Characterization of the HSP70 Gene Family and Its Expression Under Heat Stress in Non-Heading Chinese Cabbage
Abstract
1. Introduction
2. Materials and Methods
2.1. Growth and Stress Treatments
2.2. NHCC: Genome-Wide Characterization of the HSP70 Genes
2.3. Phylogenetic Analysis
2.4. Analysis of HSP70 Gene Structure and Conserved Analysis
2.5. Chromosomal Location, Duplication, and Promoter Analysis
2.6. RNA-Seq Analysis
2.7. Total RNA Isolation and Quantitative PCR Methodology
3. Results
3.1. Identification of HSP70 Genes in B. rapa ssp. chinensis
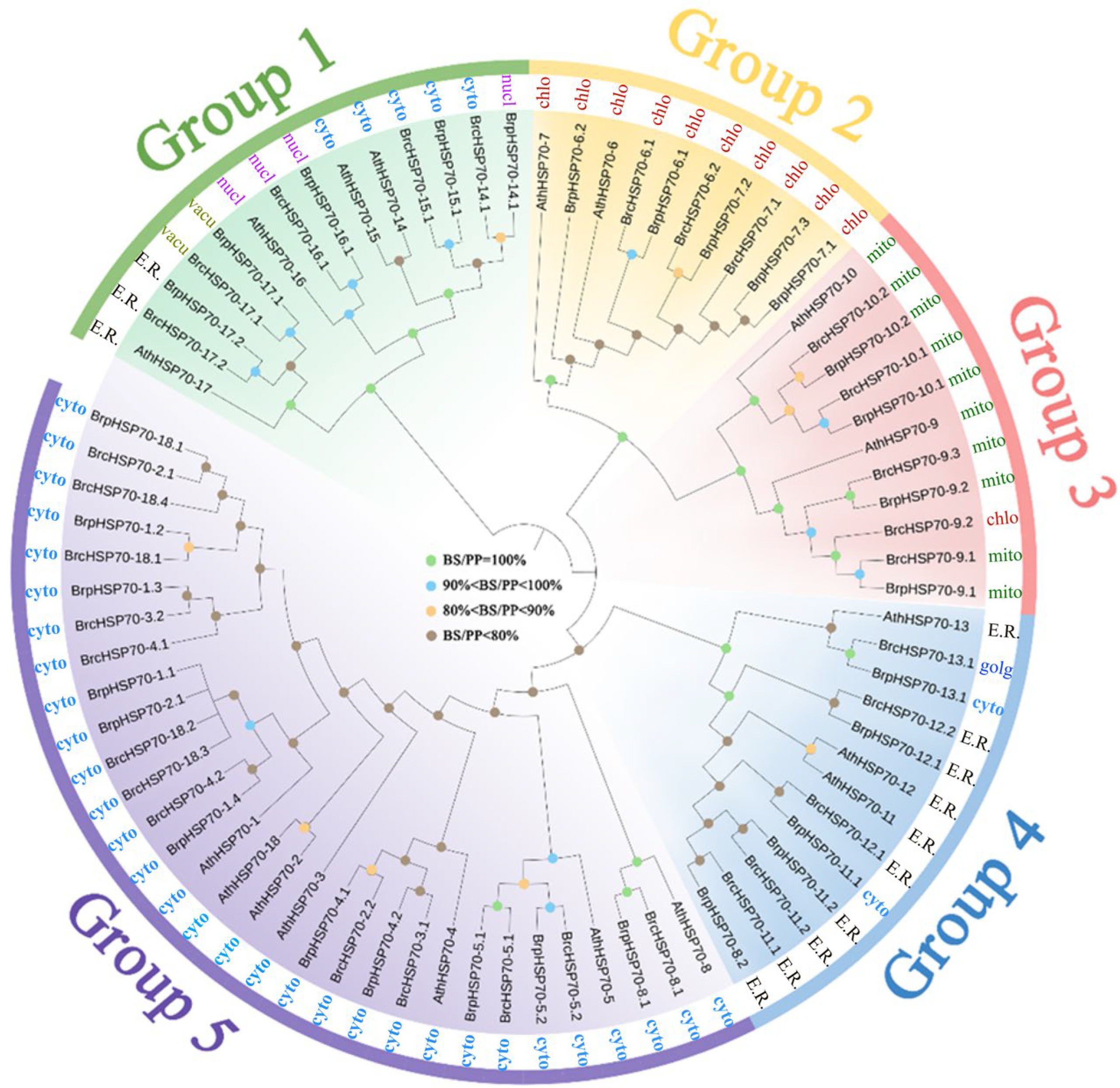
3.2. Phylogenetic Analysis of HSP70 Proteins in Brassica and Arabidopsis
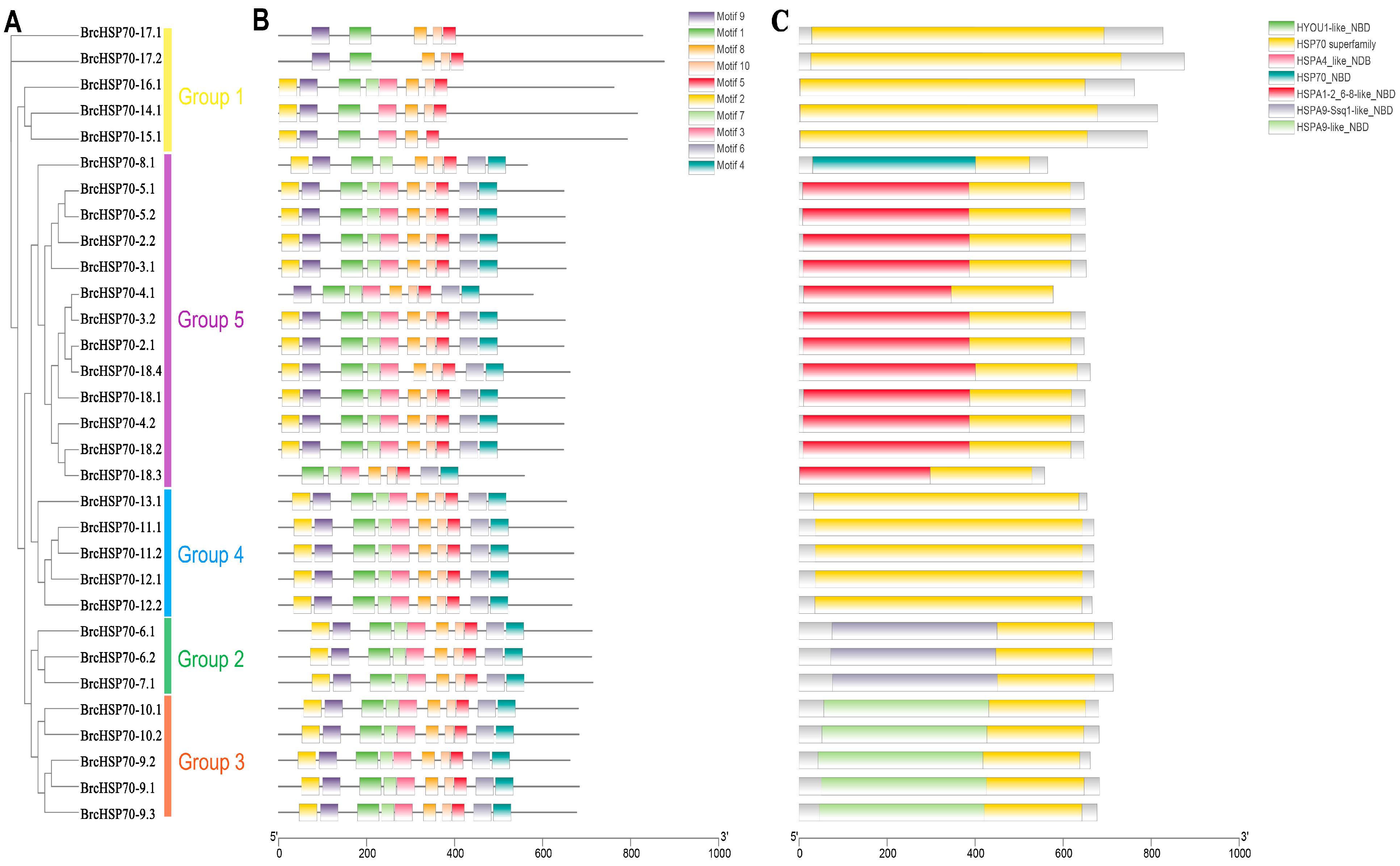
3.3. Motif, Gene Structure, and Domain Analysis of BrcHSP70s
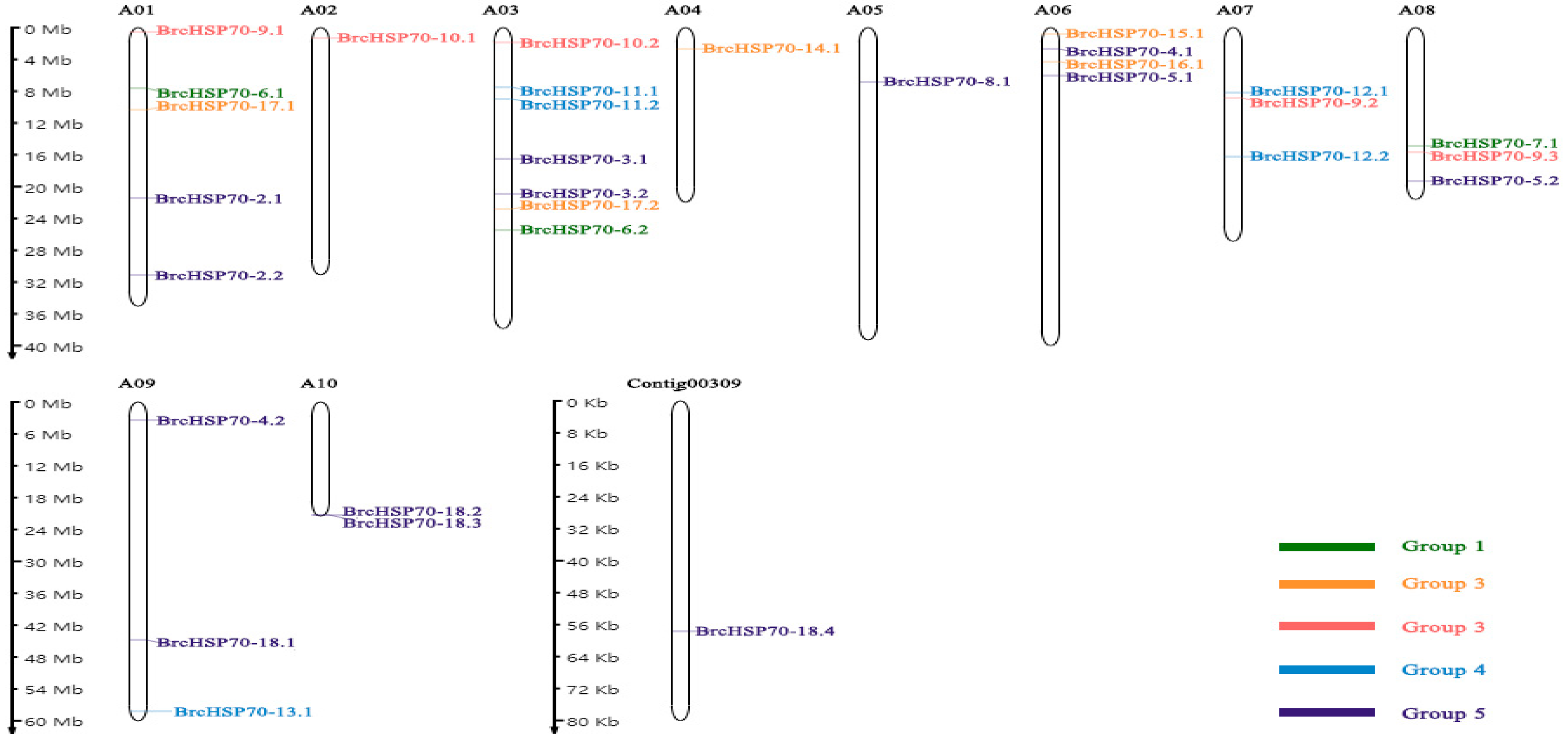
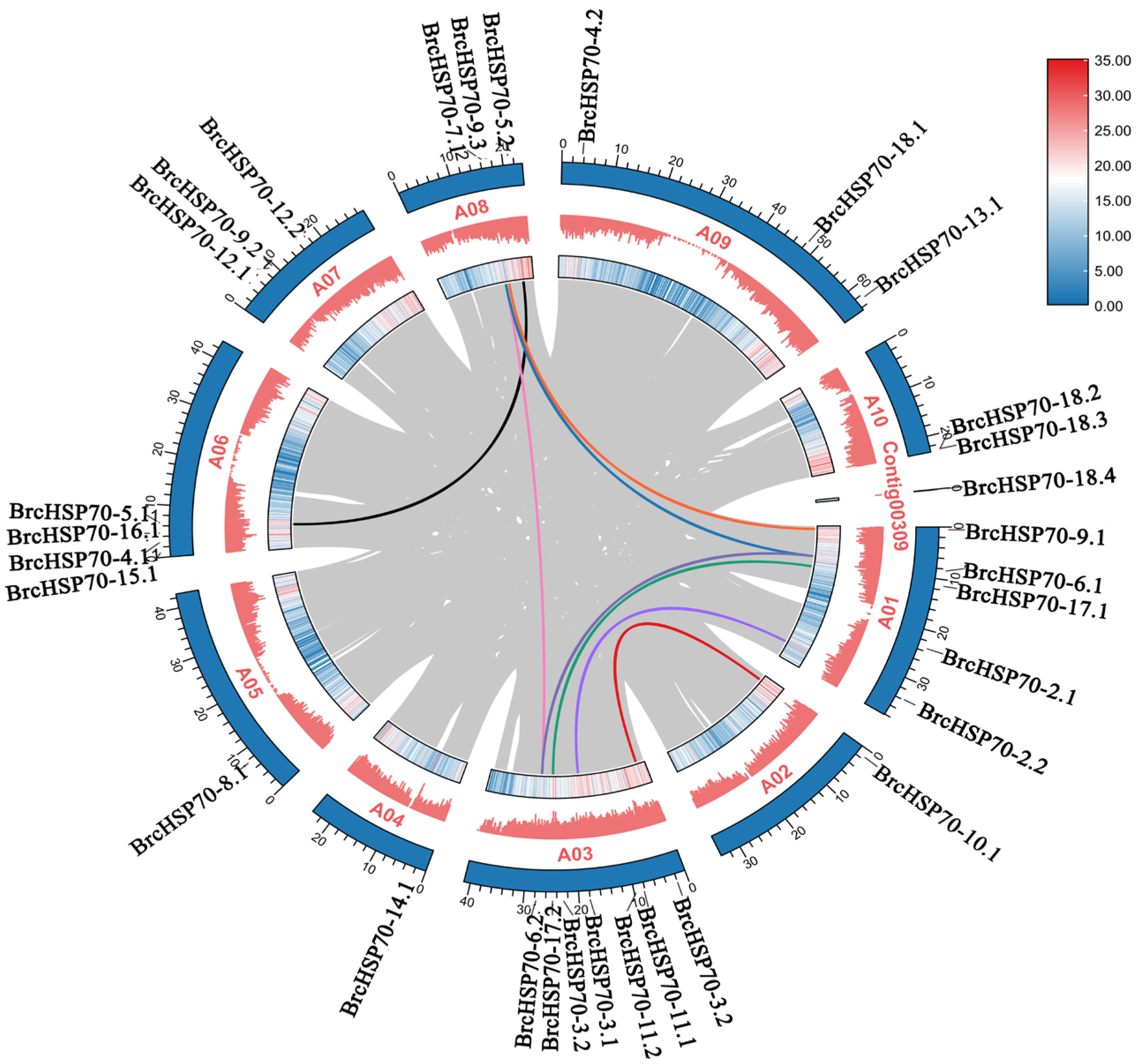
3.4. Chromosomal Distribution and Duplication Dynamics of BrcHSP70 Family
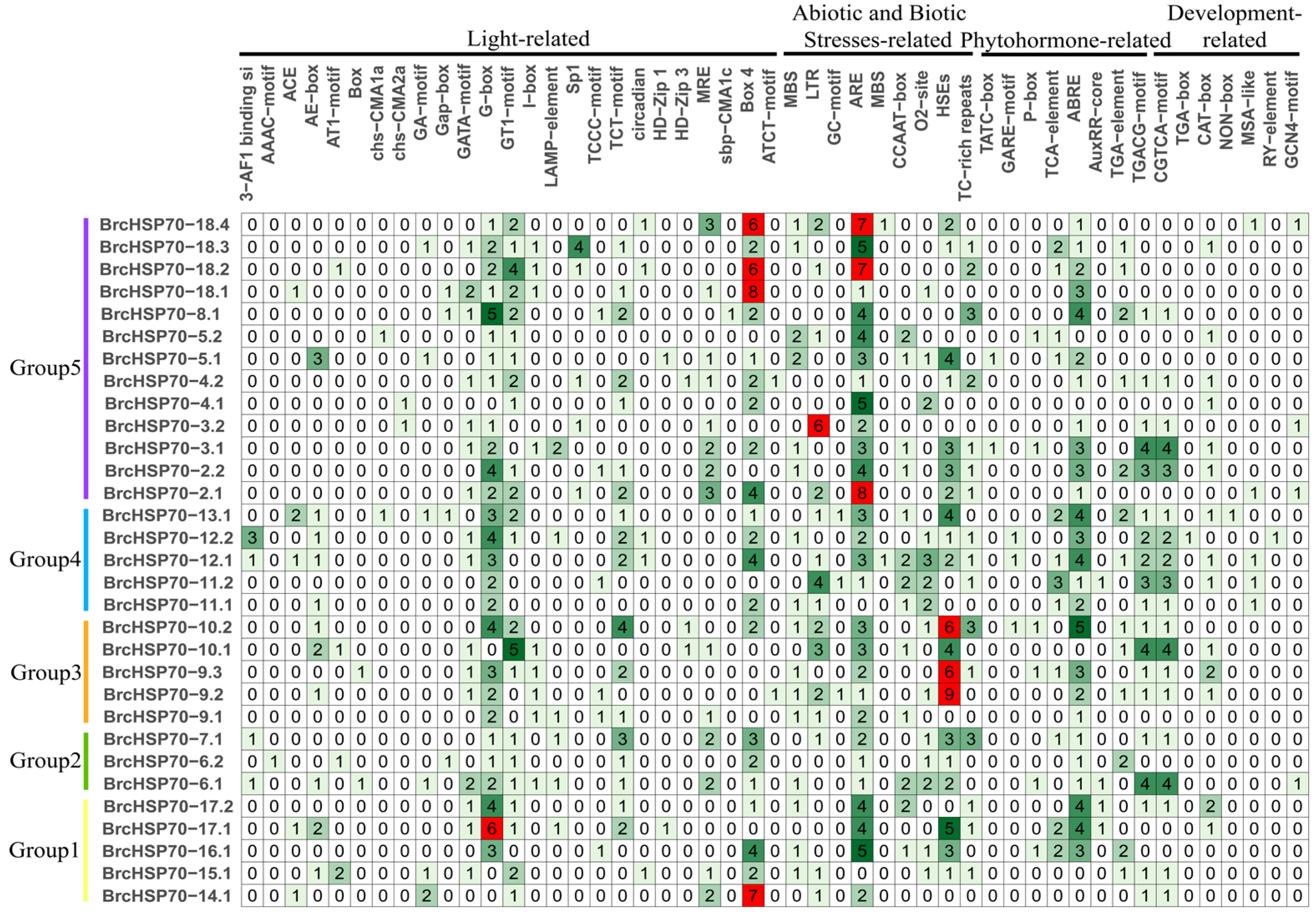
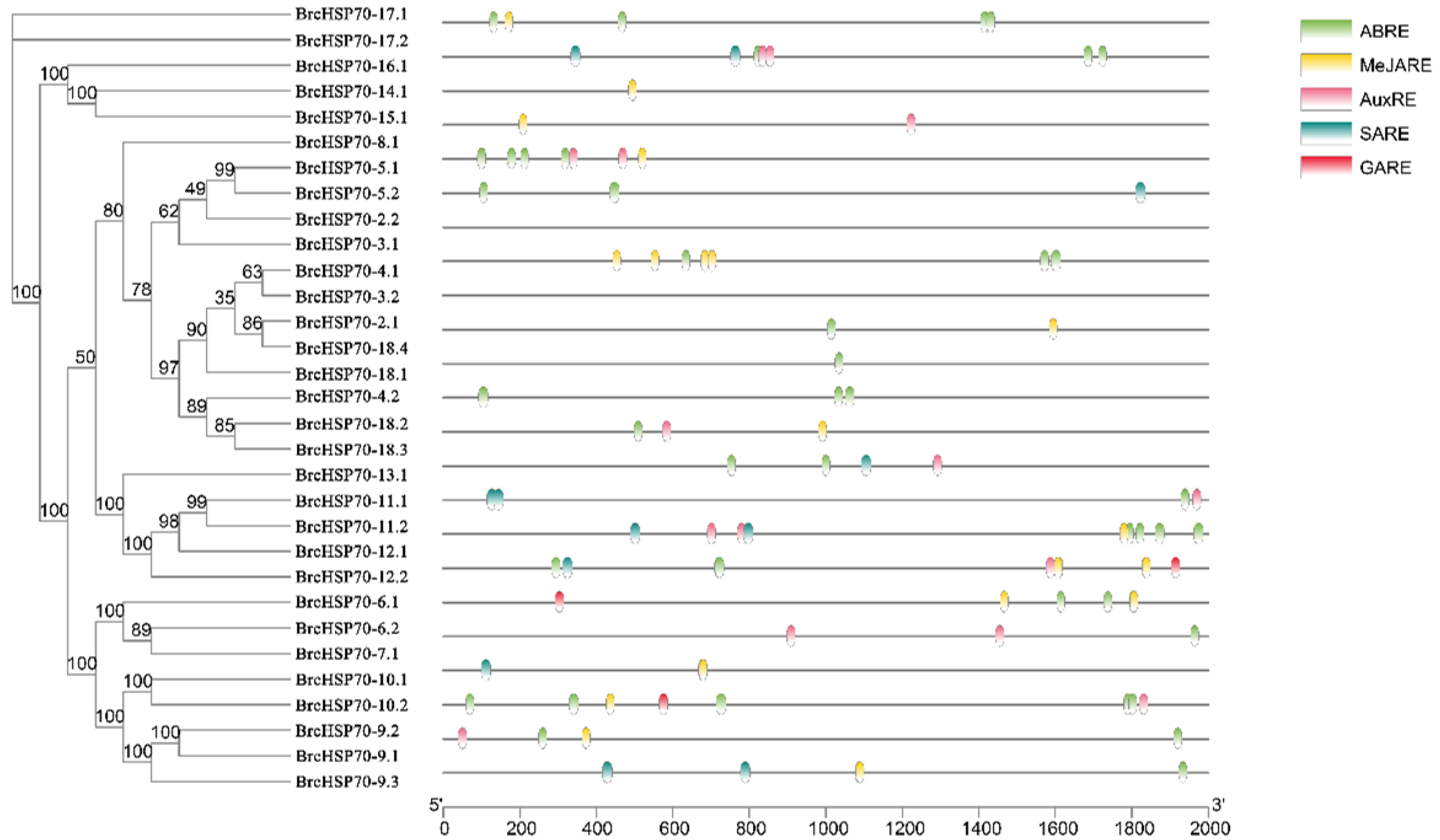
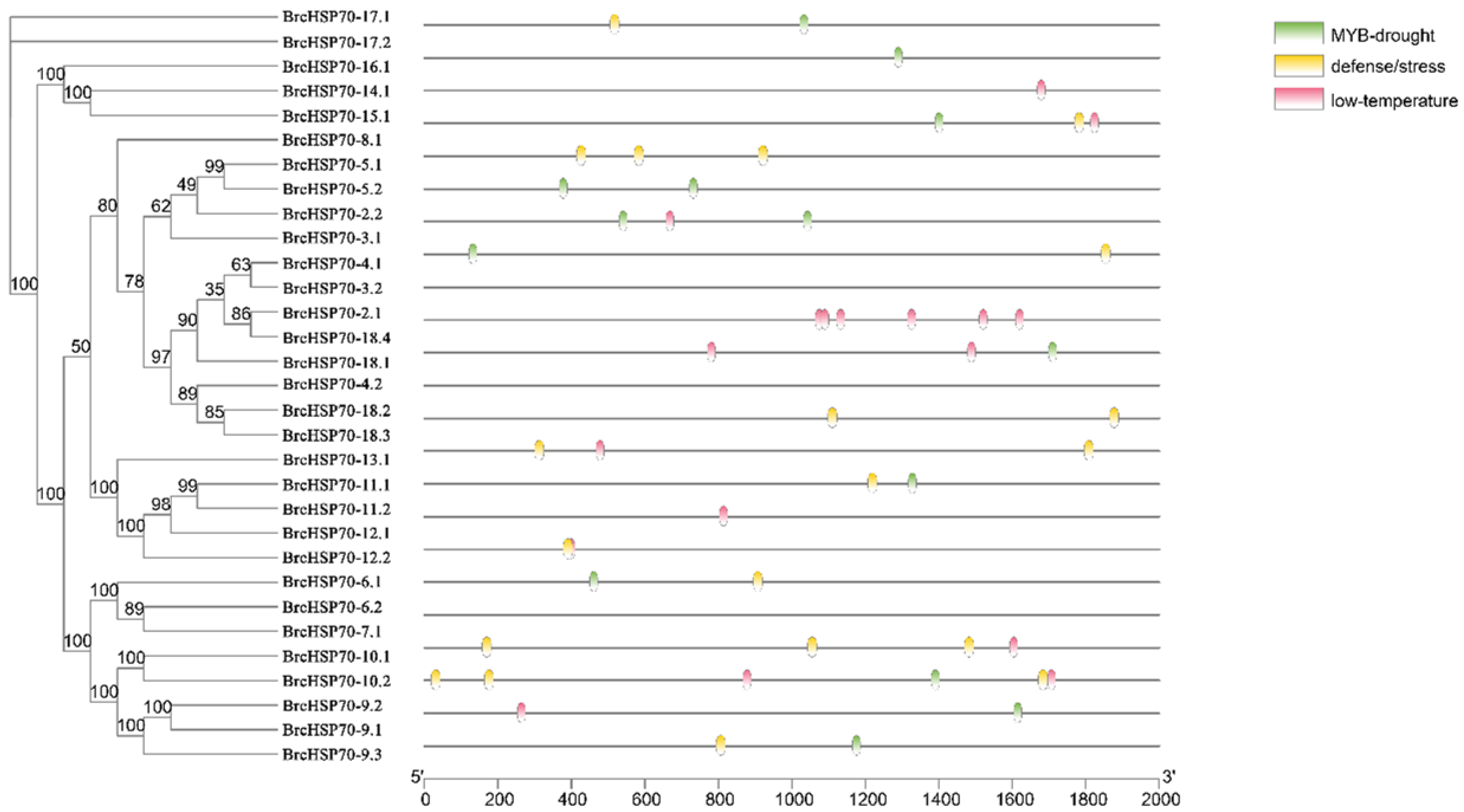
3.5. Characterization of BrcHSP70 Promoter Cis-Elements
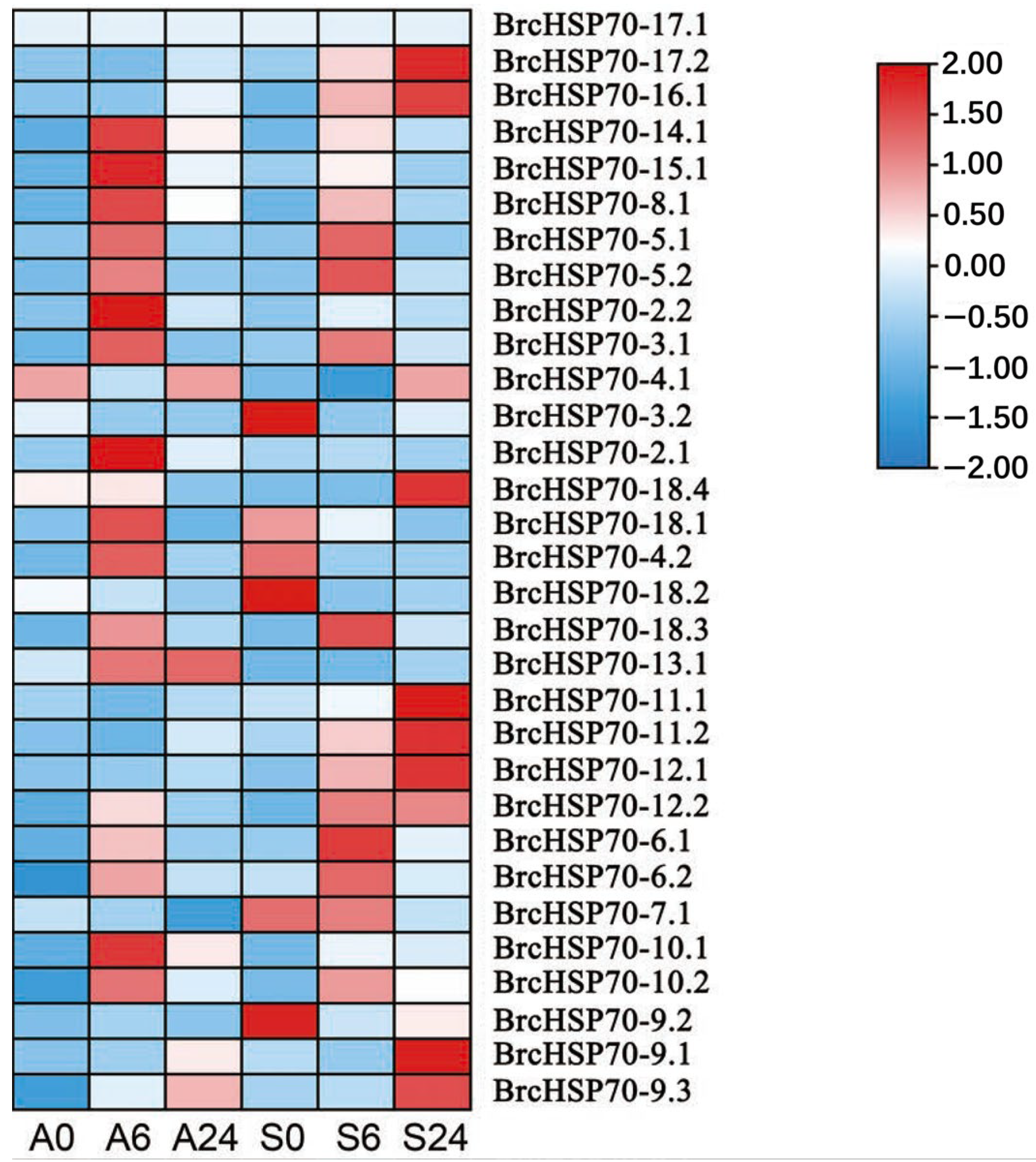
3.6. Expression Profile of the BrcHSP70 Gene Family
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ohama, N.; Sato, H.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Transcriptional Regulatory Network of Plant Heat Stress Response. Trends Plant Sci. 2017, 22, 53–65. [Google Scholar] [CrossRef]
- Chai, W.; Peng, X.; Liu, B.; Wang, J.; Zhu, Z.; Liu, Y.; Zhao, K.; Cheng, B.; Si, W.; Jiang, H. Comparative Genomics, Whole-Genome Re-sequencing and Expression Profile Analysis of Nucleobase:Cation Symporter 2 (NCS2) Genes in Maize. Front. Plant Sci. 2018, 9, 856. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Lu, Z.; Wang, L.; Jin, B. Plant Responses to Heat Stress: Physiology, Transcription, Noncoding RNAs, and Epigenetics. Int. J. Mol. Sci. 2020, 22, 117. [Google Scholar] [CrossRef] [PubMed]
- Nakashima, K.; Yamaguchi-Shinozaki, K.; Shinozaki, K. The transcriptional regulatory network in the drought response and its crosstalk in abiotic stress responses including drought, cold, and heat. Front. Plant Sci. 2014, 5, 170. [Google Scholar] [CrossRef] [PubMed]
- Kai, H.; Hirashima, K.; Matsuda, O.; Ikegami, H.; Winkelmann, T.; Nakahara, T.; Iba, K. Thermotolerant cyclamen with reduced acrolein and methyl vinyl ketone. J. Exp. Bot. 2012, 63, 4143–4150. [Google Scholar] [CrossRef]
- Sakata, T.; Oshino, T.; Miura, S.; Tomabechi, M.; Tsunaga, Y.; Higashitani, N.; Miyazawa, Y.; Takahashi, H.; Watanabe, M.; Higashitani, A. Auxins reverse plant male sterility caused by high temperatures. Proc. Natl. Acad. Sci. USA 2010, 107, 8569–8574. [Google Scholar] [CrossRef]
- Lippmann, R.; Babben, S.; Menger, A.; Delker, C.; Quint, M. Development of Wild and Cultivated Plants under Global Warming Conditions. Curr. Biol. 2019, 29, R1326–R1338. [Google Scholar] [CrossRef]
- Liu, J.; Zhang, R.; Xu, X.; Fowler, J.C.; Miller, T.E.X.; Dong, T. Effect of summer warming on growth, photosynthesis and water status in female and male Populus cathayana: Implications for sex-specific drought and heat tolerances. Tree Physiol. 2020, 40, 1178–1191. [Google Scholar] [CrossRef]
- Wahid, A.; Gelani, S.; Ashraf, M.; Foolad, M.R. Heat tolerance in plants: An overview. Environ. Exp. Bot. 2007, 61, 199–223. [Google Scholar] [CrossRef]
- Garbuz, D.G. Regulation of heat shock gene expression in response to stress. Mol. Biol. 2017, 51, 400–417. [Google Scholar] [CrossRef]
- Yan, H.; Zhang, A.; Chen, J.; He, X.; Xu, B.; Xie, G.; Miao, Z.; Zhang, X.; Huang, L. Genome-Wide Analysis of the PvHsp20 Family in Switchgrass: Motif, Genomic Organization, and Identification of Stress or Developmental-Related Hsp20s. Front. Plant Sci. 2017, 8, 1024. [Google Scholar] [CrossRef]
- Guihur, A.; Rebeaud, M.E.; Bourgine, B.; Goloubinoff, P. How do humans and plants feel the heat? Trends Plant Sci. 2022, 27, 630–632. [Google Scholar] [CrossRef] [PubMed]
- Guihur, A.; Rebeaud, M.E.; Goloubinoff, P. How do plants feel the heat and survive? Trends Biochem. Sci. 2022, 47, 824–838. [Google Scholar] [CrossRef] [PubMed]
- Mitsugi, R.; Itoh, T.; Fujiwara, R. Expression of Human DNAJ (Heat Shock Protein-40) B3 in Humanized UDP-glucuronosyltransferase 1 Mice. Int. J. Mol. Sci. 2015, 16, 14997–15008. [Google Scholar] [CrossRef] [PubMed]
- Xiong, L.; Zhu, J.K. Molecular and genetic aspects of plant responses to osmotic stress. Plant Cell Environ. 2002, 25, 131–139. [Google Scholar] [CrossRef]
- Yang, J.; Ding, H.; Kan, X. Codon usage patterns and evolution of HSP60 in birds. Int. J. Biol. Macromol. 2021, 183, 1002–1012. [Google Scholar] [CrossRef]
- Hu, C.; Yang, J.; Qi, Z.; Wu, H.; Wang, B.; Zou, F.; Mei, H.; Liu, J.; Wang, W.; Liu, Q. Heat shock proteins: Biological functions, pathological roles, and therapeutic opportunities. MedComm 2022, 3, e161. [Google Scholar] [CrossRef]
- Wang, L.; Liu, F.; Ju, L.; Xue, B.; Wang, Y.; Wang, D.; Hou, D. Genome Structures and Evolution Analysis of Hsp90 Gene Family in Brassica napus Reveal the Possible Roles of Members in Response to Salt Stress and the Infection of Sclerotinia sclerotiorum. Front. Plant Sci. 2022, 13, 854034. [Google Scholar] [CrossRef]
- Song, Z.; Pan, F.; Lou, X.; Wang, D.; Yang, C.; Zhang, B.; Zhang, H. Genome-wide identification and characterization of Hsp70 gene family in Nicotiana tabacum. Mol. Biol. Rep. 2019, 46, 1941–1954. [Google Scholar] [CrossRef]
- Mayer, M.P.; Bukau, B. Hsp70 chaperones: Cellular functions and molecular mechanism. Cell Mol. Life Sci. 2005, 62, 670–684. [Google Scholar] [CrossRef]
- Chappell, T.G.; Konforti, B.B.; Schmid, S.L.; Rothman, J.E. The ATPase core of a clathrin uncoating protein. J. Biol. Chem. 1987, 262, 746–751. [Google Scholar] [CrossRef]
- Flynn, G.C.; Chappell, T.G.; Rothman, J.E. Peptide binding and release by proteins implicated as catalysts of protein assembly. Science 1989, 245, 385–390. [Google Scholar] [CrossRef]
- Guo, M.; Xu, L.; Long, Y.; Huang, F.; Liu, T.; Li, Y.; Hou, X. BcHTT4 Inhibits Branching of Non-Heading Chinese Cabbage at the Vegetative Stage. Plants 2021, 10, 510. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Liu, G.F.; Ma, L.M.; Liu, T.K.; Zhang, C.W.; Xiao, D.; Zheng, H.K.; Chen, F.; Hou, X.L. A chromosome-level reference genome of non-heading Chinese cabbage [Brassica campestris (syn. Brassica rapa) ssp. chinensis]. Hortic. Res. 2020, 7, 212. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Li, Y.; Liu, T.; Zhang, C.; Xiao, D.; Hou, X. Non-Heading Chinese Cabbage Database: An Open-Access Platform for the Genomics of Brassica campestris (syn. Brassica rapa) ssp. chinensis. Plants 2022, 11, 5. [Google Scholar] [CrossRef]
- Ye, Y.; Ding, H.; Bi, D.; Ge, W.; Yang, J.; Han, S.; Zhang, S.; Liu, Y.; Kan, X. The ABC1K gene family in Chinese cabbage: Phylogeny, structure and function. Genet. Resour. Crop Evol. 2024, 71, 4647–4667. [Google Scholar] [CrossRef]
- Song, X.; Liu, G.; Huang, Z.; Duan, W.; Tan, H.; Li, Y.; Hou, X. Temperature expression patterns of genes and their coexpression with LncRNAs revealed by RNA-Seq in non-heading Chinese cabbage. BMC Genom. 2016, 17, 297. [Google Scholar] [CrossRef]
- Valdés-López, O.; Batek, J.; Gomez-Hernandez, N.; Nguyen, C.T.; Isidra-Arellano, M.C.; Zhang, N.; Joshi, T.; Xu, D.; Hixson, K.K.; Weitz, K.K.; et al. Soybean Roots Grown under Heat Stress Show Global Changes in Their Transcriptional and Proteomic Profiles. Front. Plant Sci. 2016, 7, 517. [Google Scholar] [CrossRef]
- Sung, D.Y.; Vierling, E.; Guy, C.L. Comprehensive expression profile analysis of the Arabidopsis Hsp70 gene family. Plant Physiol. 2001, 126, 789–800. [Google Scholar] [CrossRef]
- Sarkar, N.K.; Kundnani, P.; Grover, A. Functional analysis of Hsp70 superfamily proteins of rice (Oryza sativa). Cell Stress Chaperones 2013, 18, 427–437. [Google Scholar] [CrossRef]
- Tabusam, J.; Shi, Q.; Feng, D.; Zulfiqar, S.; Shen, S.; Ma, W.; Zhao, J. HSP70 Gene Family in Brassica rapa: Genome-Wide Identification, Characterization, and Expression Patterns in Response to Heat and Cold Stress. Cells 2022, 11, 2316. [Google Scholar] [CrossRef] [PubMed]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.L.; Tosatto, S.C.E.; Paladin, L.; Raj, S.; Richardson, L.J.; et al. Pfam: The protein families database in 2021. Nucleic Acids Res. 2021, 49, D412–D419. [Google Scholar] [CrossRef]
- Eddy, S.R. Accelerated Profile HMM Searches. PLoS Comput. Biol. 2011, 7, e1002195. [Google Scholar] [CrossRef] [PubMed]
- Finn, R.D.; Clements, J.; Eddy, S.R. HMMER web server: Interactive sequence similarity searching. Nucleic Acids Res. 2011, 39, W29–W37. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.; Wang, J.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; Gwadz, M.; Hurwitz, D.I.; Marchler, G.H.; Song, J.S. CDD/SPARCLE: The conserved domain database in 2020. Nucleic Acids Res. 2020, 48, D265–D268. [Google Scholar] [CrossRef]
- Wilkins, M.R.; Gasteiger, E.; Bairoch, A.; Sanchez, J.C.; Williams, K.L.; Appel, R.D.; Hochstrasser, D.F. Protein identification and analysis tools in the ExPASy server. Methods Mol. Biol. 1999, 112, 531–552. [Google Scholar] [CrossRef]
- Jones, T.S.; Oliveira, S.M.D.; Myers, C.J.; Voigt, C.A.; Densmore, D. Genetic circuit design automation with Cello 2.0. Nat. Protoc. 2022, 17, 1097–1113. [Google Scholar] [CrossRef]
- Chen, H.; Wang, T.; He, X.; Cai, X.; Lin, R.; Liang, J.; Wu, J.; King, G.; Wang, X. BRAD V3.0: An upgraded Brassicaceae database. Nucleic Acids Res. 2021, 50, D1432–D1441. [Google Scholar] [CrossRef]
- Zhang, D.; Gao, F.; Jakovlić, I.; Zou, H.; Zhang, J.; Li, W.X.; Wang, G.T. PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour. 2020, 20, 348–355. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive tree of life (iTOL) v3: An online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016, 44, W242–W245. [Google Scholar] [CrossRef]
- Hu, B.; Jin, J.; Guo, A.-Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME Suite. Nucleic Acids Res. 2015, 43, W39–W49. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef] [PubMed]
- Jiangtao, C.; Yingzhen, K.; Qian, W.; Yuhe, S.; Daping, G.; Jing, L.; Guanshan, L. MapGene2Chrom, a tool to draw gene physical map based on Perl and SVG languages. Yi Chuan 2015, 37, 91–97. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; Debarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Zhang, Z.; Xiao, J.; Wu, J.; Zhang, H.; Liu, G.; Wang, X.; Dai, L. ParaAT: A parallel tool for constructing multiple protein-coding DNA alignments. Biochem. Biophys. Res. Commun. 2012, 419, 779–781. [Google Scholar] [CrossRef]
- Posada, D.; Buckley, T.R. Model selection and model averaging in phylogenetics: Advantages of Akaike information criterion and Bayesian approaches over likelihood ratio tests. Syst. Biol. 2004, 53, 793–808. [Google Scholar] [CrossRef]
- Wang, D.; Zhang, Y.; Zhang, Z.; Zhu, J.; Yu, J. KaKs_Calculator 2.0: A toolkit incorporating gamma-series methods and sliding window strategies. Genom. Proteom. Bioinform. 2010, 8, 77–80. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Dai, Z.; Jia, J.; Li, X.; Zhu, H.; Kan, X.; Zhu, B. Comparative physiological, transcriptomic, and WGCNA analyses reveal the key genes and regulatory pathways associated with heat stress in non-heading Chinese cabbage. Res. Sq. 2025. preprint. [Google Scholar] [CrossRef]
- Lin, B.L.; Wang, J.S.; Liu, H.C.; Chen, R.W.; Meyer, Y.; Barakat, A.; Delseny, M. Genomic analysis of the Hsp70 superfamily in Arabidopsis thaliana. Cell Stress Chaperones 2001, 6, 201–208. [Google Scholar] [CrossRef] [PubMed]
- Ye, Q.; Yu, J.; Zhang, Z.; Hou, L.; Liu, X. VvBAP1, a Grape C2 Domain Protein, Plays a Positive Regulatory Role Under Heat Stress. Front. Plant Sci. 2020, 11, 544374. [Google Scholar] [CrossRef]
- Reeg, S.; Jung, T.; Castro, J.P.; Davies, K.J.A.; Henze, A.; Grune, T. The molecular chaperone Hsp70 promotes the proteolytic removal of oxidatively damaged proteins by the proteasome. Free Radic. Biol. Med. 2016, 99, 153–166. [Google Scholar] [CrossRef]
- Cho, E.K.; Choi, Y.J. A nuclear-localized HSP70 confers thermoprotective activity and drought-stress tolerance on plants. Biotechnol. Lett. 2009, 31, 597–606. [Google Scholar] [CrossRef]
- Büyük, İ.; Inal, B.; Ilhan, E.; Tanriseven, M.; Aras, S.; Erayman, M. Genome-wide identification of salinity responsive HSP70s in common bean. Mol. Biol. Rep. 2016, 43, 1251–1266. [Google Scholar] [CrossRef]
- Davoudi, M.; Chen, J.; Lou, Q. Genome-Wide Identification and Expression Analysis of Heat Shock Protein 70 (HSP70) Gene Family in Pumpkin (Cucurbita moschata) Rootstock under Drought Stress Suggested the Potential Role of these Chaperones in Stress Tolerance. Int. J. Mol. Sci. 2022, 23, 1918. [Google Scholar] [CrossRef]
- Jasrotia, R.S.; Jaiswal, S.; Yadav, P.K.; Raza, M.; Iquebal, M.A.; Rai, A.; Kumar, D. Genome-Wide Analysis of HSP70 Family Protein in Vigna radiata and Coexpression Analysis Under Abiotic and Biotic Stress. J. Comput. Biol. 2020, 27, 738–754. [Google Scholar] [CrossRef]
- Jung, K.H.; Gho, H.J.; Nguyen, M.X.; Kim, S.R.; An, G. Genome-wide expression analysis of HSP70 family genes in rice and identification of a cytosolic HSP70 gene highly induced under heat stress. Funct. Integr. Genom. 2013, 13, 391–402. [Google Scholar] [CrossRef]
- Lespinet, O.; Wolf, Y.I.; Koonin, E.V.; Aravind, L. The role of lineage-specific gene family expansion in the evolution of eukaryotes. Genome Res. 2002, 12, 1048–1059. [Google Scholar] [CrossRef] [PubMed]
- Rehman, A.; Atif, R.M.; Qayyum, A.; Du, X.; Hinze, L.; Azhar, M.T. Genome-wide identification and characterization of HSP70 gene family in four species of cotton. Genomics 2020, 112, 4442–4453. [Google Scholar] [CrossRef] [PubMed]
- Freeling, M. Bias in plant gene content following different sorts of duplication: Tandem, whole-genome, segmental, or by transposition. Annu. Rev. Plant. Biol. 2009, 60, 433–453. [Google Scholar] [CrossRef] [PubMed]
- Doyle, J.J.; Flagel, L.E.; Paterson, A.H.; Rapp, R.A.; Soltis, D.E.; Soltis, P.S.; Wendel, J.F. Evolutionary genetics of genome merger and doubling in plants. Annu. Rev. Genet. 2008, 42, 443–461. [Google Scholar] [CrossRef]
- Zhang, J.B.; Wang, X.P.; Wang, Y.C.; Chen, Y.H.; Luo, J.W.; Li, D.D.; Li, X.B. Genome-wide identification and functional characterization of cotton (Gossypium hirsutum) MAPKKK gene family in response to drought stress. BMC Plant Biol. 2020, 20, 217. [Google Scholar] [CrossRef]
- Li, Y.; Chen, X.; Chen, Z.; Cai, R.; Zhang, H.; Xiang, Y. Identification and Expression Analysis of BURP Domain-Containing Genes in Medicago truncatula. Front. Plant Sci. 2016, 7, 485. [Google Scholar] [CrossRef]
- Baldoni, E.; Genga, A.; Cominelli, E. Plant MYB Transcription Factors: Their Role in Drought Response Mechanisms. Int. J. Mol. Sci. 2015, 16, 15811–15851. [Google Scholar] [CrossRef]
- Yu, Y.; Ni, Z.; Chen, Q.; Qu, Y. The wheat salinity-induced R2R3-MYB transcription factor TaSIM confers salt stress tolerance in Arabidopsis thaliana. Biochem. Biophys. Res. Commun. 2017, 491, 642–648. [Google Scholar] [CrossRef]
- Wei, Q.; Luo, Q.; Wang, R.; Zhang, F.; He, Y.; Zhang, Y.; Qiu, D.; Li, K.; Chang, J.; Yang, G.; et al. A Wheat R2R3-type MYB Transcription Factor TaODORANT1 Positively Regulates Drought and Salt Stress Responses in Transgenic Tobacco Plants. Front. Plant Sci. 2017, 8, 1374. [Google Scholar] [CrossRef]
- Gao, F.; Zhou, J.; Deng, R.Y.; Zhao, H.X.; Li, C.L.; Chen, H.; Suzuki, T.; Park, S.U.; Wu, Q. Overexpression of a tartary buckwheat R2R3-MYB transcription factor gene, FtMYB9, enhances tolerance to drought and salt stresses in transgenic Arabidopsis. J. Plant Physiol. 2017, 214, 81–90. [Google Scholar] [CrossRef]
- Fujita, Y.; Yoshida, T.; Yamaguchi-Shinozaki, K. Pivotal role of the AREB/ABF-SnRK2 pathway in ABRE-mediated transcription in response to osmotic stress in plants. Physiol. Plant 2013, 147, 15–27. [Google Scholar] [CrossRef]


| Group | Gene ID | Transcript_ID | Length (aa) | Molecular Weight (KDa) | pI | RGAVY | Localization Predictor |
|---|---|---|---|---|---|---|---|
| 1 | BrcHSP70-14.1 | BraC04g004760.1 | 814 | 90.06124 | 5.2 | −0.421 | Cytoplasmic |
| BrcHSP70-15.1 | BraC06g001210.1 | 791 | 87.8087 | 5.11 | −0.409 | Cytoplasmic | |
| BrcHSP70-16.1 | BraC06g008220.1 | 761 | 84.4894 | 5.78 | −0.486 | Nuclear | |
| BrcHSP70-17.1 | BraC01g020590.1 | 826 | 92.44471 | 5.4 | −0.468 | Vacuole | |
| BrcHSP70-17.2 | BraC03g048540.1 | 875 | 97.49774 | 5.78 | −0.488 | ER | |
| 2 | BrcHSP70-6.1 | BraC01g015450.1 | 711 | 76.0055 | 5.09 | −0.325 | Chloroplast |
| BrcHSP70-6.2 | BraC03g053490.1 | 710 | 75.98278 | 5.27 | −0.313 | Chloroplast | |
| BrcHSP70-7.1 | BraC08g021270.2 | 713 | 76.37809 | 5.15 | −0.319 | Chloroplast | |
| 3 | BrcHSP70-9.1 | BraC01g001110.1 | 682 | 73.00276 | 5.56 | −0.297 | Mitochondrial |
| BrcHSP70-9.2 | BraC07g009150.1 | 661 | 71.30408 | 5.39 | −0.275 | Chloroplast | |
| BrcHSP70-9.3 | BraC08g022530.1 | 676 | 72.48936 | 5.79 | −0.281 | Mitochondrial | |
| BrcHSP70-10.1 | BraC02g003110.1 | 680 | 72.87141 | 5.82 | −0.335 | Mitochondrial | |
| BrcHSP70-10.2 | BraC03g004180.1 | 681 | 72.74525 | 5.61 | −0.331 | Mitochondrial | |
| 4 | BrcHSP70-11.1 | BraC03g016780.1 | 669 | 73.63128 | 5.08 | −0.468 | ER |
| BrcHSP70-11.2 | BraC03g019710.1 | 669 | 73.6073 | 5.08 | −0.461 | ER | |
| BrcHSP70-12.1 | BraC07g008480.1 | 669 | 73.84858 | 5.08 | −0.477 | ER | |
| BrcHSP70-12.2 | BraC07g020710.1 | 665 | 73.49322 | 5.11 | −0.466 | ER | |
| BrcHSP70-13.1 | BraC09g070470.1 | 653 | 72.52744 | 4.96 | −0.43 | Golgi apparatus | |
| 5 | BrcHSP70-2.1 | BraC01g030020.1 | 647 | 70.88335 | 5.07 | −0.404 | Cytoplasmic |
| BrcHSP70-2.2 | BraC01g043750.1 | 650 | 71.27168 | 5.13 | −0.429 | Cytoplasmic | |
| BrcHSP70-3.1 | BraC03g036010.1 | 652 | 71.38772 | 5.13 | −0.45 | Cytoplasmic | |
| BrcHSP70-3.2 | BraC03g044790.1 | 650 | 71.1766 | 5.04 | −0.42 | Cytoplasmic | |
| BrcHSP70-4.1 | BraC06g004500.1 | 577 | 63.66623 | 5.37 | −0.381 | Cytoplasmic | |
| BrcHSP70-4.2 | BraC09g006520.1 | 647 | 70.94741 | 5.04 | −0.401 | Cytoplasmic | |
| BrcHSP70-5.1 | BraC06g011730.1 | 647 | 71.04354 | 5.48 | −0.408 | Cytoplasmic | |
| BrcHSP70-5.2 | BraC08g030530.1 | 650 | 71.37063 | 5.28 | −0.435 | Cytoplasmic | |
| BrcHSP70-8.1 | BraC05g012920.2 | 564 | 60.81675 | 5.39 | 0.051 | Chloroplast | |
| BrcHSP70-18.1 | BraC09g044850.1 | 649 | 71.29577 | 5.07 | −0.386 | Cytoplasmic | |
| BrcHSP70-18.2 | BraC10g035070.1 | 646 | 70.8744 | 5.07 | −0.396 | Cytoplasmic | |
| BrcHSP70-18.3 | BraC10g035090.1 | 557 | 61.27375 | 5.07 | −0.397 | Cytoplasmic | |
| BrcHSP70-18.4 | BraCxxg008040.1 | 661 | 72.34702 | 5.11 | −0.407 | Cytoplasmic |
| Gene Pair | Ka | Ks | Ka/Ks Ratio |
|---|---|---|---|
| BrcHSP70-3.1/BrcHSP70-2.2 | 0.00832 | 1.06657 | 0.00780 |
| BrcHSP70-5.2/BrcHSP70-5.1 | 0.02688 | 0.91947 | 0.02923 |
| BrcHSP70-6.2/BrcHSP70-6.1 | 0.02643 | 0.49163 | 0.05376 |
| BrcHSP70-7.1/BrcHSP70-6.1 | 0.02860 | 0.45505 | 0.06286 |
| BrcHSP70-7.1/BrcHSP70-6.2 | 0.02001 | 0.47105 | 0.04248 |
| BrcHSP70-9.3/BrcHSP70-9.1 | 0.02838 | 0.43173 | 0.06574 |
| BrcHSP70-10.2/BrcHSP70-10.1 | 0.01798 | 0.44104 | 0.04078 |
| BrcHSP70-17.2/BrcHSP70-17.1 | 0.03107 | 0.30507 | 0.10185 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, B.; Jia, J.; Zhang, S.; Xiao, Y.; Dai, C.; Kan, X. Characterization of the HSP70 Gene Family and Its Expression Under Heat Stress in Non-Heading Chinese Cabbage. Horticulturae 2025, 11, 938. https://doi.org/10.3390/horticulturae11080938
Zhu B, Jia J, Zhang S, Xiao Y, Dai C, Kan X. Characterization of the HSP70 Gene Family and Its Expression Under Heat Stress in Non-Heading Chinese Cabbage. Horticulturae. 2025; 11(8):938. https://doi.org/10.3390/horticulturae11080938
Chicago/Turabian StyleZhu, Bo, Jingyi Jia, Sijia Zhang, Yingying Xiao, Chenwei Dai, and Xianzhao Kan. 2025. "Characterization of the HSP70 Gene Family and Its Expression Under Heat Stress in Non-Heading Chinese Cabbage" Horticulturae 11, no. 8: 938. https://doi.org/10.3390/horticulturae11080938
APA StyleZhu, B., Jia, J., Zhang, S., Xiao, Y., Dai, C., & Kan, X. (2025). Characterization of the HSP70 Gene Family and Its Expression Under Heat Stress in Non-Heading Chinese Cabbage. Horticulturae, 11(8), 938. https://doi.org/10.3390/horticulturae11080938






