Vv14-3-3ω Is a Susceptible Factor for Grapevine Downy Mildew
Abstract
1. Introduction
2. Materials and Methods
2.1. Phylogenetic Analysis
2.2. Bacterial Strains and Plasmids
2.3. Plant Materials, Growth Conditions, and Transient Gene Expression in Plants
2.4. Quantitative Reverse Transcription PCR
2.5. ROS Production Assay
2.6. Pathogenicity Assay
2.7. Western Blot Analysis
2.8. Subcellular Localization Assay
2.9. VIGS Assay in N. benthamiana
3. Results
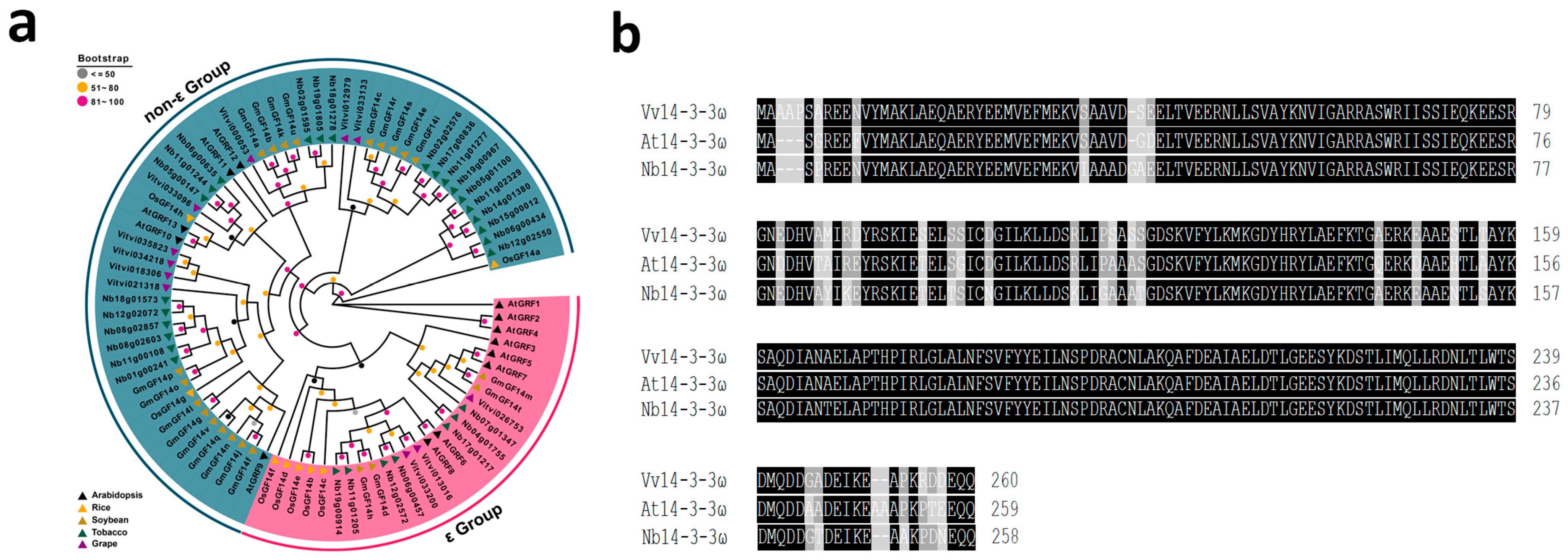
3.1. Phylogenetic Analysis of Vv14-3-3 Isoforms
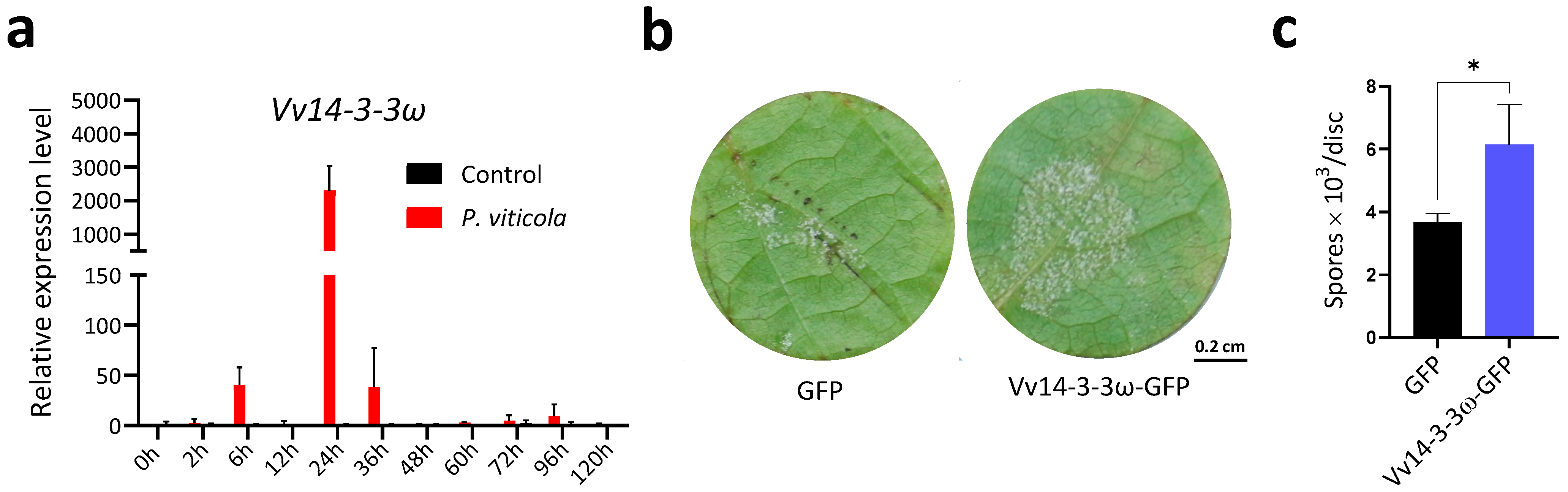
3.2. Vv14-3-3ω Promotes P. viticola Sporulation on Grape Leaves
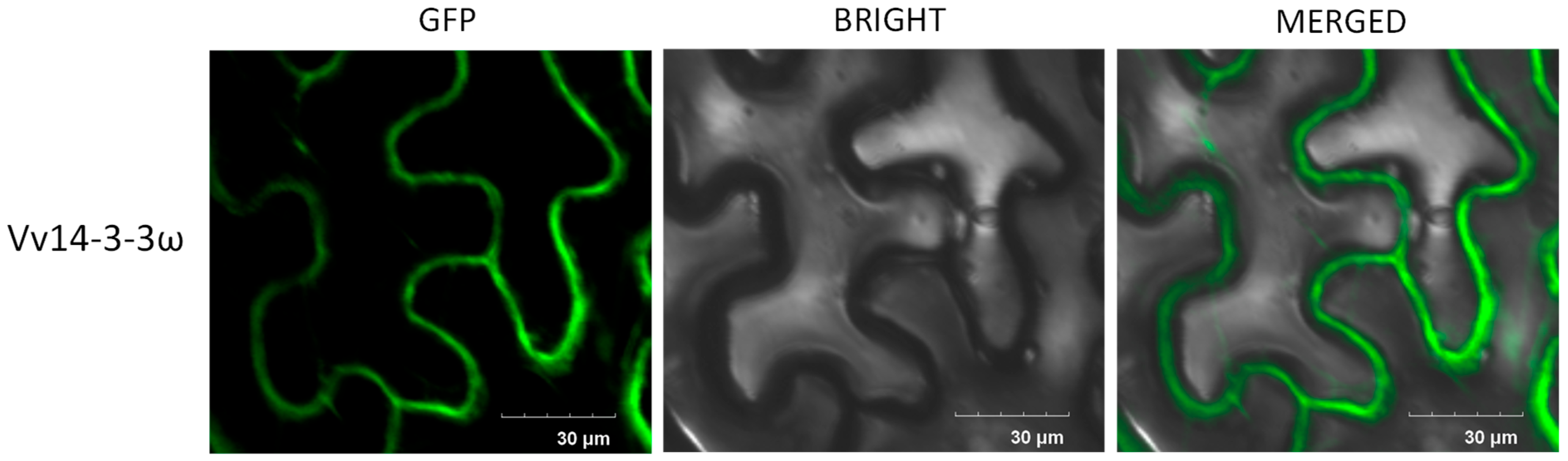
3.3. Vv14-3-3ω Is Localized to the Plasma Membrane and Cytoplasm and Enhances P. capsici Pathogenicity in N. benthamiana
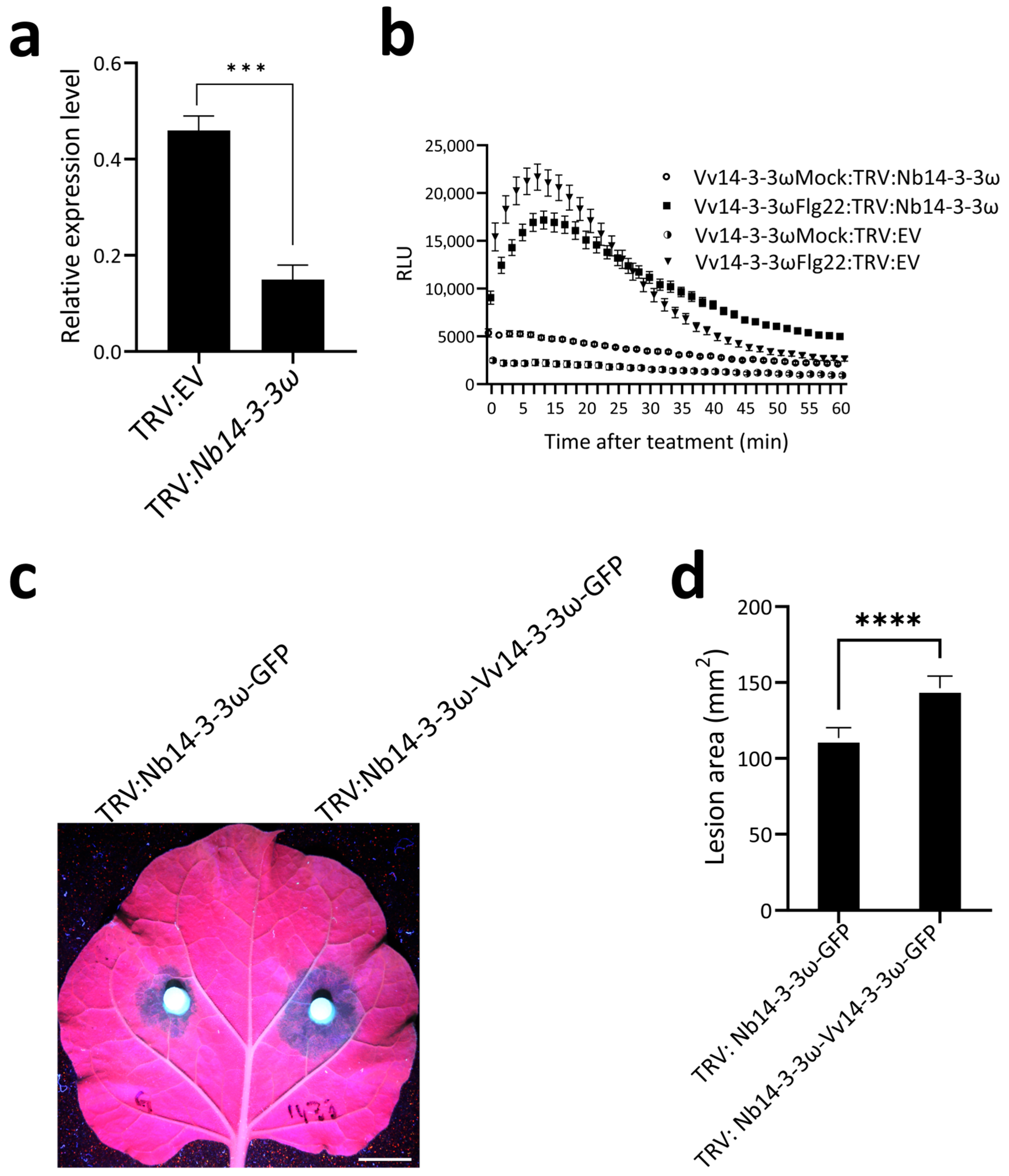
3.4. Vv14-3-3ω Suppresses Flg22-Induced Immune Responses in N. benthamiana
3.5. Overexpression of Vv14-3-3ω in Nb14-3-3ω Silenced Leaves Suppress Immunity
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Gessler, C.; Pertot, I.; Perazzolli, M. Plasmopara viticola: A review of knowledge on downy mildew of grapevine and effective disease management. Phytopathol. Mediterr. 2011, 50, 3–44. [Google Scholar]
- Wang, X.; Tu, M.; Wang, Y.; Zhang, Y.; Yin, W.; Fang, J.; Gao, M.; Li, Z.; Zhan, W.; Fang, Y.; et al. Telomere-to-telomere and gap-free genome assembly of a susceptible grapevine species (Thompson Seedless) to facilitate grape functional genomics. Hortic. Res. 2023, 11, uhad260. [Google Scholar] [CrossRef]
- Dussert, Y.; Mazet, I.D.; Couture, C.; Gouzy, J.; Piron, M.C.; Kuchly, C.; Bouchez, O.; Rispe, C.; Mestre, P.; Delmotte, F. A high-quality grapevine downy mildew genome assembly reveals rapidly evolving and lineage-specific putative host adaptation genes. Genome Biol. Evol. 2019, 11, 954–969. [Google Scholar] [CrossRef]
- Rizzo, D.M.; Garbelotto, M.; Hansen, E.M. Phytophthora ramorum: Integrative research and management of an emerging pathogen in California and Oregon forests. Annu. Rev. Phytopathol. 2005, 43, 309–335. [Google Scholar] [CrossRef] [PubMed]
- Haas, B.J.; Kamoun, S.; Zody, M.C.; Jiang, R.H.; Handsaker, R.E.; Cano, L.M.; Grabherr, M.; Kodira, C.D.; Raffaele, S.; Torto-Alalibo, T.; et al. Genome sequence and analysis of the Irish potato famine pathogen Phytophthora infestans. Nature 2009, 461, 393–398. [Google Scholar] [CrossRef] [PubMed]
- Depotter, J.R.; Doehlemann, G. Target the core: Durable plant resistance against filamentous plant pathogens through effector recognition. Pest Manag. Sci. 2020, 76, 426–431. [Google Scholar] [CrossRef]
- He, Q.; McLellan, H.; Boevink, P.C.; Birch, P.R. All roads lead to susceptibility: The many modes of action of fungal and oomycete intracellular effectors. Plant Commun. 2020, 1, 100050. [Google Scholar] [CrossRef]
- Li, Q.; Wang, J.; Bai, T.; Zhang, M.; Jia, Y.; Shen, D.; Zhang, M.; Dou, D. A Phytophthora capsici effector suppresses plant immunity via interaction with EDS1. Mol. Plant Pathol. 2020, 21, 502–511. [Google Scholar] [CrossRef]
- Li, T.; Wang, Q.; Feng, R.; Li, L.; Ding, L.; Fan, G.; Li, W.; Du, Y.; Zhang, M.; Huang, G.; et al. Negative regulators of plant immunity derived from cinnamyl alcohol dehydrogenases are targeted by multiple Phytophthora Avr3a-like effectors. New Phytol. 2019, nph.16139. [Google Scholar] [CrossRef]
- Moore, B.W. Specific acidic proteins of the nervous system. Physiol. Biochem. Asp. Neverous Integr. 1967, 343–349. [Google Scholar]
- Camoni, L.; Visconti, S.; Aducci, P.; Marra, M. 14-3-3 proteins in plant hormone signaling: Doing several things at once. Front. Plant Sci. 2018, 9, 297. [Google Scholar] [CrossRef]
- Lozano-Durán, R.; Robatzek, S. 14-3-3 proteins in plant-pathogen interactions. Mol. Plant-Microbe Interact. 2015, 28, 511–518. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.; Ylä-Anttila, P.; Sandalova, T.; Sun, R.; Achour, A.; Masucci, M.G. 14-3-3 scaffold proteins mediate the inactivation of trim25 and inhibition of the type I interferon response by herpesvirus deconjugases. PLoS Pathog. 2019, 15, e1008146. [Google Scholar] [CrossRef] [PubMed]
- Harada, K.-I.; Furuita, K.; Yamashita, E.; Taoka, K.I.; Tsuji, H.; Fujiwara, T.; Nakagawa, A.; Kojima, C. Crystal structure of potato 14-3-3 protein St14f revealed the importance of helix I in StFDL1 recognition. Sci. Rep. 2022, 12, 11596. [Google Scholar] [CrossRef] [PubMed]
- Knetsch, M.L.; van Heusden, G.P.H.; Ennis, H.L.; Shaw, D.R.; Epskamp, S.J.; Snaar-Jagalska, B.E. Isolation of a Dictyostelium discoideum 14-3-3 homologue. Biochim. Biophys. Acta Mol. Cell Res. 1997, 1357, 243–248. [Google Scholar] [CrossRef]
- Lalle, M.; Salzano, A.M.; Crescenzi, M.; Pozio, E. The Giardia duodenalis 14-3-3 protein is post-translationally modified by phosphorylation and polyglycylation of the C-terminal tail. J. Biol. Chem. 2006, 281, 5137–5148. [Google Scholar] [CrossRef]
- Ferl, R.J.; Manak, M.S.; Reyes, M.F. The 14-3-3s. Genome Biol. 2002, 3, reviews3010-1. [Google Scholar] [CrossRef]
- Cheng, C.; Wang, Y.; Chai, F.; Li, S.; Xin, H.; Liang, Z. Genome-wide identification and characterization of the 14–3-3 family in Vitis vinifera L. during berry development and cold-and heat-stress response. BMC Genom. 2018, 19, 579. [Google Scholar] [CrossRef]
- Keicher, J.; Jaspert, N.; Weckermann, K.; Möller, C.; Throm, C.; Kintzi, A.; Oecking, C. Arabidopsis 14-3-3 epsilon members contribute to polarity of PIN auxin carrier and auxin transport-related development. elife 2017, 6, e24336. [Google Scholar] [CrossRef]
- Hermeking, H. The 14-3-3 cancer connection. Nat. Rev. Cancer 2003, 3, 931–943. [Google Scholar] [CrossRef]
- Oh, C.-S.; Martin, G.B. Tomato 14-3-3 protein TFT7 interacts with a MAP kinase kinase to regulate immunity-associated programmed cell death mediated by diverse disease resistance proteins. J. Biol. Chem. 2011, 286, 14129–14136. [Google Scholar] [CrossRef]
- Kim, J.-G.; Li, X.; Roden, J.A.; Taylor, K.W.; Aakre, C.D.; Su, B.; Lalonde, S.; Kirik, A.; Chen, Y.; Baranage, G.; et al. Xanthomonas T3S effector XopN suppresses PAMP-triggered immunity and interacts with a tomato atypical receptor-like kinase and TFT1. Plant Cell 2009, 21, 1305–1323. [Google Scholar] [CrossRef]
- Mukhtar, M.S.; Carvunis, A.R.; Dreze, M.; Epple, P.; Steinbrenner, J.; Moore, J.; Tasan, M.; Galli, M.; Hao, T.; Nishimura, M.T.; et al. Independently evolved virulence effectors converge onto hubs in a plant immune system network. Science 2011, 333, 596–601. [Google Scholar] [CrossRef] [PubMed]
- Seo, Y.-E.; Yan, X.; Choi, D.; Mang, H. Phytophthora infestans RxLR effector PITG06478 hijacks 14-3-3 to suppress PMA activity leading to necrotrophic cell death. Mol. Plant Microbe Interact. 2023, 36, 150–158. [Google Scholar] [CrossRef] [PubMed]
- Evangelisti, E.; Guyon, A.; Shenhav, L.; Schornack, S. FIRE mimics a 14-3-3-binding motif to promote Phytophthora palmivora infection. Mol. Plant Microbe Interact. 2023, 36, 315–322. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Ling, L.; Jiang, Z.; Tan, W.; Liu, Z.; Wu, L.; Zhao, Y.; Xia, S.; Ma, J.; Wang, G.; et al. Genome-wide identification and expression analysis of the 14-3-3 gene family in soybean (Glycine max). PeerJ 2019, 7, e7950. [Google Scholar] [CrossRef]
- Chen, W.; Yan, M.; Chen, S.; Sun, J.; Wang, J.; Meng, D.; Li, J.; Zhang, L.; Guo, L. The complete genome assembly of Nicotiana benthamiana reveals the genetic and epigenetic landscape of centromeres. Nat. Plants 2024, 10, 1928–1943. [Google Scholar] [CrossRef]
- Shi, X.; Cao, S.; Wang, X.; Huang, S.; Wang, Y.; Liu, Z.; Liu, W.; Leng, X.; Peng, Y.; Wang, N.; et al. The complete reference genome for grapevine (Vitis vinifera L.) genetics and breeding. Hortic. Res. 2023, 10, uhad061. [Google Scholar] [CrossRef]
- Shidore, T.; Broeckling, C.D.; Kirkwood, J.S.; Long, J.J.; Miao, J.; Zhao, B.; Leach, J.E.; Triplett, L.R. The effector AvrRxo1 phosphorylates NAD in planta. PLoS Pathog. 2017, 13, e1006442. [Google Scholar] [CrossRef]
- Ma, T.; Chen, S.; Liu, J.; Fu, P.; Wu, W.; Song, S.; Gao, Y.; Ye, W.; Lu, J. Plasmopara viticola effector PvRXLR111 stabilizes VvWRKY40 to promote virulence. Mol. Plant Pathol. 2021, 22, 231–242. [Google Scholar] [CrossRef]
- Lan, X.; Liu, Y.; Song, S.; Yin, L.; Xiang, J.; Qu, J.; Lu, J. Plasmopara viticola effector PvRXLR131 suppresses plant immunity by targeting plant receptor-like kinase inhibitor BKI1. Mol. Plant Pathol. 2019, 20, 765–783. [Google Scholar] [CrossRef]
- Visconti, S.; D’Ambrosio, C.; Fiorillo, A.; Arena, S.; Muzi, C.; Zottini, M.; Aducci, P.; Marra, M.; Scaloni, A.; Camoni, L. Overexpression of 14-3-3 proteins enhances cold tolerance and increases levels of stress-responsive proteins of Arabidopsis plants. Plant Sci. 2019, 289, 110215. [Google Scholar] [CrossRef] [PubMed]
- Rosenquist, M.; Alsterfjord, M.; Larsson, C.; Sommarin, M. Data mining the Arabidopsis genome reveals fifteen 14-3-3 genes. Expression is demonstrated for two out of five novel genes. Plant Physiol. 2001, 127, 142–149. [Google Scholar] [CrossRef] [PubMed]
- DeLille, J.M.; Sehnke, P.C.; Ferl, R.J. The Arabidopsis 14-3-3 family of signaling regulators. Plant Physiol. 2001, 126, 35–38. [Google Scholar] [CrossRef] [PubMed]
- Yao, Y.; Du, Y.; Jiang, L.; Liu, J.Y. Molecular analysis and expression patterns of the 14-3-3 gene family from Oryza sativa. BMB Rep. 2007, 40, 349–357. [Google Scholar] [CrossRef]
- Xu, W.F.; Shi, W. Expression profiling of the 14-3-3 gene family in response tosalt stress and potassium and iron deficiencies in young tomato (Solanum lycopersicum) roots: Analysi sby real-time RT-PCR. Ann. Bot. 2006, 98, 965–974. [Google Scholar] [CrossRef]
- Li, X.; Dhaubhadel, S. Soybean 14-3-3 gene family: Identification and molecular characterization. Planta 2011, 233, 569–582. [Google Scholar] [CrossRef]
- Wilson, R.S.; Swatek, K.N.; Thelen, J.J. Regulation of the regulators: Post-translational modifications, subcellular, and spatiotemporal distribution of plant 14-3-3 proteins. Front. Plant Sci. 2016, 7, 611. [Google Scholar] [CrossRef]
- Henriksson, M.L.; Francis, M.S.; Peden, A.; Aili, M.; Stefansson, K.; Palmer, R.; Aitken, A.; Hallberg, B. A nonphosphorylated 14-3-3 binding motif on exoenzyme S that is functional in vivo. Eur. J. Biochem. 2002, 269, 4921–4929. [Google Scholar] [CrossRef]
- Würtele, M.; Jelich-Ottmann, C.; Wittinghofer, A.; Oecking, C. Structural view of a fungal toxin acting on a 14-3-3 regulatory complex. EMBO J. 2003, 22, 987–994. [Google Scholar] [CrossRef]
- Lu, W.; Deng, F.; Jia, J.; Chen, X.; Li, J.; Wen, Q.; Li, T.; Meng, Y.; Shan, W. The Arabidopsis thaliana gene AtERF019 negatively regulates plant resistance to Phytophthora parasitica by suppressing PAMP-triggered immunity. Mol. Plant Pathol. 2020, 21, 1179–1193. [Google Scholar] [CrossRef]
- Jones, J.D.; Dangl, J.L. The plant immune system. Nature 2006, 444, 323–329. [Google Scholar] [CrossRef]
- Turnbull, D.; Yang, L.; Naqvi, S.; Breen, S.; Welsh, L.; Stephens, J.; Morris, J.; Boevink, P.C.; Hedley, P.E.; Zhan, J.; et al. RXLR effector AVR2 up-regulates a brassinosteroid-responsive bHLH transcription factor to suppress immunity. Plant Physiol. 2017, 174, 356–369. [Google Scholar] [CrossRef]
- Schornack, S.; Huitema, E.; Cano, L.M.; Bozkurt, T.O.; Oliva, R.; Van Damme, M.; Schwizer, S.; Raffaele, S.; Chaparro-Garcia, A.; Farrer, R.; et al. Ten things to know about oomycete effectors. Mol. Plant Pathol. 2009, 10, 795–803. [Google Scholar] [CrossRef]
- Kamoun, S. A catalogue of the effector secretome of plant pathogenic oomycetes. Annu. Rev. Phytopathol. 2006, 44, 41–60. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Li, Y.; Hu, Y.; Yang, Y.; Zhang, W.; He, M.; Li, X.; Zhang, C.; Kong, F.; Liu, X.; et al. EDS1-interacting J protein 1 is an essential negative regulator of plant innate immunity in Arabidopsis. Plant Cell 2021, 33, 153–171. [Google Scholar] [CrossRef] [PubMed]
- Mengarelli, D.A.; Roldán Tewes, L.; Balazadeh, S.; Zanor, M.I. FITNESS acts as a negative regulator of immunity and influences the plant reproductive output after Pseudomonas syringae infection. Front. Plant Sci. 2021, 12, 606791. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Babar, Z.; Khan, A.; Liu, J.; Fu, P.; Lu, J. Vv14-3-3ω Is a Susceptible Factor for Grapevine Downy Mildew. Horticulturae 2025, 11, 1199. https://doi.org/10.3390/horticulturae11101199
Babar Z, Khan A, Liu J, Fu P, Lu J. Vv14-3-3ω Is a Susceptible Factor for Grapevine Downy Mildew. Horticulturae. 2025; 11(10):1199. https://doi.org/10.3390/horticulturae11101199
Chicago/Turabian StyleBabar, Zainib, Asaf Khan, Jiaqi Liu, Peining Fu, and Jiang Lu. 2025. "Vv14-3-3ω Is a Susceptible Factor for Grapevine Downy Mildew" Horticulturae 11, no. 10: 1199. https://doi.org/10.3390/horticulturae11101199
APA StyleBabar, Z., Khan, A., Liu, J., Fu, P., & Lu, J. (2025). Vv14-3-3ω Is a Susceptible Factor for Grapevine Downy Mildew. Horticulturae, 11(10), 1199. https://doi.org/10.3390/horticulturae11101199





