Comparative Genomic Analysis of Wild Cymbidium Species from Fujian Using Whole-Genome Resequencing
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection and Whole-Genome Resequencing of Wild Cymbidium Species
2.2. Variant Detection and Annotation
2.3. Genetic Evolution Analysis
3. Results
3.1. Whole-Genome Resequencing and Variant Detection of Cymbidium Species in Fujian
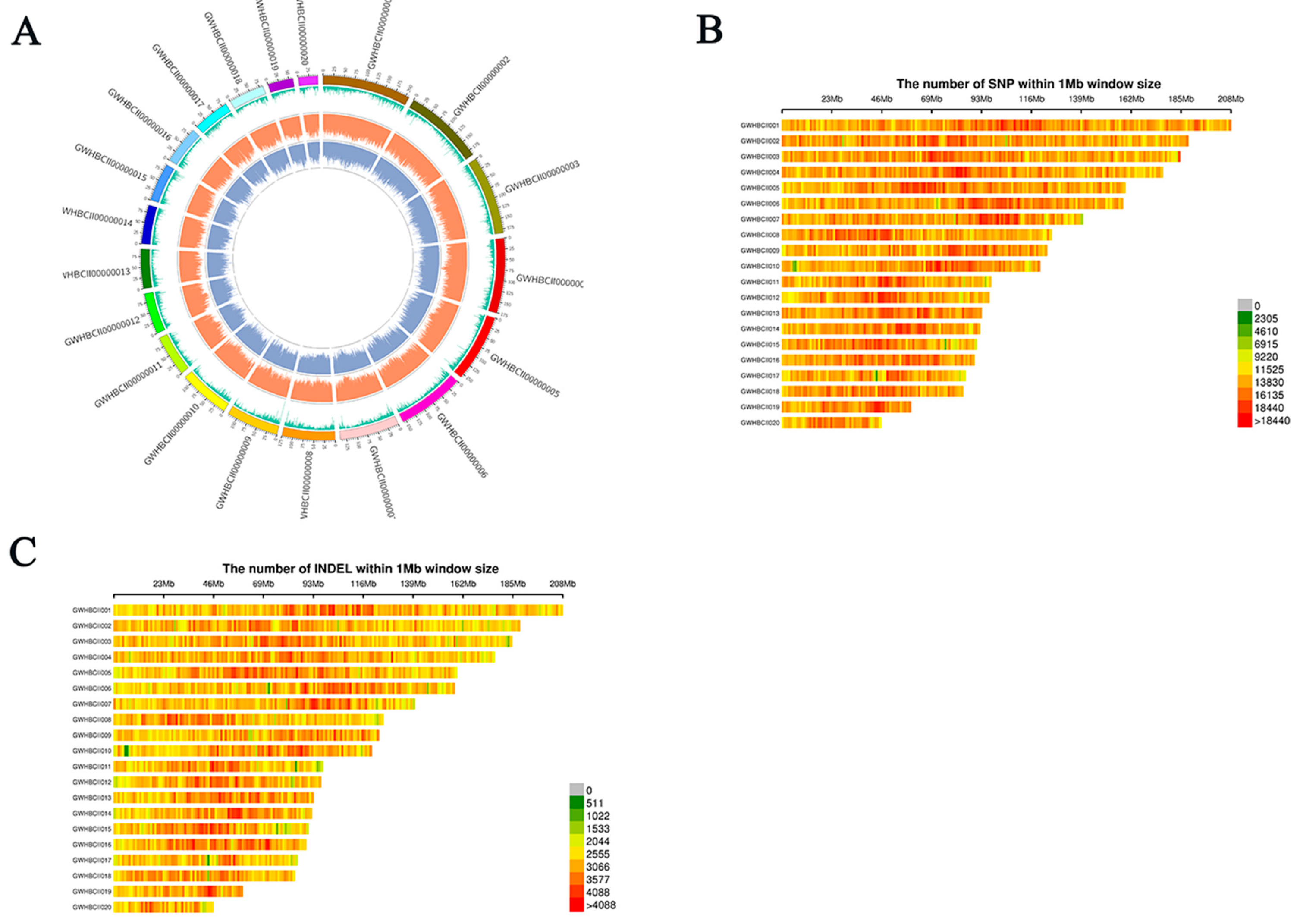
3.2. Genomic Distribution of Indel and SNP Variants in Cymbidium Species from Fujian
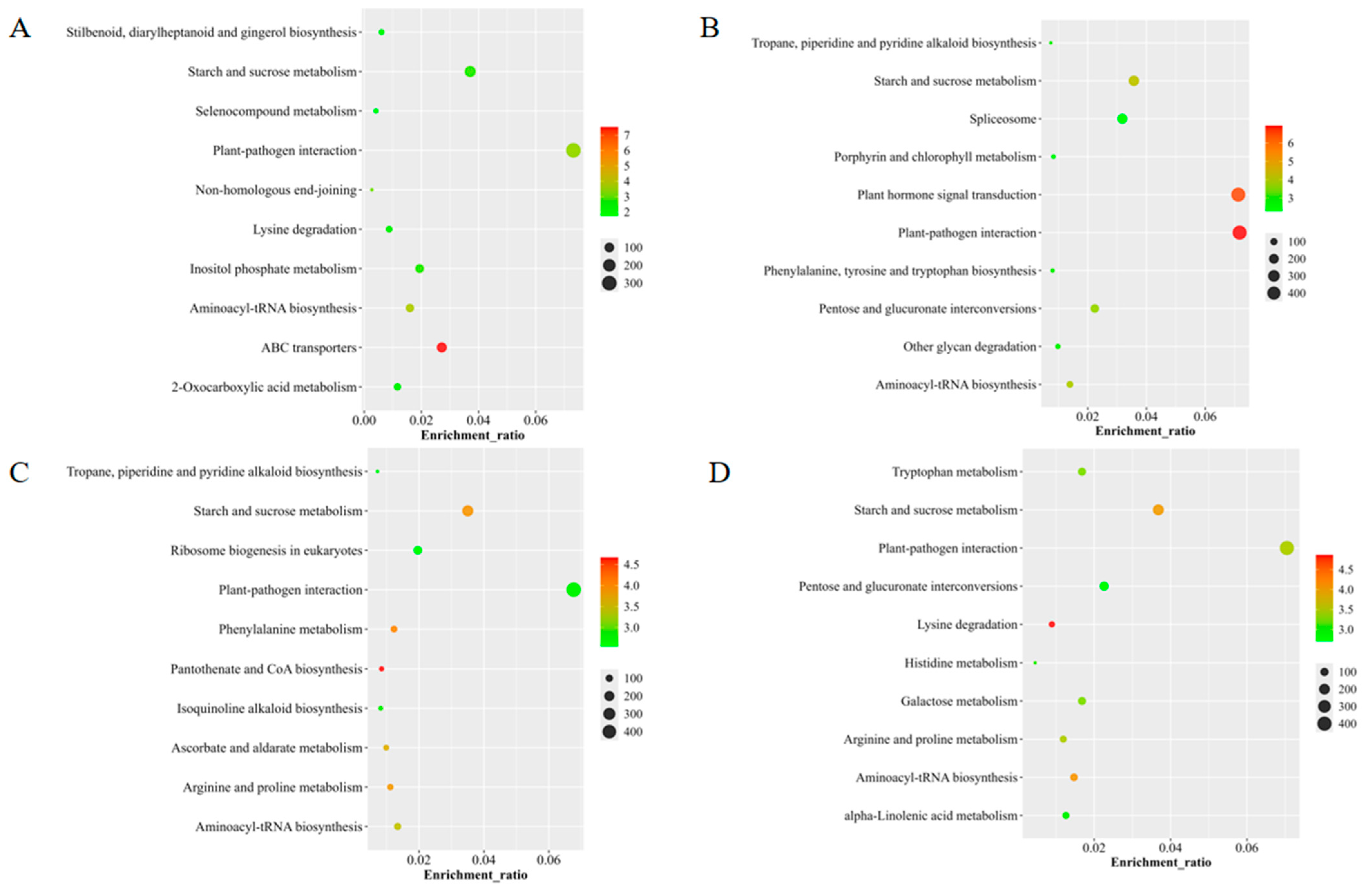
3.3. Functional Characterization of Variant Genes in Four Wild Cymbidium Species from Fujian Based on KEGG Enrichment Analysis
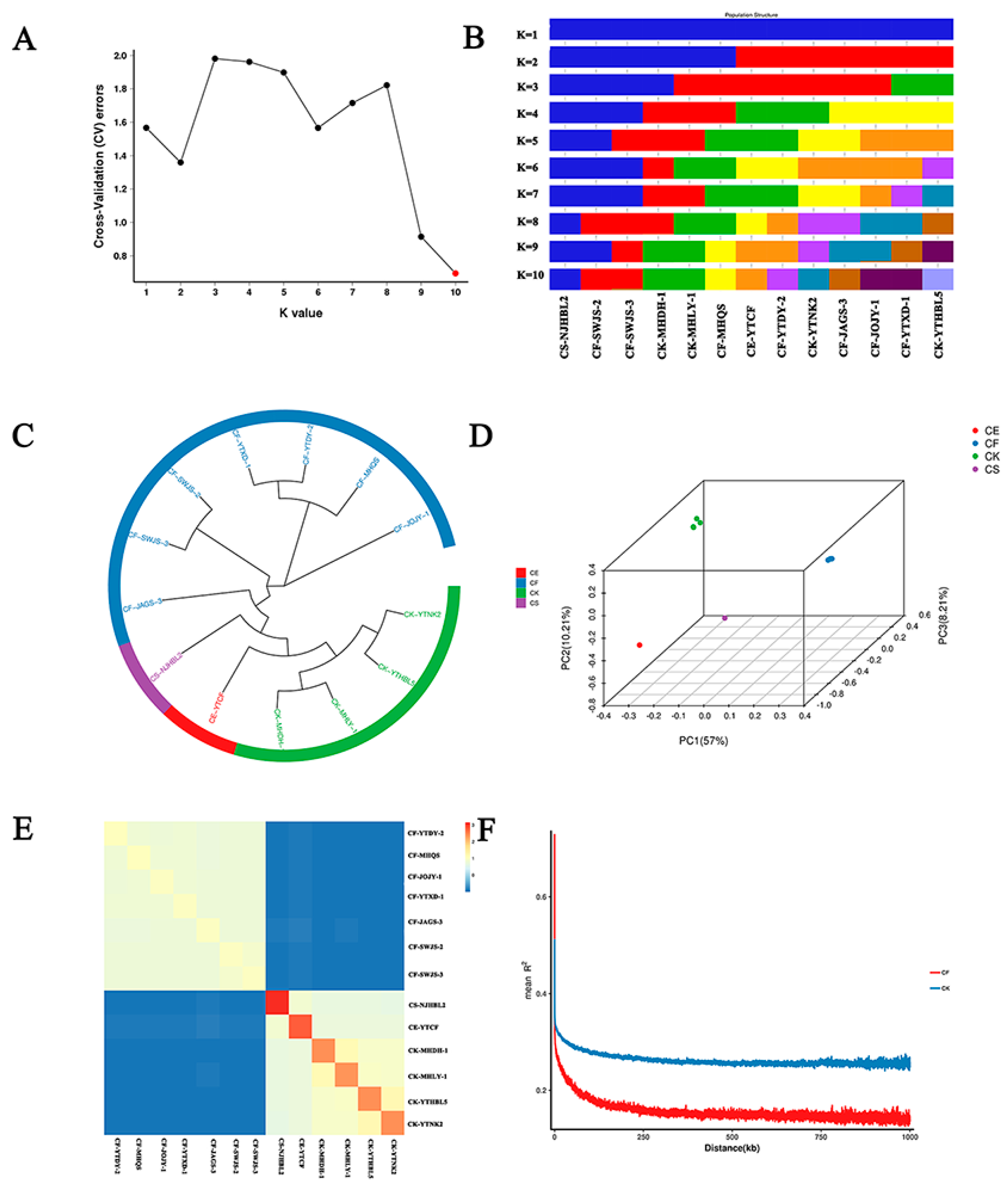
3.4. Population Structure and Genetic Evolution Analysis of Four Wild Cymbidium Species in Fujian
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Fay, M.F.; Pailler, T.; Dixon, K.W. Orchid conservation: Making the links. Ann. Bot. 2015, 116, 377–379. [Google Scholar] [CrossRef]
- Zhang, M.; Zhang, X.; Ge, C.; Chen, B. Danxiaorchis mangdangshanensis (Orchidaceae, Epidendroideae), a new species from central Fujian Province based on morphological and genomic data. PhytoKeys 2022, 212, 37–55. [Google Scholar] [CrossRef]
- Liu, J.; Lan, S.; He, B.; Liang, Y. Bulbophyllum pingnanense (Orchidaceae, Epidendroideae, Dendrobiinae), a new species from Fujian, China. PhytoKeys 2016, 65, 107–112. [Google Scholar] [CrossRef]
- Fay, M.F. Orchid conservation: How can we meet the challenges in the twenty-first century? Bot. Stud. 2018, 59, 16. [Google Scholar] [CrossRef] [PubMed]
- Chhina, A.K.; Abhari, N.; Mooers, A.; Lewthwaite, J.M.M. Linking the spatial and genomic structure of adaptive potential for conservation management: A review. Genome 2024, 67, 403–423. [Google Scholar] [CrossRef] [PubMed]
- Meienberg, J.; Bruggmann, R.; Oexle, K.; Matyas, G. Clinical sequencing: Is WGS the better WES? Hum. Genet. 2016, 135, 359–362. [Google Scholar] [CrossRef]
- Tysklind, N.; Blanc-Jolivet, C.; Mader, M.; Meyer-Sand, B.R.V.; Paredes-Villanueva, K.; Honorio Coronado, E.N.; García-Dávila, C.R.; Sebbenn, A.M.; Caron, H.; Troispouxet, V.; et al. Development of nuclear and plastid SNP and INDEL markers for population genetic studies and timber traceability of Carapa species. Conserv. Genet. Resour. 2019, 11, 337–339. [Google Scholar] [CrossRef]
- Liu, S.; Xu, Y.; Yang, K.; Huang, Y.; Lu, Z.; Chen, S.; Gao, X.; Xiao, G.; Chen, P.; Zeng, X.; et al. Origin and de novo domestication of sweet orange. Nat. Genet. 2025, 57, 754–762. [Google Scholar] [CrossRef]
- Kong, W.; Kong, X.; Xia, Z.; Li, X.; Wang, F.; Shan, R.; Chen, Z.; You, X.; Zhao, Y.; Hu, Y.; et al. Genomic analysis of 1,325 Camellia accessions sheds light on agronomic and metabolic traits for tea plant improvement. Nat. Genet. 2025, 57, 997–1007. [Google Scholar] [CrossRef]
- Cai, J.; Liu, X.; Vanneste, K.; Proost, S.; Tsai, W.; Liu, K.; Chen, L.-J.; He, Y.; Xu, Q.; Bian, C.; et al. The genome sequence of the orchid Phalaenopsis equestris. Nat. Genet. 2015, 47, 65–72. [Google Scholar] [CrossRef]
- Yang, L.; Zhou, R.; Xue, T.; Yuan, Y. Evolutionary Conservation and Expression Patterns of Flavonoid Biosynthetic Genes in Dendrobium catenatum. Russ. J. Plant Physiol. 2023, 70, 157. [Google Scholar] [CrossRef]
- Liang, H.; Wang, X.; Chen, W.; Hu, S.; Yang, F.; Yang, X. Spatial genetic structure of terrestrial orchid Cymbidium faberi in the Qinling Mountains revealed by microsatellite loci. Plant Syst. Evol. 2021, 307, 5. [Google Scholar] [CrossRef]
- Wang, H.; Lu, J.; Hu, X.; Liu, J. Genetic variation and cultivar identification in Cymbidium ensifolium. Plant Syst. Evol. 2011, 293, 101–110. [Google Scholar] [CrossRef]
- Sun, Y.; Chen, G.; Huang, J.; Liu, D.; Xue, F.; Chen, X.; Chen, S.-Q.; Liu, C.-G.; Liu, H.; Ma, H.; et al. The Cymbidium goeringii genome provides insight into organ development and adaptive evolution in orchids. Ornam. Plant Res. 2021, 1, 10. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, B.; Zhang, S.; Wong, C.E.; Liang, Q.; Pang, S.; Wu, Y.; Zhao, M.; Yu, H. Pangeneric genome analyses reveal the evolution and diversity of the orchid genus Dendrobium. Nat. Plants 2025, 11, 421–437. [Google Scholar] [CrossRef]
- Hoffmeister, T.S.; Vet, L.E.M.; Biere, A.; Holsinger, K.; Filser, J. Ecological and Evolutionary Consequences of Biological Invasion and Habitat Fragmentation. Ecosystems 2005, 8, 657–667. [Google Scholar] [CrossRef]
- Yang, F.; Gao, J.; Li, J.; Wei, Y.; Xie, Q.; Jin, J.; Lu, C.; Zhu, W.; Wong, S.-M.; Zhu, G. The China orchid industry: Past and future perspectives. Ornam. Plant Res. 2024, 4, e002. [Google Scholar] [CrossRef]
- Schenk, J.J.; Becklund, L.E.; Carey, S.J.; Fabre, P.P. What is the “modified” CTAB protocol? Characterizing modifications to the CTAB DNA extraction protocol. Appl. Plant Sci. 2023, 11, e11517. [Google Scholar] [CrossRef]
- Dong, S.; Li, X.; Liu, Q.; Zhu, T.; Tian, A.; Chen, N.; Tu, X. Comparative genomics uncovers evolutionary drivers of locust migratory adaptation. BMC Genom. 2025, 26, 203. [Google Scholar] [CrossRef]
- Zhu, K.; Du, P.; Xiong, J.; Ren, X.; Sun, C.; Tao, Y.; Ding, Y.; Xu, Y.; Meng, H.; Wang, C.-C.; et al. Comparative Performance of the MGISEQ-2000 and Illumina X-Ten Sequencing Platforms for Paleogenomics. Front. Genet. 2021, 12, 745508. [Google Scholar] [CrossRef]
- Ai, Y.; Li, Z.; Sun, W.; Chen, J.; Zhang, D.; Ma, L.; Chen, M.-K.; Zheng, Q.-D.; Liu, J.-F.; Jiang, Y.-T.; et al. The Cymbidium genome reveals the evolution of unique morphological traits. Hortic. Res. 2021, 8, 255. [Google Scholar] [CrossRef] [PubMed]
- Jung, Y.; Han, D. BWA-MEME: BWA-MEM emulated with a machine learning approach. Bioinformatics 2022, 38, 2404–2413. [Google Scholar] [CrossRef]
- Van der Auwera Geraldine, A.; Carneiro, M.O.; Hartl, C.; Poplin, R.; Del Angel, G.; Levy-Moonshine, A.; Jordan, T.; Shakir, K.; Roazen, D.; Thibault, J.; et al. From FastQ data to high confidence variant calls: The Genome Analysis Toolkit best practices pipeline. Curr. Protoc. Bioinform. 2013, 43, 11.10.1–11.10.33. [Google Scholar] [CrossRef]
- Bu, D.; Luo, H.; Huo, P.; Wang, Z.; Zhang, S.; He, Z.; Wu, Y.; Zhao, L.; Liu, J.; Guo, J.; et al. KOBAS-i: Intelligent prioritization and exploratory visualization of biological functions for gene enrichment analysis. Nucleic Acids Res. 2021, 49, W317–W325. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Lawson, D.J.; Van Dorp, L.; Falush, D. A tutorial on how not to over-interpret STRUCTURE and ADMIXTURE bar plots. Nat. Commun. 2018, 9, 3258. [Google Scholar] [CrossRef]
- Herrando-Pérez, S.; Tobler, R.; Huber, C.D. Smartsnp, an r package for fast multivariate analyses of big genomic data. Methods Ecol. Evol. 2021, 12, 2084–2093. [Google Scholar] [CrossRef]
- Ong, J.; MacGregor, S. Implementing MR-PRESSO and GCTA-GSMR for pleiotropy assessment in Mendelian randomization studies from a practitioner’s perspective. Genet. Epidemiol. 2019, 43, 609–616. [Google Scholar] [CrossRef]
- Zhang, C.; Dong, S.; Xu, J.; He, W.; Yang, T. PopLDdecay: A fast and effective tool for linkage disequilibrium decay analysis based on variant call format files. Bioinformatics 2019, 35, 1786–1788. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.; Muse, S.V. PowerMarker: An integrated analysis environment for genetic marker analysis. Bioinformatics 2005, 21, 2128–2129. [Google Scholar] [CrossRef] [PubMed]
- Peakall, R.; Smouse, P.E. GenAlEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research—An update. Bioinformatics 2012, 28, 2537–2539. [Google Scholar] [CrossRef] [PubMed]
- Danecek, P.; Auton, A.; Abecasis, G.; Albers, C.A.; Banks, E.; DePristo, M.A.; Handsaker, R.E.; Lunter, G.; Marth, G.T.; Sherry, S.T.; et al. The variant call format and VCFtools. Bioinformatics 2011, 27, 2156–2158. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.C.; Chow, C.C.; Tellier, L.C.; Vattikuti, S.; Purcell, S.M.; Lee, J.J. Second-generation PLINK: Rising to the challenge of larger and richer datasets. Gigascience 2015, 4, 7. [Google Scholar] [CrossRef]
- Yang, X.; Dong, X.; Li, K.; Zhang, Y.; Xu, C.; Huo, H.; Tian, L.; Xu, J.; Liu, C.; Qi, D.; et al. The Genetic Diversity, Population Structure, and Historical Dynamics of Wild Pyrus Species on the Yunnan–Kweichow Plateau. Horticulturae 2025, 11, 106. [Google Scholar] [CrossRef]
- Li, X.; Gao, W.; Guo, H.; Zhang, X.; Fang, D.D.; Lin, Z. Development of EST-based SNP and InDel markers and their utilization in tetraploid cotton genetic mapping. BMC Genom. 2014, 15, 1046. [Google Scholar] [CrossRef]
- Feng, H.; Du, Q.; Jiang, Y.; Jia, Y.; He, T.; Wang, Y.; Chapman, B.; Yu, J.; Zhang, H.; Gu, M.; et al. Hordeum I genome unlocks adaptive evolution and genetic potential for crop improvement. Nat. Plants 2025, 11, 438–4352. [Google Scholar] [CrossRef]
- Nocchi, G.; Whiting, J.R.; Yeaman, S. Repeated global adaptation across plant species. Proc. Natl. Acad. Sci. USA 2024, 121, e2406832121. [Google Scholar] [CrossRef]
- Li, X.; Li, W.; Di, C.; Xie, M.; Jin, L.; Huang, C.; Wu, D. Development of Genic Simple Sequence Repeat Panels for Population Classification of Chinese Cymbidium Species. J. Am. Soc. Hortic. Sci. 2016, 141, 125–130. [Google Scholar] [CrossRef]
- Chen, X.; Peng, D.; Lan, S.; Chen, J.; Fu, W. Characterization of the complete chloroplast genome of Paphiopedilum micranthum, an Endangered orchid in China. Mitochondrial DNA Part B Resour. 2020, 5, 115–116. [Google Scholar] [CrossRef]
- Li, X.; Xu, W.; Chowdhury, M.R.; Jin, F. Comparative proteomic analysis of labellum and inner lateral petals in Cymbidium ensifolium flowers. Int. J. Mol. Sci. 2014, 15, 19877–19897. [Google Scholar] [CrossRef]
- Yang, F.; Gao, J.; Wei, Y.; Ren, R.; Zhang, G.; Lu, C.-Q.; Jin, J.-P.; Ai, Y.; Wang, Y.-Q.; Chen, L.-J.; et al. The genome of Cymbidium sinense revealed the evolution of orchid traits. Plant Biotechnol. J. 2021, 19, 2501–2516. [Google Scholar] [CrossRef]
- Zhou, Z.; Ying, Z.; Wu, Z.; Yang, Y.; Fu, S.; Xu, W.; Yao, L.; Zeng, A.; Huang, J.; Lan, S.; et al. Anthocyanin Genes Involved in the Flower Coloration Mechanisms of Cymbidium kanran. Front. Plant Sci. 2021, 12, 737815. [Google Scholar] [CrossRef]
- Ren, R.; Wei, Y.; Ahmad, S.; Jin, J.; Gao, J.; Lu, C.; Zhu, G.; Yang, F. Identification and Characterization of NPR1 and PR1 Homologs in Cymbidium orchids in Response to Multiple Hormones, Salinity and Viral Stresses. Int. J. Mol. Sci. 2020, 21, 1977. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Hu, X.; Liu, J.; Wang, H. Genetic diversity and population structure of 151 Cymbidium sinense cultivars. Afr. J. Wood Sci. For. 2013, 1, 104–114. [Google Scholar]
- Wu, Q.; Dong, S.; Zhao, Y.; Yang, L.; Qi, X.; Ren, Z.; Dong, S.; Cheng, J. Genetic diversity, population genetic structure and gene flow in the rare and endangered wild plant Cypripedium macranthos revealed by genotyping-by-sequencing. BMC Plant Biol. 2023, 23, 254. [Google Scholar] [CrossRef]
- Morrell, P.L.; Buckler, E.S.; Ross-Ibarra, J. Crop genomics: Advances and applications. Nat. Rev Genet. 2012, 13, 85–96. [Google Scholar] [CrossRef]
- Huang, X.; Wei, X.; Sang, T.; Zhao, Q.; Feng, Q.; Zhao, Y.; Li, C.; Zhu, C.; Lu, T.; Zhang, Z.; et al. Genome-wide association studies of 14 agronomic traits in rice landraces. Nat. Genet. 2010, 42, 961–967. [Google Scholar] [CrossRef]
- Yan, J.; Shah, T.; Warburton, M.L.; Buckler, E.S.; McMullen, M.D.; Crouch, J. Genetic characterization and linkage disequilibrium estimation of a global maize collection using SNP markers. PLoS ONE 2009, 4, e8451. [Google Scholar] [CrossRef]
- Liu, J.; Huang, J.; Guo, H.; Lan, L.; Wang, H.; Xu, Y.; Yang, X.; Li, W.; Tong, H.; Xiao, Y.; et al. The Conserved and Unique Genetic Architecture of Kernel Size and Weight in Maize and Rice. Plant Physiol. 2017, 175, 774–785. [Google Scholar] [CrossRef]
- Geppert, C.; Perazza, G.; Wilson, R.J.; Bertolli, A.; Prosser, F.; Melchiori, G.; Marini, L. Consistent population declines but idiosyncratic range shifts in Alpine orchids under global change. Nat. Commun. 2020, 11, 5835. [Google Scholar] [CrossRef] [PubMed]

| Sample ID | Species | Total Reads | Q30 (%) | GC (%) | Mapped (%) | Coverage_1X (%) |
|---|---|---|---|---|---|---|
| CE-YTCF | C. ensifolium | 182,181,610 | 97.98 | 34.81 | 99.26 | 78.63 |
| CF-JAGS-3 | C. floribundum | 179,063,054 | 98.03 | 34.38 | 96.98 | 60.93 |
| CF-JOJY-1 | C. floribundum | 182,426,590 | 97.98 | 35.62 | 94.52 | 60.44 |
| CF-MHQS | C. floribundum | 179,429,906 | 98.33 | 35.36 | 97.60 | 60 |
| CF-SWJS-2 | C. floribundum | 177,184,418 | 98.02 | 34.85 | 97.64 | 59.82 |
| CF-SWJS-3 | C. floribundum | 179,751,908 | 97.86 | 33.64 | 97.26 | 60.55 |
| CF-YTDY-2 | C. floribundum | 182,340,752 | 98.30 | 34.66 | 97.73 | 60.06 |
| CF-YTXD-1 | C. floribundum | 180,713,392 | 97.78 | 34.40 | 96.86 | 60.46 |
| CK-MHDH-1 | C. kanran | 182,680,742 | 97.90 | 35.38 | 99.14 | 74.49 |
| CK-MHLY-1 | C. kanran | 182,416,930 | 98.06 | 34.94 | 99.26 | 74.42 |
| CK-YTHBL5 | C. kanran | 181,081,042 | 97.82 | 35.18 | 99.14 | 73.90 |
| CK-YTNK2 | C. kanran | 176,434,476 | 98.29 | 38.11 | 98.83 | 70.35 |
| CS-NJHBL2 | C. sinense | 181,733,734 | 98.07 | 34.95 | 98.82 | 74.01 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, X.; Chen, B.; El-Kassaby, Y.A.; Zhang, J.; Zhang, L.; Liu, S.; Huang, Y.; Li, J.; Lin, Z.; Xie, W.; et al. Comparative Genomic Analysis of Wild Cymbidium Species from Fujian Using Whole-Genome Resequencing. Horticulturae 2025, 11, 944. https://doi.org/10.3390/horticulturae11080944
Xu X, Chen B, El-Kassaby YA, Zhang J, Zhang L, Liu S, Huang Y, Li J, Lin Z, Xie W, et al. Comparative Genomic Analysis of Wild Cymbidium Species from Fujian Using Whole-Genome Resequencing. Horticulturae. 2025; 11(8):944. https://doi.org/10.3390/horticulturae11080944
Chicago/Turabian StyleXu, Xinyu, Bihua Chen, Yousry A. El-Kassaby, Juan Zhang, Lanqi Zhang, Sijia Liu, Yu Huang, Junnan Li, Zhiyong Lin, Weiwei Xie, and et al. 2025. "Comparative Genomic Analysis of Wild Cymbidium Species from Fujian Using Whole-Genome Resequencing" Horticulturae 11, no. 8: 944. https://doi.org/10.3390/horticulturae11080944
APA StyleXu, X., Chen, B., El-Kassaby, Y. A., Zhang, J., Zhang, L., Liu, S., Huang, Y., Li, J., Lin, Z., Xie, W., Wu, J., Lai, Z., Huang, X., Huang, J., Wu, W., & Shen, L. (2025). Comparative Genomic Analysis of Wild Cymbidium Species from Fujian Using Whole-Genome Resequencing. Horticulturae, 11(8), 944. https://doi.org/10.3390/horticulturae11080944






