Transcriptome Analysis Reveals Potential Mechanism of Regulating Fruit Shape of ‘Laiyang Cili’ Pear with Calyx Excision Treatment
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. Differentially Expressed Genes Screening
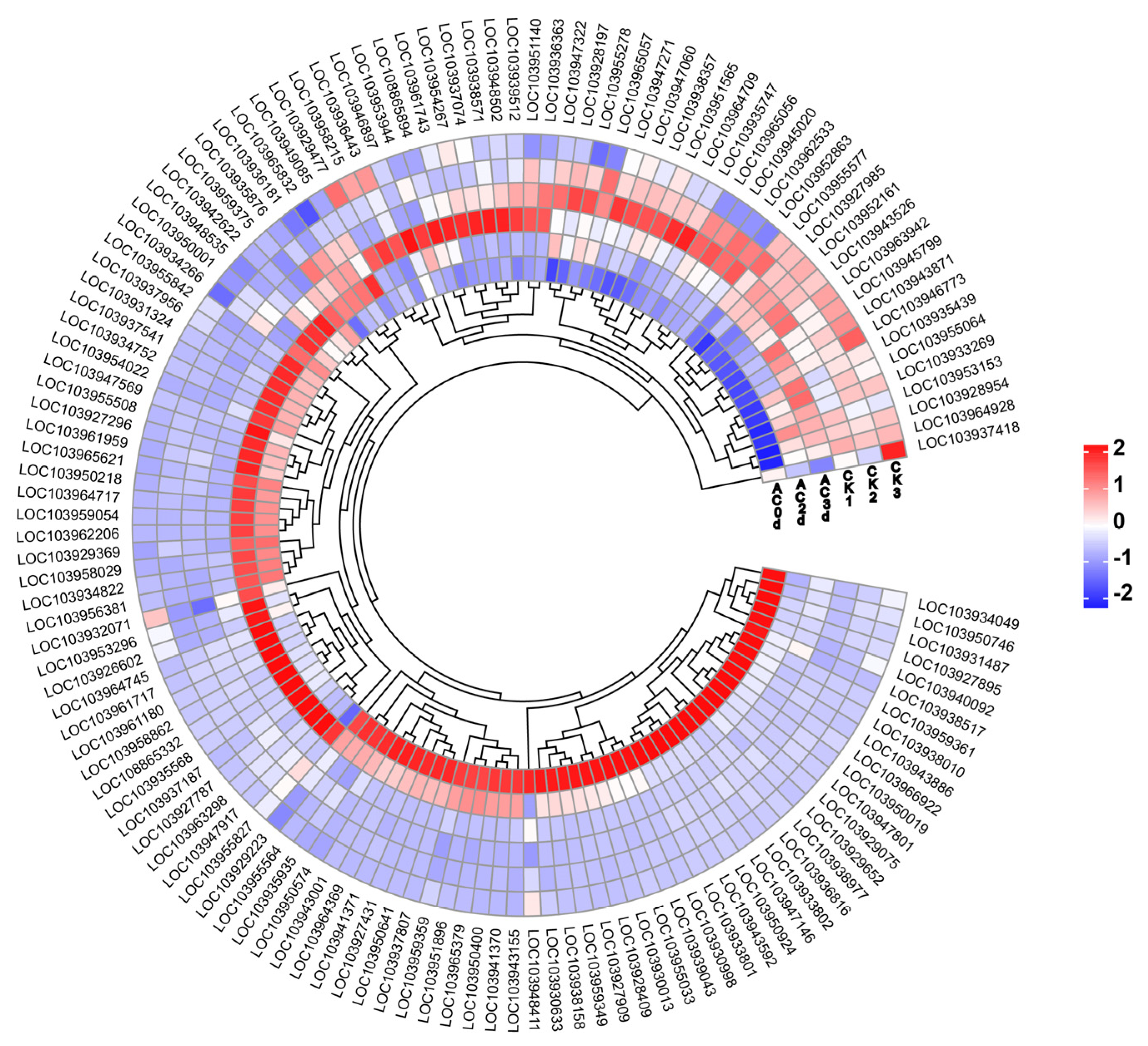
2.3. The Heatmap Analysis of DEGs Ranked Top 30 of Each Group
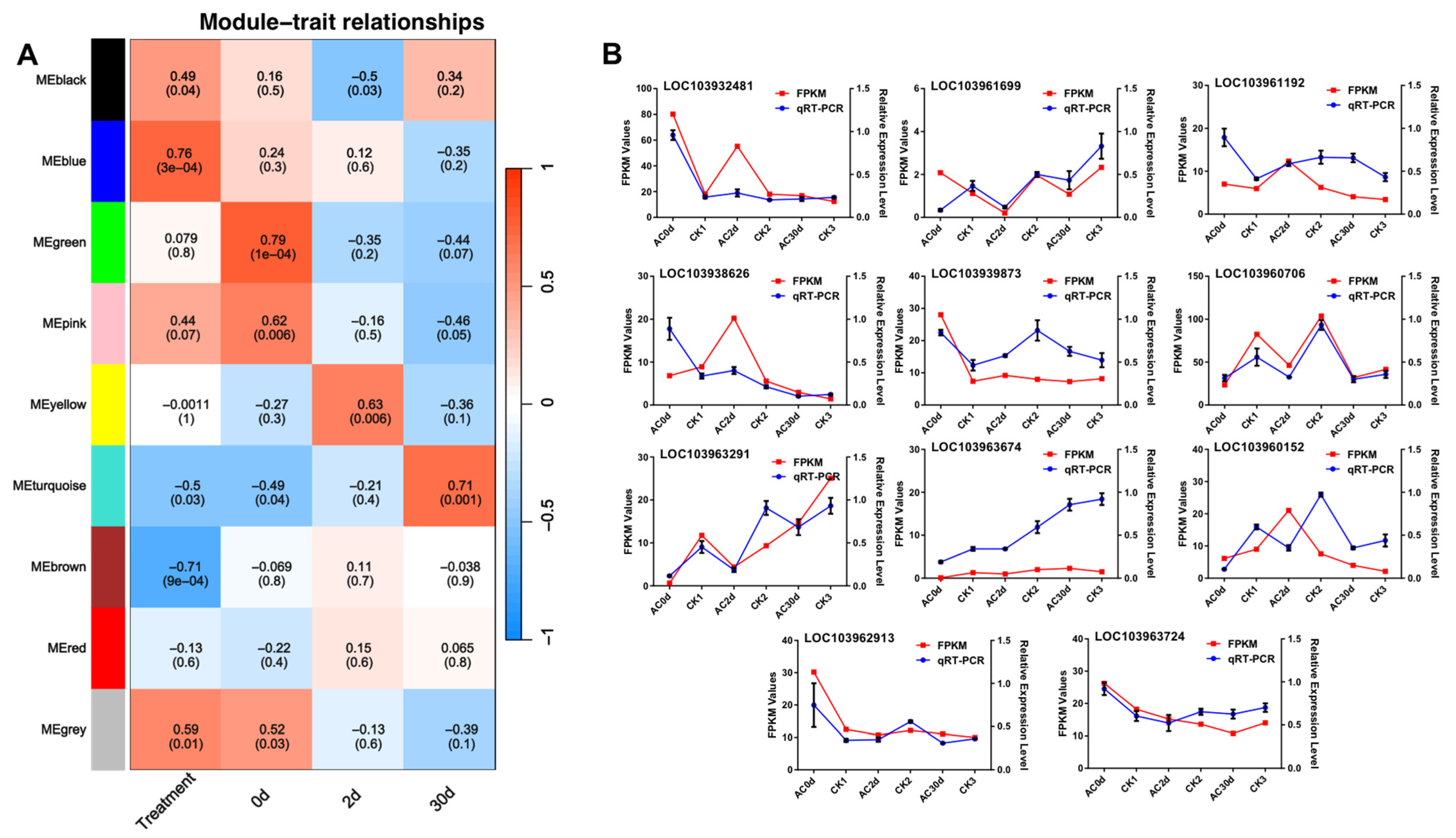
2.4. WGCNA Analysis
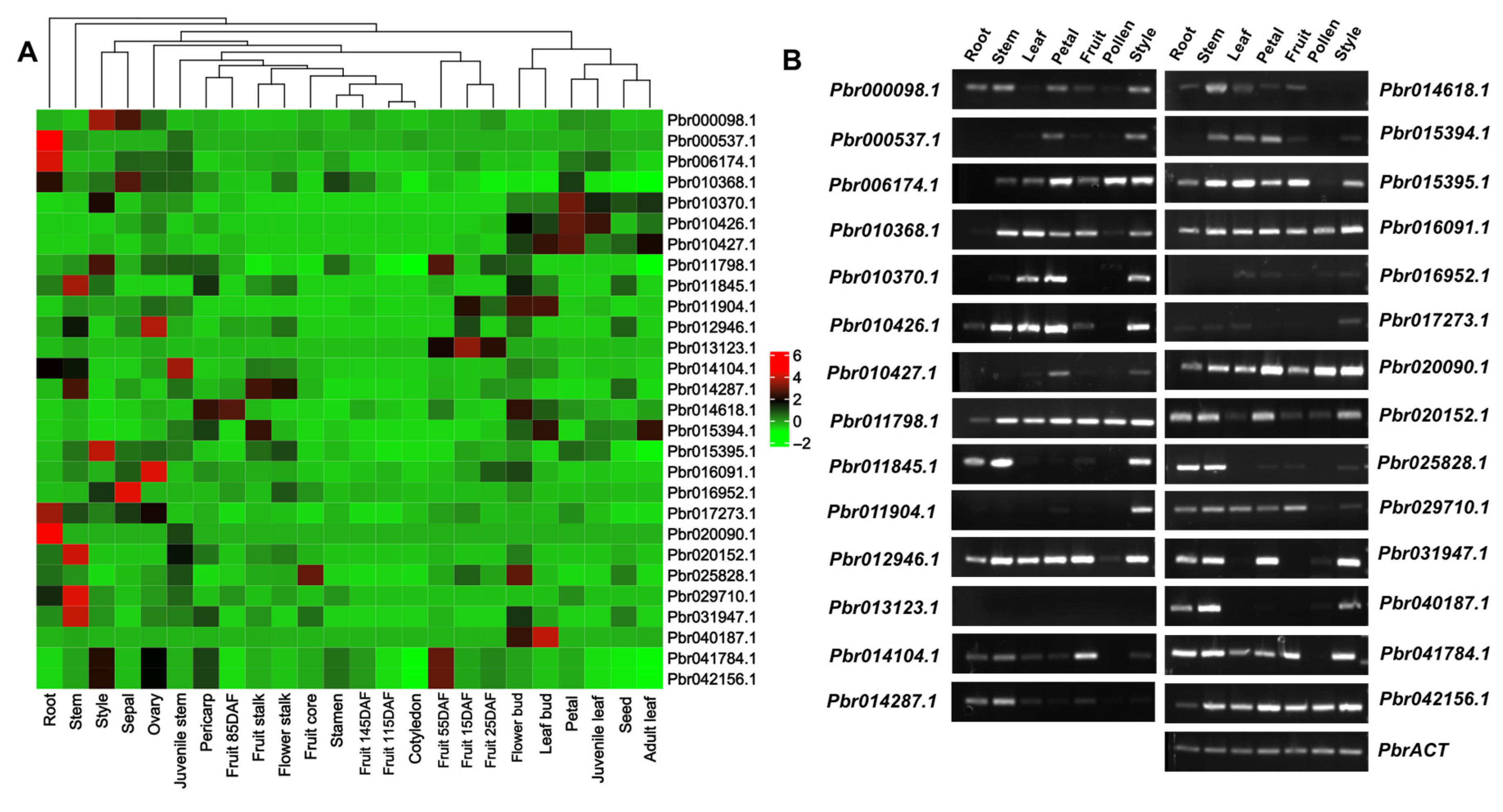
2.5. Identification and Tissue Expression Analysis of PbrOFP from Pear Genome
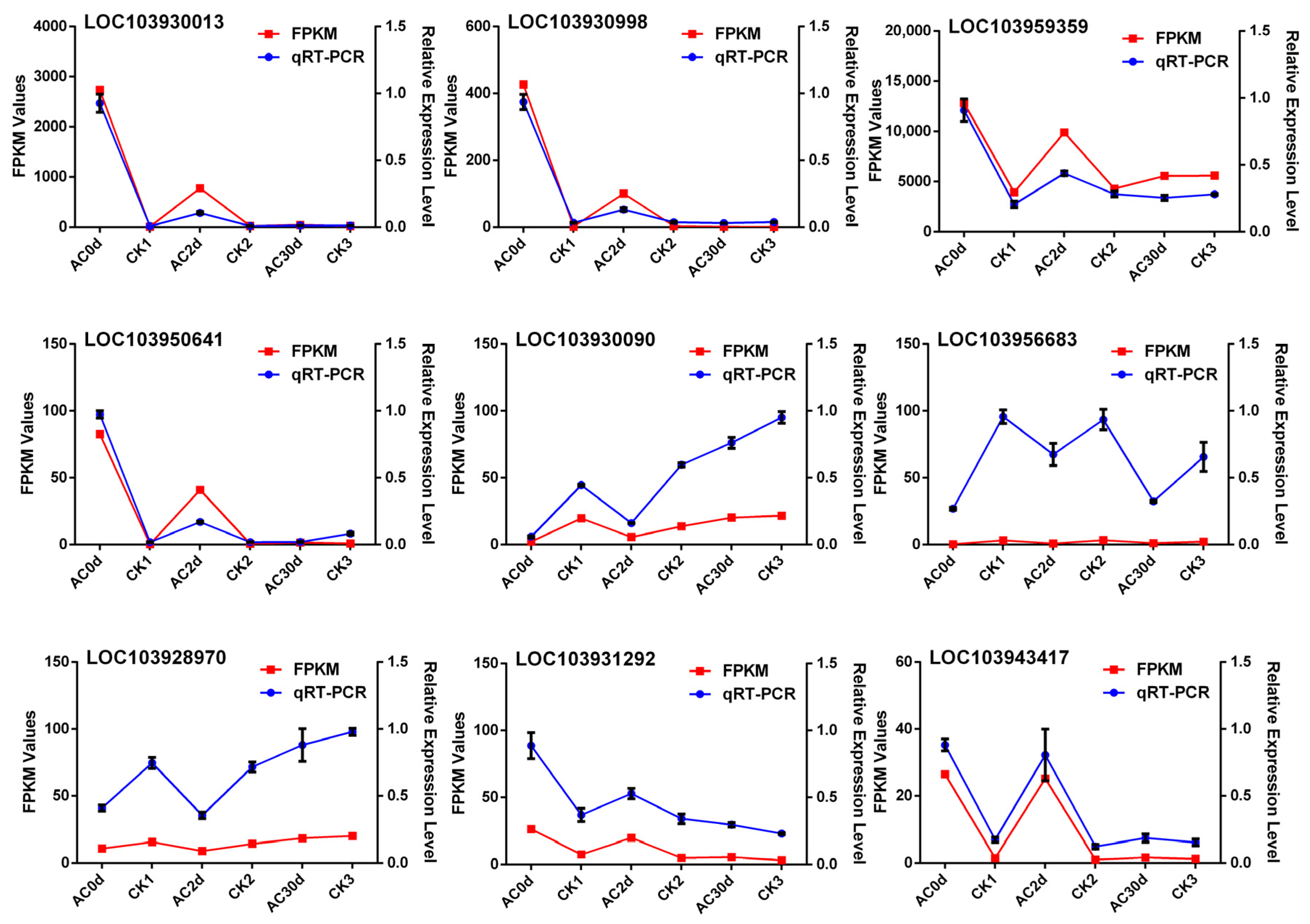
2.6. Reverse Transcription-Quantitative Real-Time Polymerase Chain Reaction (RT-qPCR) and RT-PCR Analysis
2.7. Statistical Analysis
3. Results
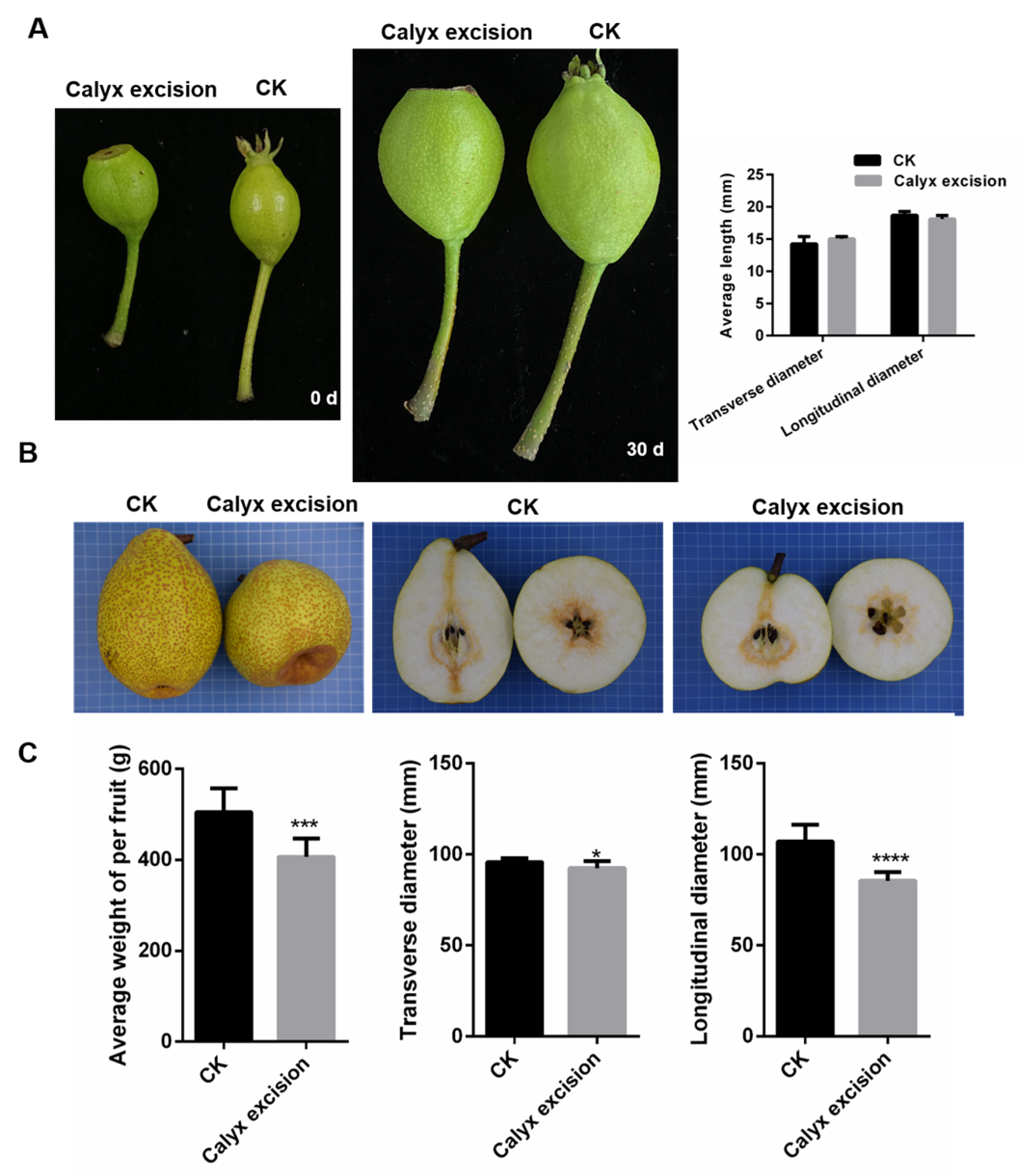
3.1. Effect of Calyx Excision on Fruit Quality of ‘Laiyang Cili’
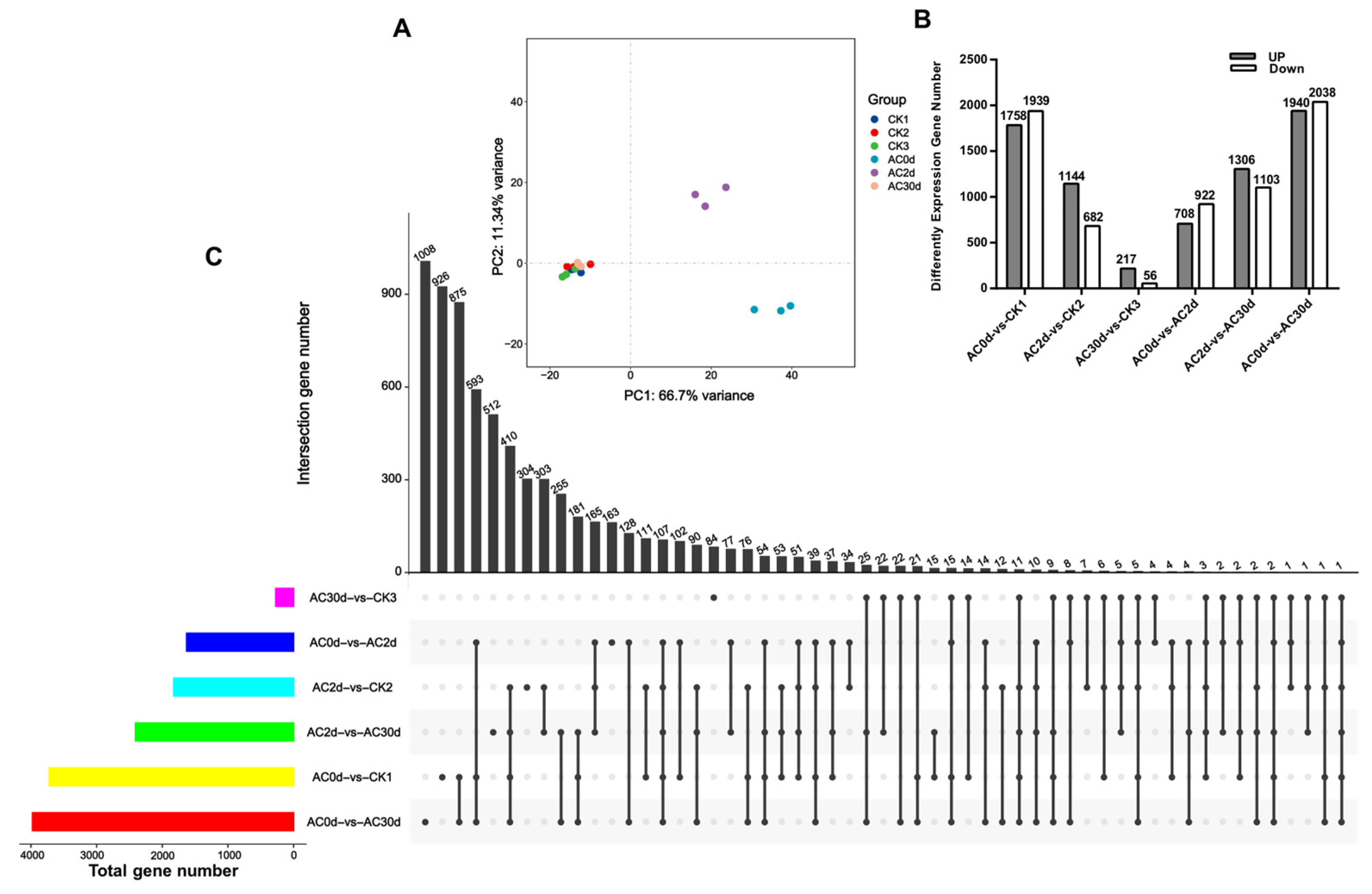
3.2. Screening and Analysis of Differentially Expressed Genes (DEGs)
3.3. Expression Level Analysis of DEGs
3.4. GO and KEGG Analysis of DEGs
3.5. Expression Trend Analysis of the Candidate Genes
3.6. Identification of Potential Key Genes for Pear Shape Regulation
3.7. OFP Family Gene Expression Patterns Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| AC0d | After calyx excision treatment (12 h) |
| AC2d | After calyx excision treatment (2 days) |
| AC30d | After calyx excision treatment (30 days) |
| ARF3 | Auxin response factor 3 |
| AFLP | Amplified fragment length polymorphism |
| bHLH | Basic helix–loop–helix |
| CaM | Calmodulin |
| DEGs | Differentially Expressed Genes |
| DAF | Days after flowering |
| ER-PM | Endoplasmic Reticulum–Plasma Membrane |
| FPKM | Fragments Per Kilobase of Exon Model Per Million Mapped Fragment |
| GA | Gibberellins |
| GO | Gene Ontology |
| IAA17 | Auxin-responsive protein 17 |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| MYB62 | Myeloblastosis 62 |
| OEP62 | Outer envelope pore protein 62 |
| OFP | OVATE Family Protein |
| PP2C 51 | Protein phosphatase 2C 51 |
| PME11 | Pectin methylesterase 11 |
| PYRC1 | Major allergen Pyr c 1 |
| QTL | Quantitative trait loci |
| RT-PCR | Reverse Transcription Polymerase Chain Reaction |
| RT-qPCR | Reverse Transcription-quantitative real-time Polymerase Chain Reaction |
| SUN | SUN domain-containing protein |
| SAUR50 | Auxin-responsive protein 50 |
| SRAP | Sequence-related amplified polymorphism |
| SSR | Simple sequence repeat |
| SD | Standard deviation |
| TIP1-3 | Aquaporin TIP1-3 |
| TGA4 | Transcription factor |
| TRM | TONNEAU1 Recruiting Motif |
| WGCNA | Weighted gene co-expression network analysis |
References
- Rodríguez, G.R.; Muños, S.; Anderson, C.; Sim, S.C.; Michel, A.; Causse, M.; Gardener, B.B.; Francis, D.; Van der Knaap, E. Distribution of SUN, OVATE, LC, and FAS in the tomato germplasm and the relationship to fruit shape diversity. Plant Physiol. 2011, 156, 275–285. [Google Scholar] [CrossRef]
- Van der Knaap, E.; Chakrabarti, M.; Chu, Y.H.; Clevenger, J.P.; Illa-berenguer, E.; Huang, Z.J.; Keyhaninejad, N.; Mu, Q.; Sun, L.; Wang, Y.P.; et al. What lies beyond the eye: The molecular mechanisms regulating tomato fruit weight and shape. Front. Plant Sci. 2014, 5, 227. [Google Scholar] [CrossRef]
- Pan, Y.P.; Wang, Y.H.; McGregor, C.; Liu, S.; Luan, F.S.; Gao, M.L.; Weng, Y.Q. Genetic architecture of fruit size and shape variation in cucurbits: A comparative perspective. Theor. Appl. Genet. 2020, 133, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Xiao, H.; Jiang, N.; Schaffner, E.; Stockinger, E.J.; Van der Knaap, E. A retrotransposon-mediated gene duplication underlies morphological variation of tomato fruit. Science 2008, 319, 1527–1530. [Google Scholar] [CrossRef] [PubMed]
- Zang, J.; Klemm, S.; Pain, C.; Duckney, P.; Bao, Z.R.; Stamm, G.; Bürstenbinder, K.; Hussey, P.J.; Wang, P.W. A novel plant actin-microtubule bridging complex regulates cytoskeletal and ER structure at ER-PM contact sites. Curr. Biol. 2021, 31, 1251–1260. [Google Scholar] [CrossRef] [PubMed]
- Levy, M.; Wang, Q.M.; Kaspi, R.; Parrella, M.P.; Abel, S. Arabidopsis IQD1, a novel calmodulin-binding nuclear protein, stimulates glucosinolate accumulation and plant defense. Plant J. 2005, 43, 79–96. [Google Scholar] [CrossRef] [PubMed]
- Bürstenbinder, K.; Möller, B.; Plötner, R.; Stamm, G.; Hause, G.; Mitra, D.; Abel, S. The IQD family of calmodulin-binding proteins links calcium signaling to microtubules, membrane subdomains, and the nucleus. Plant Physiol. 2017, 173, 1692–1708. [Google Scholar] [CrossRef]
- Pan, Y.P.; Liang, X.J.; Gao, M.L.; Liu, H.Q.; Meng, H.W.; Weng, Y.Q.; Cheng, Z.H. Round fruit shape in WI7239 cucumber is controlled by two interacting quantitative trait loci with one putatively encoding a tomato SUN homolog. Theor. Appl. Genet. 2017, 130, 573–586. [Google Scholar] [CrossRef]
- Wendrich, J.R.; Yang, B.J.; Mijnhout, P.; Xue, H.W.; Rybel, B.D.; Weijers, D. IQD proteins integrate auxin and calcium signaling to regulate microtubule dynamics during Arabidopsis development. BioRxiv 2018, 275560. [Google Scholar] [CrossRef]
- Liu, J.P.; Eck, J.V.; Cong, B.; Tanksley, S.D. A new class of regulatory genes underlying the cause of pear-shaped tomato fruit. Proc. Natl. Acad. Sci. USA 2002, 99, 13302–13306. [Google Scholar] [CrossRef]
- Hackbusch, J.; Richter, K.; Salamini, F.; Uhrig, J.F. A central role of Arabidopsis thaliana ovate family proteins in networking and subcellular localization of 3-aa loop extension homeodomain proteins. Proc. Natl. Acad. Sci. USA 2005, 102, 4908–4912. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.C.; Chang, Y.; Guo, J.J.; Chen, J.G. Arabidopsis Ovate Family Protein 1 is a transcriptional repressor that suppresses cell elongation. Plant J. 2007, 50, 858–872. [Google Scholar] [CrossRef]
- Ma, J.; Li, C.C.; Zong, M.; Qiu, Y.H.; Liu, Y.M.; Huang, Y.T.; Xie, Y.L.; Zhang, H.J.; Wang, J.S. CmFSI8/CmOFP13 encoding an OVATE family protein controls fruit shape in melon. J. Exp. Bot. 2022, 73, 1370–1384. [Google Scholar] [CrossRef]
- Wang, Y.-K.; Chang, W.-C.; Liu, P.-P.; Hsiao, M.-K.; Lin, C.-T.; Lin, S.-M.; Pan, R.-L. Ovate family protein 1 as a plant Ku70 interacting protein involving in DNA double-strand break repair. Plant Mol. Biol. 2010, 74, 453–466. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Zhang, B.; Keyhaninejad, N.; Rodríguez, G.R.; Kim, H.J.; Chakrabarti, M.; Illa-Berenguer, E.; Taitano, N.K.; Gonzalo, M.J.; Díaz, A.; et al. A common genetic mechanism underlies morphological diversity in fruits and other plant organs. Nat. Commun. 2018, 9, 4734. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Cao, K.; Deng, C.; Li, Y.; Zhu, G.R.; Fang, W.C.; Chen, C.W.; Wang, X.W.; Wu, J.L.; Guan, L.P.; et al. An integrated peach genome structural variation map uncovers genes associated with fruit traits. Genome Biol. 2020, 21, 1–19. [Google Scholar] [CrossRef]
- Zhou, H.; Ma, R.J.; Gao, L.; Zhang, J.Y.; Zhang, A.D.; Zhang, X.J.; Ren, F.; Zhang, W.H.; Liao, L.; Yang, Q.R.; et al. A 1.7-Mb chromosomal inversion downstream of a PpOFP1 gene is responsible for flat fruit shape in peach. Plant Biotechnol. J. 2021, 19, 192–205. [Google Scholar] [CrossRef] [PubMed]
- Zhao, P.P.; Wang, F.P.; Deng, Y.J.; Zhong, F.J.; Tian, P.; Lin, D.B.; Deng, J.H.; Zhang, Y.X.; Huang, T.B. Sly-miR159 regulates fruit morphology by modulating GA biosynthesis in tomato. Plant Biotechnol. J. 2022, 20, 833–845. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.G.; Chen, G.P.; Guo, X.H.; Yin, W.C.; Yu, X.H.; Hu, J.T.; Hu, Z.L. Overexpression of SlPRE2, an atypical bHLH transcription factor, affects plant morphology and fruit pigment accumulation in tomato. Sci. Rep. 2017, 7, 5786. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.G.; Liang, H.L.; Chen, G.P.; Li, F.F.; Wang, Y.S.; Liao, C.G.; Hu, Z.L. The bHLH transcription factor SlPRE2 regulates tomato fruit development and modulates plant response to gibberellin. Plant Cell Rep. 2019, 38, 1053–1064. [Google Scholar] [CrossRef]
- Li, J.; Gong, J.; Zhang, L.C.; Shen, H.; Chen, G.P.; Xie, Q.L.; Hu, Z.L. Overexpression of SlPRE5, an atypical bHLH transcription factor, affects plant morphology and chlorophyll accumulation in tomato. J. Plant Physiol. 2022, 273, 153698. [Google Scholar] [CrossRef]
- Zhang, R.-P.; Wu, J.; Li, X.G.; Khan, M.A.; Chen, H.; Korban, S.S.; Zhang, S.L. An AFLP, SRAP, and SSR genetic linkage map and identification of QTLs for fruit traits in pear (Pyrus L.). Plant Mol. Biol. Report. 2013, 31, 678–687. [Google Scholar] [CrossRef]
- Xue, Z.Y.; Yang, R.; Wang, Y.D.; Ma, Y.C.; Lin, Y.J.; Li, Z.G.; Song, Y.Q.; Feng, X.X.; Li, L.L. Candidate gene mining of GA-mediated regulation of pear fruit shape. Hortic. Environ. Biotechnol. 2024, 65, 403–412. [Google Scholar] [CrossRef]
- Xue, Z. Identification and Expression Characteristics Analysis of Pear Fruit Shape Related Genes. Master’s Thesis, Shanxi Agricultural University, Taiyuan, China, 2023. [Google Scholar]
- Teng, Y. Pyrus: Origin, Diversity and Cultivar Differentiation; Science Press: Beijing, China, 2024; pp. 149–151. [Google Scholar]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Araki, M.; Goto, S.; Hattori, M.; Hirakawa, M.; Itoh, M.; Katayama, T.; Kawashima, S.; Okuda, S.; Tokimatsu, T.; et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008, 36, 480–484. [Google Scholar] [CrossRef]
- Anders, S.; Huber, W. Differential Expression of RNA-Seq Data at the Gene Level—The DESeq Package; European Molecular Biology Laboratory: Heidelberg, Germany, 2012. [Google Scholar]
- Zhang, B.; Horvath, S. A General Framework for Weighted Gene Co-Expression Network Analysis. Stat. Appl. Genet. Mol. Biol. 2005, 4, 1. [Google Scholar] [CrossRef] [PubMed]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Su, L.Y.; Bassa, C.; Audran, C.; Mila, I.; Cheniclet, C.; Chevalier, C.; Buzayen, M.; Roustan, J.-P.; Chervin, C. The auxin Sl-IAA17 transcriptional repressor controls fruit size via the regulation of endoreduplication-related cell expansion. Plant Cell Physiol. 2014, 55, 1969–1976. [Google Scholar] [CrossRef]
- Wang, S.; Cheng, D.W.; Gu, H.; Li, M.; He, S.S.; Li, F.F.; Gu, S.C.; Chen, J.Y. Effects of plant growth regulators on the seedless rate and fruit quality of ‘shine muscat’ grape. J. Fruit Sci. 2019, 36, 1675–1682. (In Chinese) [Google Scholar]
- Wang, Y.P.; Clevenger, J.P.; Illa-Berenguer, E.; Meulia, T.; Van der Knaap, E.; Sun, L. A comparison of sun, ovate, fs8.1 and auxin application on tomato fruit shape and gene expression. Plant Cell Physiol. 2019, 60, 1067–1081. [Google Scholar] [CrossRef]
- Jong, M.D.; Wolters-Arts, M.; García-Martínez, J.L.; Mariani, C.; Vriezen, W.H. The Solanum lycopersicum AUXIN RESPONSE FACTOR 7 (SlARF7) mediates cross-talk between auxin and gibberellin signalling during tomato fruit set and development. J. Exp. Bot. 2011, 62, 617–626. [Google Scholar] [CrossRef]
- Hu, J.H.; Israeli, A.; Ori, N.M.; Sun, T.-P. The Interaction between DELLA and ARF/IAA Mediates Crosstalk between Gibberellin and Auxin Signaling to Control Fruit Initiation in Tomato. Plant Cell 2018, 30, 1710–1728. [Google Scholar] [CrossRef] [PubMed]
- Schüssler, M.D.; Alexandesson, E.; Bienert, G.P.; Kicjhey, T.; Laursen, K.H.; Johanson, U.; Kjellbom, P.; Schjoerring, J.K.; Jahn, T.P. The effects of the loss of TIP1; 1 and TIP1; 2 aquaporins in Arabidopsis thaliana. Plant J. 2008, 56, 756–767. [Google Scholar] [CrossRef]
- Bienert, G.P.; Møller, A.L.B.; Kristiansen, K.A.; Schulz, A.; Møller, I.M.; Schjoerring, J.K.; Jahn, T.P. Specific aquaporins facilitate the diffusion of hydrogen peroxide across membranes. J. Biol. Chem. 2007, 282, 1183–1192. [Google Scholar] [CrossRef]
- Lin, W.L.; Peng, Y.H.; Li, G.W.; Arora, R.; Tang, Z.C.; Su, W.A.; Cai, W.M. Isolation and functional characterization of PgTIP1, a hormone-autotrophic cells-specific tonoplast aquaporin in ginseng. J. Exp. Bot. 2007, 58, 947–956. [Google Scholar] [CrossRef]
- Peng, Y.H.; Lin, W.L.; Cai, W.M.; Arora, R. Overexpression of a Panax ginseng tonoplast aquaporin alters salt tolerance, drought tolerance and cold acclimation ability in transgenic Arabidopsis plants. Planta 2007, 226, 729–740. [Google Scholar] [CrossRef] [PubMed]
- Van der Knaap, E.; Lippman, Z.B.; Tanksley, S.D. Extremely elongated tomato fruit controlled by four quantitative trait loci with epistatic interactions. Theor. Appl. Genet. 2002, 104, 241–247. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jiao, H.; Chang, Y.; Chen, Q.; Xu, C.; Guan, Q.; Wei, S. Transcriptome Analysis Reveals Potential Mechanism of Regulating Fruit Shape of ‘Laiyang Cili’ Pear with Calyx Excision Treatment. Horticulturae 2025, 11, 939. https://doi.org/10.3390/horticulturae11080939
Jiao H, Chang Y, Chen Q, Xu C, Guan Q, Wei S. Transcriptome Analysis Reveals Potential Mechanism of Regulating Fruit Shape of ‘Laiyang Cili’ Pear with Calyx Excision Treatment. Horticulturae. 2025; 11(8):939. https://doi.org/10.3390/horticulturae11080939
Chicago/Turabian StyleJiao, Huijun, Yaojun Chang, Qiming Chen, Chaoran Xu, Qiuzhu Guan, and Shuwei Wei. 2025. "Transcriptome Analysis Reveals Potential Mechanism of Regulating Fruit Shape of ‘Laiyang Cili’ Pear with Calyx Excision Treatment" Horticulturae 11, no. 8: 939. https://doi.org/10.3390/horticulturae11080939
APA StyleJiao, H., Chang, Y., Chen, Q., Xu, C., Guan, Q., & Wei, S. (2025). Transcriptome Analysis Reveals Potential Mechanism of Regulating Fruit Shape of ‘Laiyang Cili’ Pear with Calyx Excision Treatment. Horticulturae, 11(8), 939. https://doi.org/10.3390/horticulturae11080939





