed]
- Diao, J.; Gu, W.; Jiang, Z.; Wang, J.; Zou, H.; Zong, C.; Ma, L. Comprehensive analysis of universal stress protein family genes and their expression in Fusarium oxysporum response of Populus davidiana × P. alba var. pyramidalis Louche based on the transcriptome. Int. J. Mol. Sci. 2023, 24, 5405. [Google Scholar] [CrossRef]
- Song, Y.; Ma, B.; Feng, X.; Guo, Q.; Zhou, L.; Zhang, X.; Zhang, C. Genome-wide analysis of the universal stress protein gene family in blueberry and their transcriptional responses to UV-B irradiation and abscisic acid. Int. J. Mol. Sci. 2023, 24, 16819. [Google Scholar] [CrossRef]
- Gross, J.; Cho, W.; Lezhneva, L.; Falk, J.; Krupinska, K.; Shinozaki, K.; Seki, M.; Herrmann, R.; Meurer, J. A plant locus essential for phylloquinone (Vitamin K1) biosynthesis originated from a fusion of four Eubacterial Genes. J. Biol. Chem. 2006, 281, 17189–17196. [Google Scholar] [CrossRef]
- Wang, L.; Li, Q.; Zhang, A.; Zhou, W.; Jiang, R.; Yang, Z.; Yang, H.; Qin, X.; Ding, S.; Lu, Q.; et al. The phytol phosphorylation pathway is essential for the biosynthesis of phylloquinone, which is required for photosystem I stability in Arabidopsis. Mol. Plant 2017, 10, 183–196. [Google Scholar] [CrossRef][Green Version]
- van Oostende, C.; Widhalm, J.; Furt, F.; Ducluzeau, A.; Basset, G. Chapter 6-vitamin K1 (phylloquinone): Function, enzymes and genes. Adv. Bot. Res. 2011, 59, 229–261. [Google Scholar] [CrossRef]
- Li, N.; He, Q.; Wang, J.; Wang, B.; Zhao, J.; Huang, S.; Yang, T.; Tang, Y.; Yang, S.; Aisimutuola, P.; et al. Super-pangenome analyses highlight genomic diversity and structural variation across wild and cultivated tomato species. Nat. Genet. 2023, 55, 852–860. [Google Scholar] [CrossRef]
- Zhang, R.; Wang, F.; Zhang, J.; Shang, H.; Liu, L.; Wang, H.; Zhao, G.; Shen, H.; Yan, Y.H. Dating whole genome duplication in Ceratopteris thalictroides and potential adaptive values of retained gene duplicates. Int. J. Mol. Sci. 2019, 20, 1926. [Google Scholar] [CrossRef]
- Wang, P.; Liu, W.; Han, C.; Wang, S.; Bai, M.; Song, C. Reactive oxygen species: Multidimensional regulators of plant adaptation to abiotic stress and development. J. Integr. Plant Biol. 2024, 66, 330–367. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Yang, Z.; Zhang, Z.; Li, Y.; Guo, J.; Liu, L.; Wang, C.; Fan, H.; Wang, B.; Han, G. Root hair development and adaptation to abiotic stress. J. Agric. Food Chem. 2023, 71, 9573–9598. [Google Scholar] [CrossRef] [PubMed]
- Jung, Y.; Melencion, S.; Lee, E.; Park, J.; Alinapon, C.; Oh, H.; Yun, D.; Chi, Y.; Lee, S. Universal stress protein exhibits a redox-dependent chaperone function in Arabidopsis and enhances plant tolerance to heat shock and oxidative stress. Front. Plant Sci. 2015, 6, 1141. [Google Scholar] [CrossRef] [PubMed]
- Melencion, S.; Chi, Y.; Pham, T.; Paeng, S.; Wi, S.; Lee, C.; Ryu, S.; Koo, S.; Lee, S. RNA chaperone function of a universal stress protein in Arabidopsis confers enhanced cold stress tolerance in plants. Int. J. Mol. Sci. 2017, 18, 2546. [Google Scholar] [CrossRef]
- Bhuria, M.; Goel, P.; Kumar, S.; Singh, A. The promoter of AtUSP is co-regulated by phytohormones and abiotic stresses in Arabidopsis thaliana. Front. Plant Sci. 2016, 7, 1957. [Google Scholar] [CrossRef]
- Maqbool, A.; Zahur, M.; Husnain, T.; Riazuddin, S. GUSP1 and GUSP2, two drought-responsive genes in Gossypium arboreum have homology to universal stress proteins. Plant Mol. Biol. Rep. 2009, 27, 109–114. [Google Scholar] [CrossRef]
- Hafeez, M.; Khan, M.; Sarwar, B.; Hassan, S.; Ali, Q.; Husnain, T.; Rashid, B. Mutant Gossypium universal stress protein-2 (GUSP-2) gene confers resistance to various abiotic stresses in E. coli BL-21 and CIM-496-Gossypium hirsutum. Sci. Rep. 2021, 11, 20466. [Google Scholar] [CrossRef]
- Hu, P.; Zhao, D. Cloning and functional analysis of universal stress protein gene EuUSP1 in Eucommia ulmoides. Plant Physiol. J. 2024, 60, 130–140. [Google Scholar] [CrossRef]
- Oudelaar, A.; Higgs, D.R. The relationship between genome structure and function. Nat. Rev. Genet. 2021, 22, 154–168. [Google Scholar] [CrossRef]
- Zhou, M.; Ma, J.; Zhao, Y.; Wei, Y.; Tang, Y.; Wu, Y. Improvement of drought and salt tolerance in Arabidopsis and Lotus corniculatus by overexpression of a novel DREB transcription factor from Populus euphratica. Gene 2012, 506, 10–17. [Google Scholar] [CrossRef]
- Zhang, D.; Zhou, H.; Zhang, Y.; Zhao, Y.; Zhang, Y.; Feng, X.; Lin, H. Diverse roles of MYB transcription factors in plants. J. Integr. Plant Biol. 2025, 67, 539–562. [Google Scholar] [CrossRef]
- Alabd, A.; Cheng, H.; Ahmad, M.; Wu, X.; Peng, L.; Wang, L.; Yang, S.; Bai, S.; Ni, J.; Teng, Y. ABRE-BINDING FACTOR3-WRKY DNA-BINDING PROTEIN44 module promotes salinity-induced malate accumulation in pear. Plant Physiol. 2023, 192, 1982–1996. [Google Scholar] [CrossRef]
- Han, Q.; Chen, K.; Yan, D.; Hao, G.; Qi, J.; Wang, C.; Dirk, L.; Bruce Downie, A.; Gong, J.; Wang, J.; et al. ZmDREB2A regulates ZmGH3.2 and ZmRAFS, shifting metabolism towards seed aging tolerance over seedling growth. Plant J. 2020, 104, 268–282. [Google Scholar] [CrossRef] [PubMed]
- She, M.; Zheng, D.; Zhang, S.; Ke, Z.; Wu, Z.; Zou, H.; Zhang, Z. Functional analysis of maize GRAS transcription factor gene ZmGRAS72 in response to drought and salt stresses. Agric. Commun. 2024, 2, 100054. [Google Scholar] [CrossRef]
- Li, Z.; Huang, Y.; Shen, Z.; Wu, M.; Huang, M.; Hong, S.; Xu, L.; Zang, Y. Advances in functional studies of plant MYC transcription factors. Theor. Appl. Genet. 2024, 137, 195. [Google Scholar] [CrossRef]
- Wu, T.; Al-Mamun, H.; Edwards, D.; Batley, J.; Dolatabadian, A. Genome-wide identification and prediction of disease resistance genes in Hirschfeldia incana. Agric. Commun. 2024, 2, 100049. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).