Identification and Characterization of bZIP Gene Family Combined Transcriptome Analysis Revealed Their Functional Roles on Abiotic Stress and Anthocyanin Biosynthesis in Mulberry (Morus alba)
Abstract
1. Introduction
2. Material and Methods
2.1. Acquisition of Sequencing Data
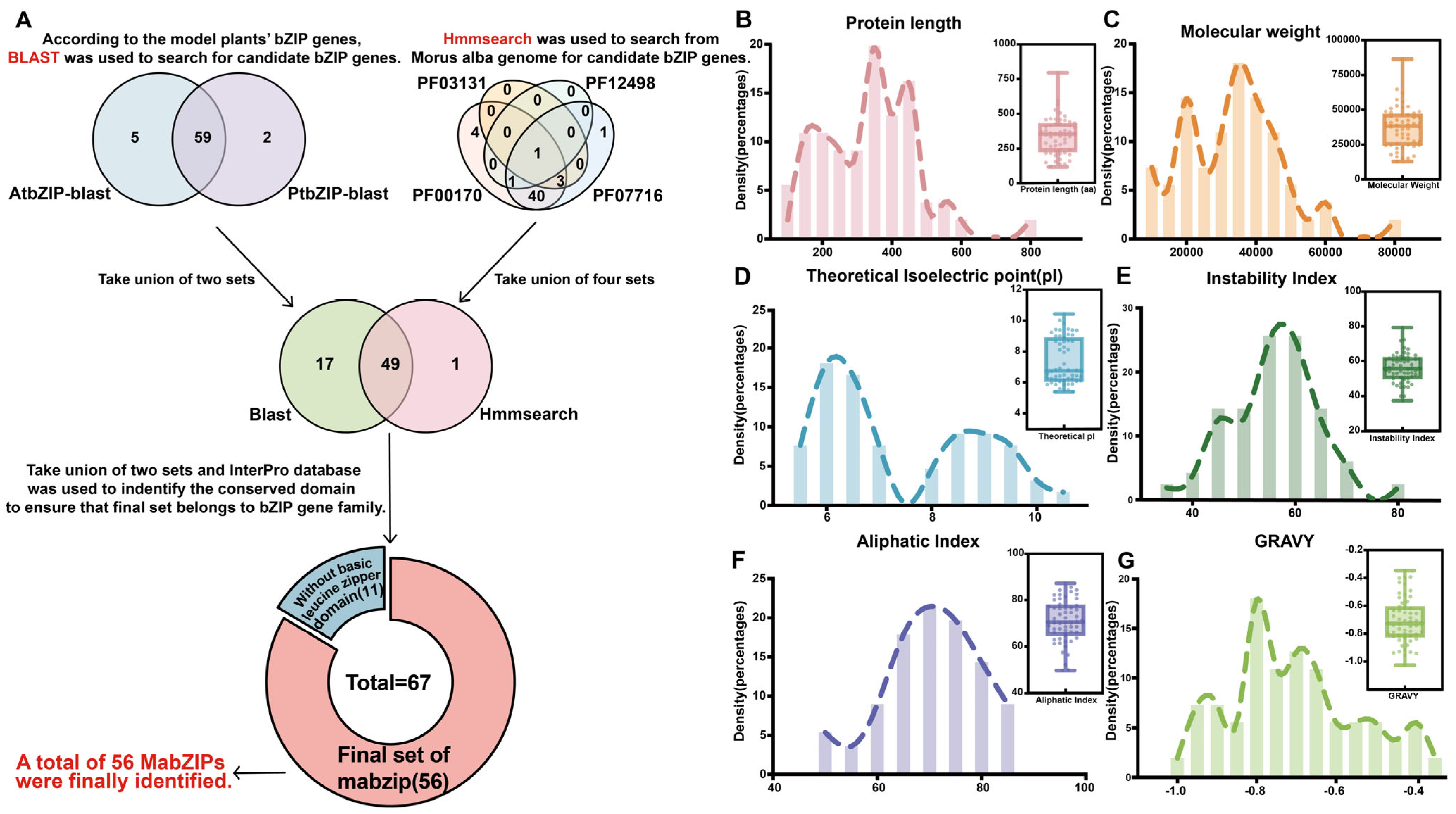
2.2. Identification of Mulberry bZIP Transcription Factors and Anthocyanin Biosynthesis Genes
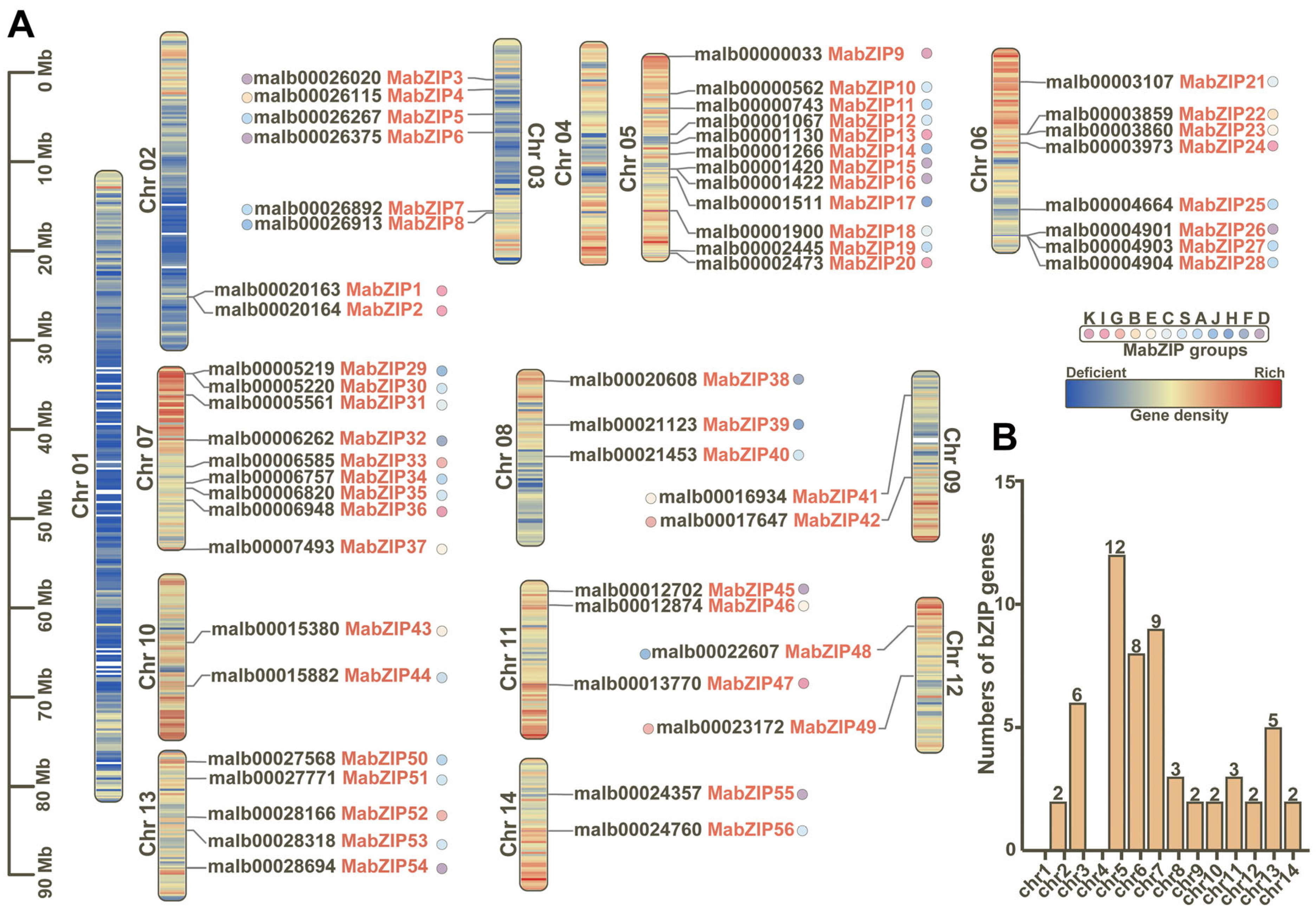
2.3. Classification, Chromosome Distribution, and Sequences Analysis on the bZIP Members
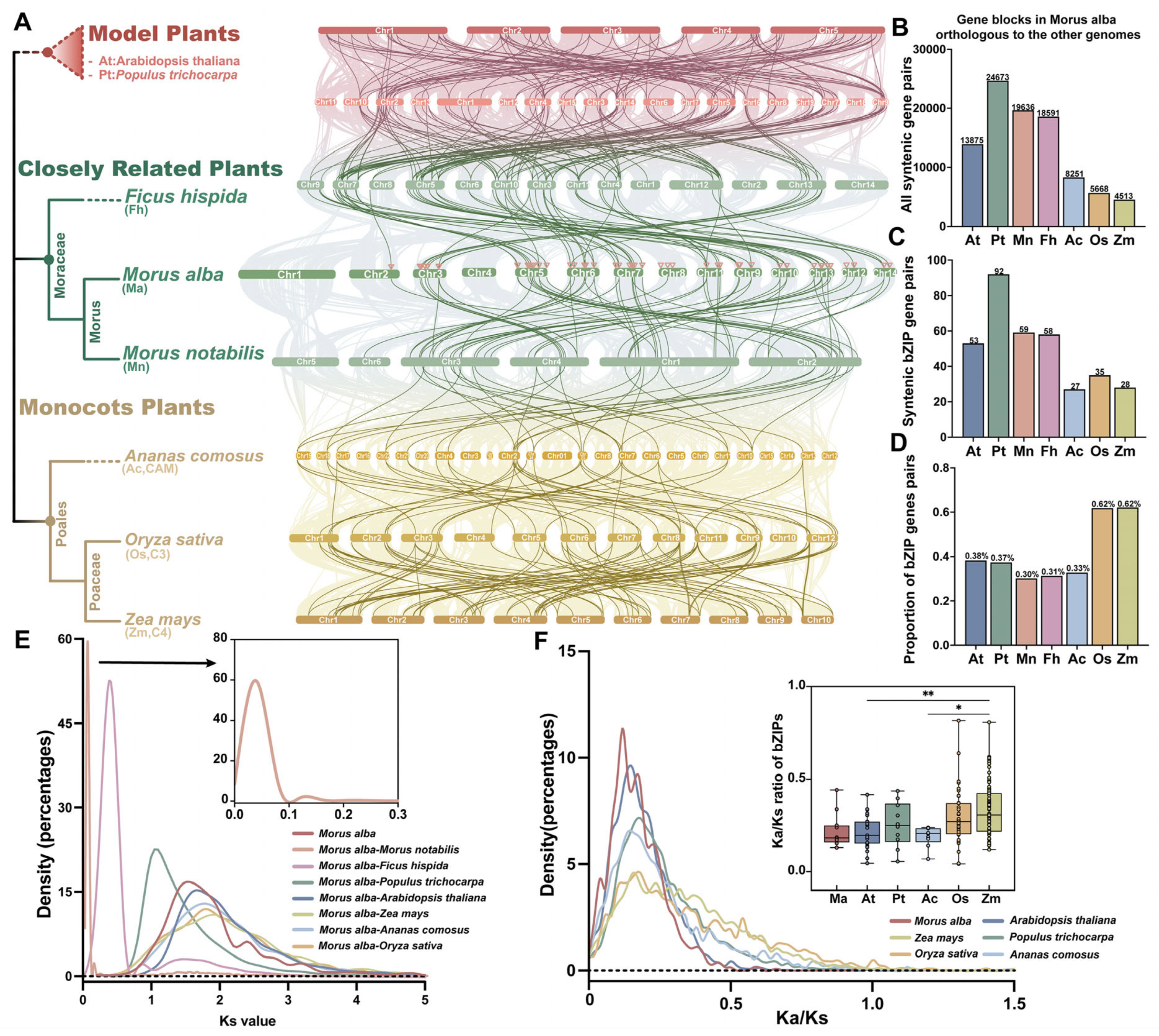
2.4. Collinearity Analysis and Promoter Cis-Acting Elements Prediction
2.5. Gene Expression Analysis, PPI Network, GO Enrichment Analysis
2.6. Plant Materials, Treatments, and Quantitative Real-Time PCR
3. Results
3.1. Identification and Physicochemical Properties Analysis of MabZIP Members in Morus alba
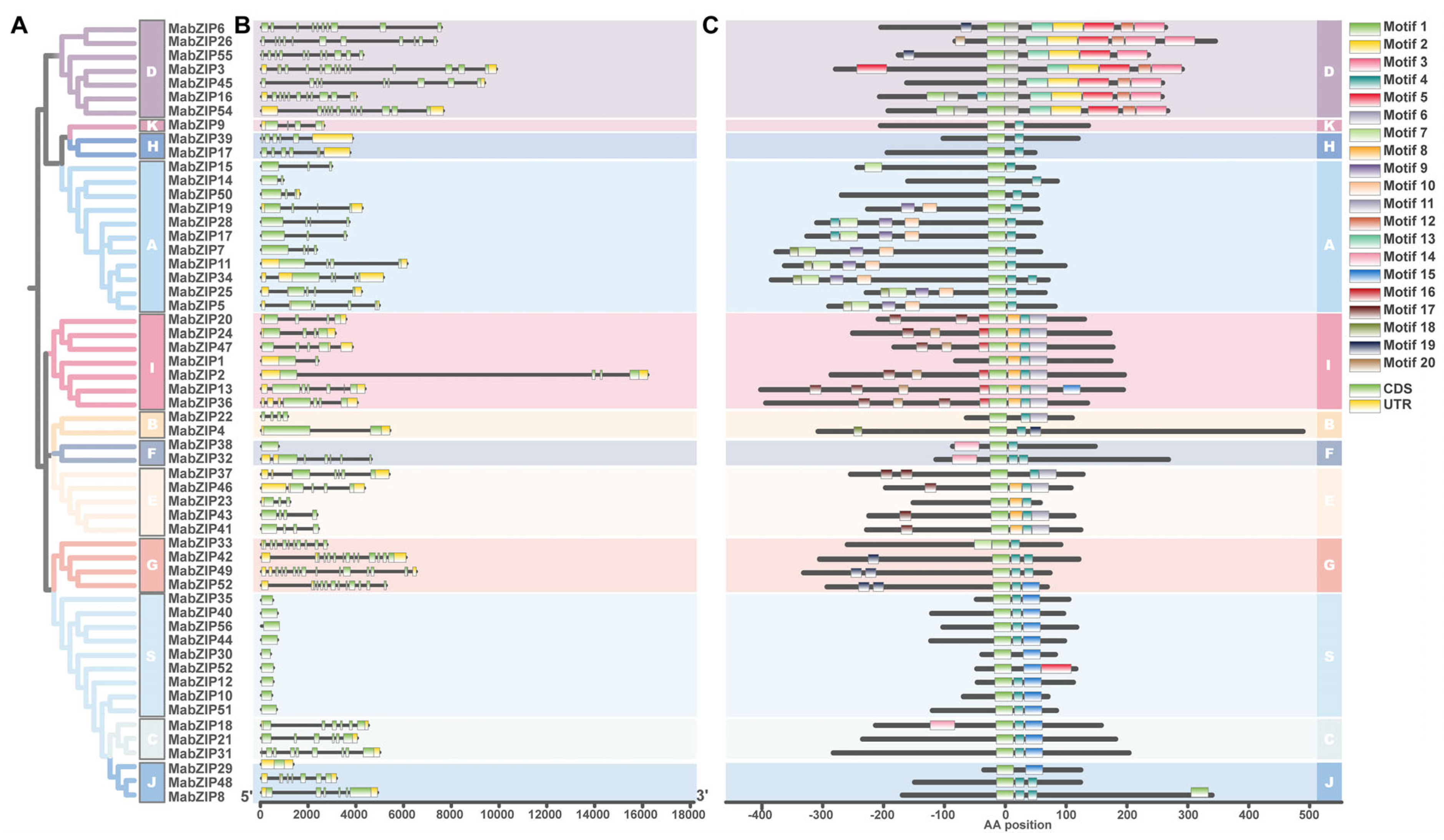
3.2. Chromosome Distribution and Phylogenetic Relationship Analysis of bZIP Genes
3.3. Conserved Motif and Gene Structure Analysis of MabZIP Genes
3.4. Collinearity and Evolutionary Analysis of bZIP Genes in Mulberry
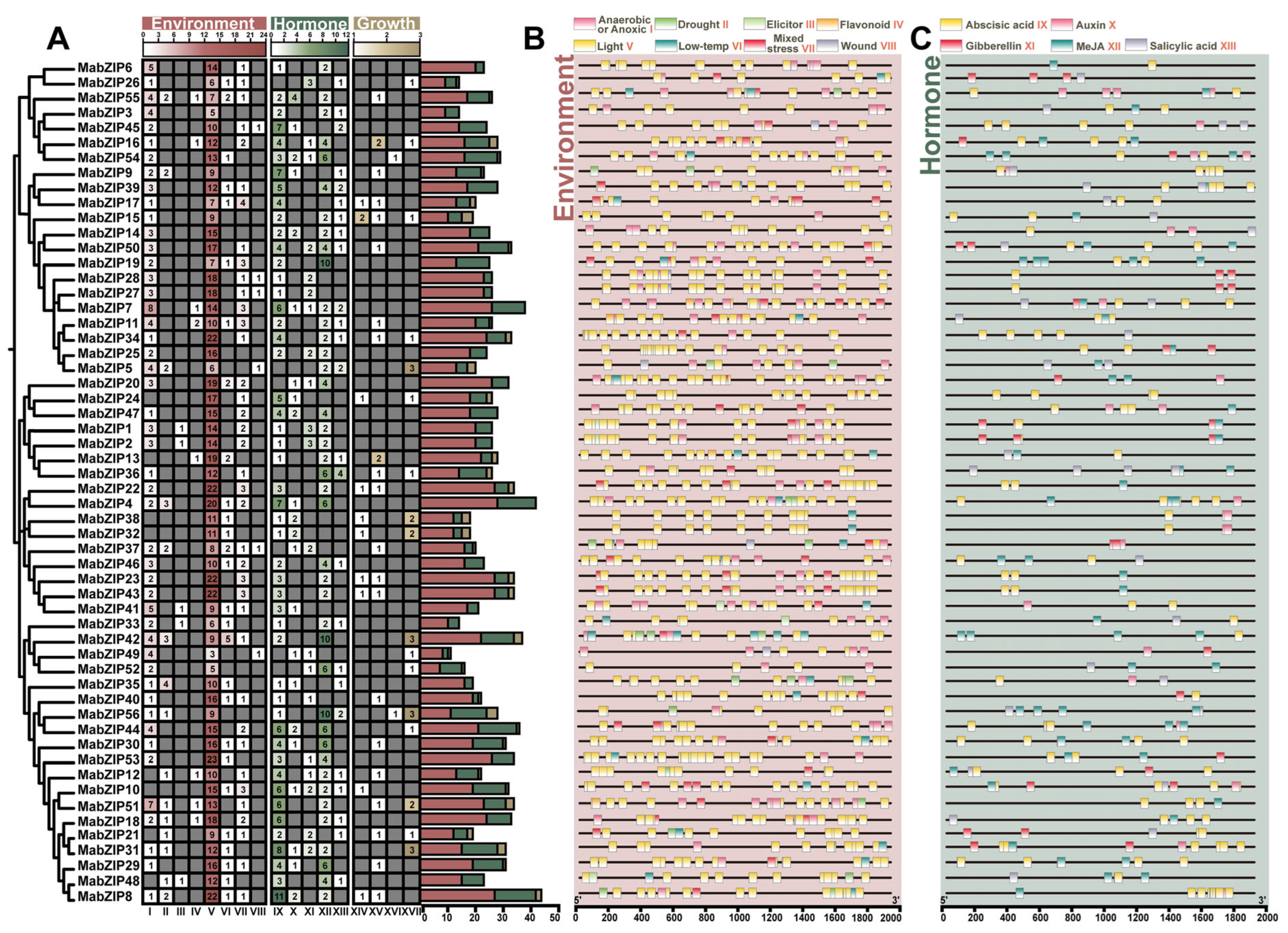
3.5. The Cis-Acting Elements Analysis of the bZIP Genes Promoter in Mulberry
3.6. Gene Expression in Response to Heat, Salt–Alkaline, and High Light Stress
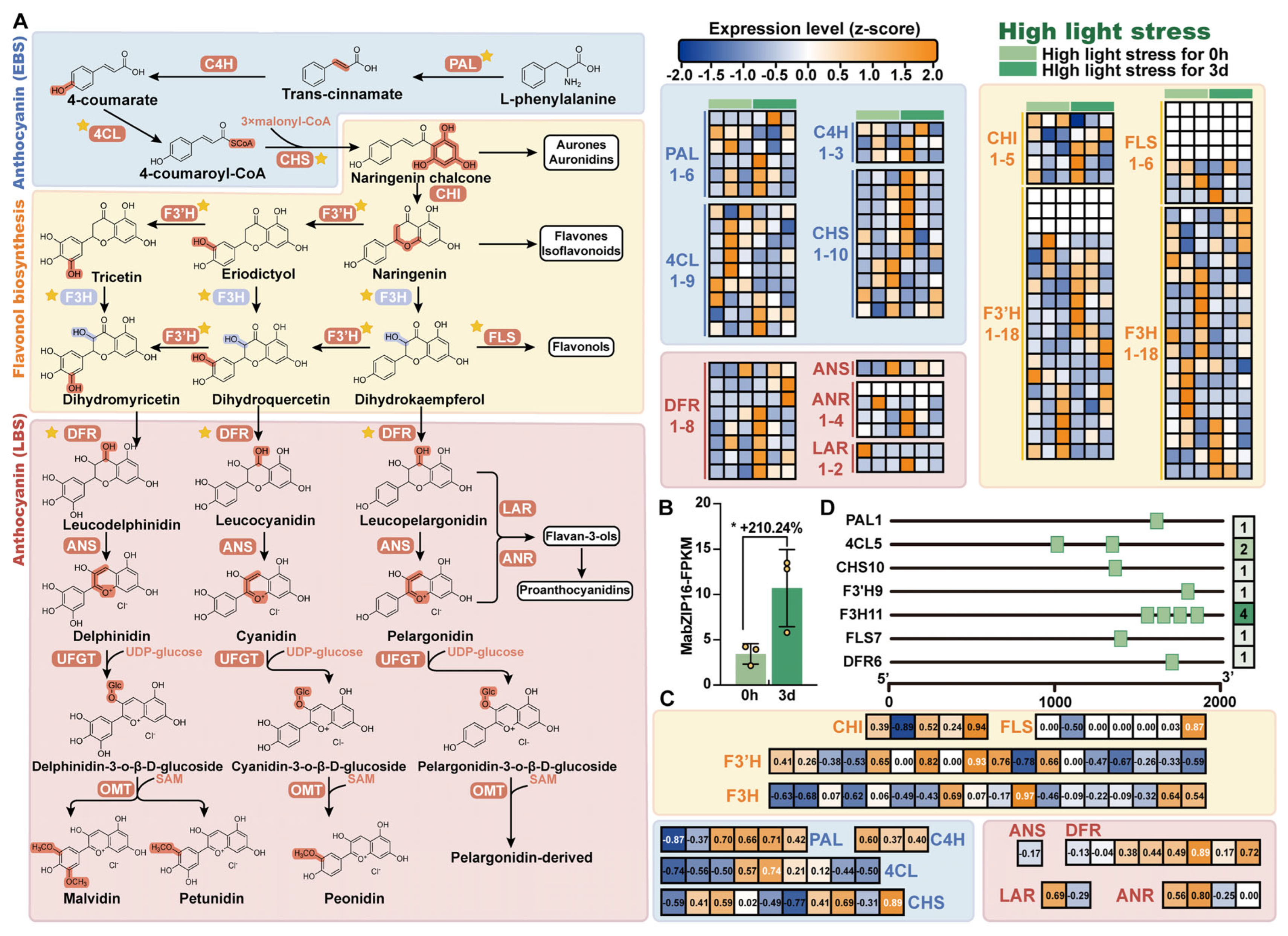
3.7. Identification of MabZIP Genes Involved in Anthocyanin Biosynthesis
3.8. Validation of Candidate Genes by Quantitive Real-Time PCR
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Jakoby, M.; Weisshaar, B.; Droge-Laser, W.; Vicente-Carbajosa, J.; Tiedemann, J.; Kroj, T.; Parcy, F. bZIP transcription factors in Arabidopsis. Trends Plant Sci. 2002, 7, 106–111. [Google Scholar] [CrossRef] [PubMed]
- Landschulz, W.H.; Johnson, P.F.; McKnight, S.L. The leucine zipper: A hypothetical structure common to a new class of DNA binding proteins. Science 1988, 240, 1759–1764. [Google Scholar] [CrossRef] [PubMed]
- Schutze, K.; Harter, K.; Chaban, C. Post-translational regulation of plant bZIP factors. Trends Plant Sci. 2008, 13, 247–255. [Google Scholar] [CrossRef] [PubMed]
- Droge-Laser, W.; Snoek, B.L.; Snel, B.; Weiste, C. The Arabidopsis bZIP transcription factor family-an update. Curr. Opin. Plant Biol. 2018, 45, 36–49. [Google Scholar] [CrossRef]
- Zhao, K.; Chen, S.; Yao, W.; Cheng, Z.; Zhou, B.; Jiang, T. Genome-wide analysis and expression profile of the bZIP gene family in poplar. BMC Plant Biol. 2021, 21, 122. [Google Scholar] [CrossRef]
- Fang, K.; Yao, X.; Tian, Y.; He, Y.; Lin, Y.; Lei, W.; Peng, S.; Pan, G.; Shi, H.; Zhang, D.; et al. Ubiquitin-specific protease UBP14 stabilizes HY5 by deubiquitination to promote photomorphogenesis in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2024, 121, e2404883121. [Google Scholar] [CrossRef]
- Gangappa, S.N.; Botto, J.F. The Multifaceted Roles of HY5 in Plant Growth and Development. Mol. Plant 2016, 9, 1353–1365. [Google Scholar] [CrossRef]
- Cackett, L.; Luginbuehl, L.H.; Schreier, T.B.; Lopez-Juez, E.; Hibberd, J.M. Chloroplast development in green plant tissues: The interplay between light, hormone, and transcriptional regulation. New Phytol. 2022, 233, 2000–2016. [Google Scholar] [CrossRef]
- Xu, X.; Chi, W.; Sun, X.; Feng, P.; Guo, H.; Li, J.; Lin, R.; Lu, C.; Wang, H.; Leister, D.; et al. Convergence of light and chloroplast signals for de-etiolation through ABI4-HY5 and COP1. Nat. Plants 2016, 2, 16066. [Google Scholar] [CrossRef]
- Ali, Z.; Sun, Y.; Ma, Z.; Zheng, Y.; Liu, Y. VvHY5 and VvBEE1 antagonistically control resveratrol biosynthesis to mitigate high light-induced damage in grapevine. J. Integr. Plant Biol. 2025, 67, 993–1008. [Google Scholar] [CrossRef]
- Brown, B.A.; Jenkins, G.I. UV-B signaling pathways with different fluence-rate response profiles are distinguished in mature Arabidopsis leaf tissue by requirement for UVR8, HY5, and HYH. Plant Physiol. 2008, 146, 576–588. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; Xu, J.; Lin, R.; Song, J.; Shao, S.; Yu, J.; Zhou, Y. Light-induced HY5 Functions as a Systemic Signal to Coordinate the Photoprotective Response to Light Fluctuation. Plant Physiol. 2020, 184, 1181–1193. [Google Scholar] [CrossRef] [PubMed]
- Giri, M.K.; Singh, N.; Banday, Z.Z.; Singh, V.; Ram, H.; Singh, D.; Chattopadhyay, S.; Nandi, A.K. GBF1 differentially regulates CAT2 and PAD4 transcription to promote pathogen defense in Arabidopsis thaliana. Plant J 2017, 91, 802–815. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.Y.; Ma, N.; Sun, P.; Zhang, F.J.; Li, L.; Li, H.; Zhang, S.; Wang, X.F.; You, C.X.; Zhang, Z. Fungal invasion-induced accumulation of salicylic acid promotes anthocyanin biosynthesis through MdNPR1-MdTGA2.2 module in apple fruits. Plant J. 2024, 119, 1859–1879. [Google Scholar] [CrossRef]
- Pieterse, C.M.; Van Loon, L.C. NPR1: The spider in the web of induced resistance signaling pathways. Curr. Opin. Plant Biol. 2004, 7, 456–464. [Google Scholar] [CrossRef]
- Lu, C.; Liu, X.; Tang, Y.; Fu, Y.; Zhang, J.; Yang, L.; Li, P.; Zhu, Z.; Dong, P. A comprehensive review of TGA transcription factors in plant growth, stress responses, and beyond. Int. J. Biol. Macromol. 2024, 258, 128880. [Google Scholar] [CrossRef]
- Zavaliev, R.; Dong, X. NPR1, a key immune regulator for plant survival under biotic and abiotic stresses. Mol. Cell 2024, 84, 131–141. [Google Scholar] [CrossRef]
- Behringer, C.; Bartsch, K.; Schaller, A. Safeners recruit multiple signalling pathways for the orchestrated induction of the cellular xenobiotic detoxification machinery in Arabidopsis. Plant Cell Environ. 2011, 34, 1970–1985. [Google Scholar] [CrossRef]
- Mueller, S.; Hilbert, B.; Dueckershoff, K.; Roitsch, T.; Krischke, M.; Mueller, M.J.; Berger, S. General detoxification and stress responses are mediated by oxidized lipids through TGA transcription factors in Arabidopsis. Plant Cell 2008, 20, 768–785. [Google Scholar] [CrossRef]
- Kim, S.Y.; Ma, J.; Perret, P.; Li, Z.; Thomas, T.L. Arabidopsis ABI5 subfamily members have distinct DNA-binding and transcriptional activities. Plant Physiol. 2002, 130, 688–697. [Google Scholar] [CrossRef]
- Skubacz, A.; Daszkowska-Golec, A.; Szarejko, I. The Role and Regulation of ABI5 (ABA-Insensitive 5) in Plant Development, Abiotic Stress Responses and Phytohormone Crosstalk. Front. Plant Sci. 2016, 7, 1884. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Yang, B.; Li, J.; Wang, Y.; Tao, R.; Yang, F.; Wu, X.; Yan, X.; Ahmad, M.; Shen, J.; et al. ABA-responsive ABRE-BINDING FACTOR3 activates DAM3 expression to promote bud dormancy in Asian pear. Plant Cell Environ. 2020, 43, 1360–1375. [Google Scholar] [CrossRef] [PubMed]
- Dietrich, K.; Weltmeier, F.; Ehlert, A.; Weiste, C.; Stahl, M.; Harter, K.; Droge-Laser, W. Heterodimers of the Arabidopsis transcription factors bZIP1 and bZIP53 reprogram amino acid metabolism during low energy stress. Plant Cell 2011, 23, 381–395. [Google Scholar] [CrossRef] [PubMed]
- Droge-Laser, W.; Weiste, C. The C/S(1) bZIP Network: A Regulatory Hub Orchestrating Plant Energy Homeostasis. Trends Plant Sci. 2018, 23, 422–433. [Google Scholar] [CrossRef]
- Hartmann, L.; Pedrotti, L.; Weiste, C.; Fekete, A.; Schierstaedt, J.; Gottler, J.; Kempa, S.; Krischke, M.; Dietrich, K.; Mueller, M.J.; et al. Crosstalk between Two bZIP Signaling Pathways Orchestrates Salt-Induced Metabolic Reprogramming in Arabidopsis Roots. Plant Cell 2015, 27, 2244–2260. [Google Scholar] [CrossRef]
- Gai, S.; Du, B.; Xiao, Y.; Zhang, X.; Turupu, M.; Yao, Q.; Wang, X.; Yan, Y.; Li, T. bZIP Transcription Factor PavbZIP6 Regulates Anthocyanin Accumulation by Increasing Abscisic Acid in Sweet Cherry. Int. J. Mol. Sci. 2024, 25, 10207. [Google Scholar] [CrossRef]
- Xu, Z.; Wang, J.; Ma, Y.; Wang, F.; Wang, J.; Zhang, Y.; Hu, X. The bZIP transcription factor SlAREB1 regulates anthocyanin biosynthesis in response to low temperature in tomato. Plant J. 2023, 115, 205–219. [Google Scholar] [CrossRef]
- An, J.P.; Yao, J.F.; Xu, R.R.; You, C.X.; Wang, X.F.; Hao, Y.J. Apple bZIP transcription factor MdbZIP44 regulates abscisic acid-promoted anthocyanin accumulation. Plant Cell Environ. 2018, 41, 2678–2692. [Google Scholar] [CrossRef]
- Liu, C.C.; Chi, C.; Jin, L.J.; Zhu, J.; Yu, J.Q.; Zhou, Y.H. The bZip transcription factor HY5 mediates CRY1a-induced anthocyanin biosynthesis in tomato. Plant Cell Environ. 2018, 41, 1762–1775. [Google Scholar] [CrossRef]
- Shin, D.H.; Choi, M.; Kim, K.; Bang, G.; Cho, M.; Choi, S.B.; Choi, G.; Park, Y.I. HY5 regulates anthocyanin biosynthesis by inducing the transcriptional activation of the MYB75/PAP1 transcription factor in Arabidopsis. FEBS Lett. 2013, 587, 1543–1547. [Google Scholar] [CrossRef]
- Stracke, R.; Favory, J.J.; Gruber, H.; Bartelniewoehner, L.; Bartels, S.; Binkert, M.; Funk, M.; Weisshaar, B.; Ulm, R. The Arabidopsis bZIP transcription factor HY5 regulates expression of the PFG1/MYB12 gene in response to light and ultraviolet-B radiation. Plant Cell Environ. 2010, 33, 88–103. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Xu, P.; Chen, G.; Wu, J.; Liu, Z.; Lian, H. FvbHLH9 Functions as a Positive Regulator of Anthocyanin Biosynthesis by Forming a HY5-bHLH9 Transcription Complex in Strawberry Fruits. Plant Cell Physiol. 2020, 61, 826–837. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Wei, J.; Li, S.; Li, J.; Cao, H.; Huang, D.; Zhang, D.; Zhang, Z.; Gao, T.; Zhang, Y.; et al. MdHY5 positively regulates cold tolerance in apple by integrating the auxin and abscisic acid pathways. New Phytol. 2025, 246, 2155–2173. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Chen, G.; Ma, B.; Zhong, C.; He, N. Metabolic Profiling and Transcriptome Analysis of Mulberry Leaves Provide Insights into Flavonoid Biosynthesis. J. Agric. Food Chem. 2020, 68, 1494–1504. [Google Scholar] [CrossRef]
- Li, H.; Yang, Z.; Zeng, Q.; Wang, S.; Luo, Y.; Huang, Y.; Xin, Y.; He, N. Abnormal expression of bHLH3 disrupts a flavonoid homeostasis network, causing differences in pigment composition among mulberry fruits. Hortic. Res. 2020, 7, 83. [Google Scholar] [CrossRef]
- Liu, C.; Hu, J.; Fan, W.; Zhu, P.; Cao, B.; Zheng, S.; Xia, Z.; Zhu, Y.; Zhao, A. Heterotrimeric G-protein gamma subunits regulate ABA signaling in response to drought through interacting with PP2Cs and SnRK2s in mulberry (Morus alba L.). Plant Physiol. Biochem. 2021, 161, 210–221. [Google Scholar] [CrossRef]
- Ma, L.; Ma, Y.; Gao, Q.; Liu, S.; Zhu, Z.; Shi, X.; Dai, F.; Reis, R.L.; Kundu, S.C.; Cai, K.; et al. Mulberry Leaf Lipid Nanoparticles: A Naturally Targeted CRISPR/Cas9 Oral Delivery Platform for Alleviation of Colon Diseases. Small 2024, 20, e2307247. [Google Scholar] [CrossRef]
- He, N.; Zhang, C.; Qi, X.; Zhao, S.; Tao, Y.; Yang, G.; Lee, T.H.; Wang, X.; Cai, Q.; Li, D.; et al. Draft genome sequence of the mulberry tree Morus notabilis. Nat. Commun. 2013, 4, 2445. [Google Scholar] [CrossRef]
- Jiao, F.; Luo, R.; Dai, X.; Liu, H.; Yu, G.; Han, S.; Lu, X.; Su, C.; Chen, Q.; Song, Q.; et al. Chromosome-Level Reference Genome and Population Genomic Analysis Provide Insights into the Evolution and Improvement of Domesticated Mulberry (Morus alba). Mol. Plant 2020, 13, 1001–1012. [Google Scholar] [CrossRef]
- Liu, L.; Chao, N.; Yidilisi, K.; Kang, X.; Cao, X. Comprehensive analysis of the MYB transcription factor gene family in Morus alba. BMC Plant Biol. 2022, 22, 281. [Google Scholar] [CrossRef]
- Ma, B.; Luo, Y.; Jia, L.; Qi, X.; Zeng, Q.; Xiang, Z.; He, N. Genome-wide identification and expression analyses of cytochrome P450 genes in mulberry (Morus notabilis). J. Integr. Plant Biol. 2014, 56, 887–901. [Google Scholar] [CrossRef] [PubMed]
- Hufford, M.B.; Seetharam, A.S.; Woodhouse, M.R.; Chougule, K.M.; Ou, S.; Liu, J.; Ricci, W.A.; Guo, T.; Olson, A.; Qiu, Y.; et al. De novo assembly, annotation, and comparative analysis of 26 diverse maize genomes. Science 2021, 373, 655–662. [Google Scholar] [CrossRef] [PubMed]
- Ming, R.; VanBuren, R.; Wai, C.M.; Tang, H.; Schatz, M.C.; Bowers, J.E.; Lyons, E.; Wang, M.L.; Chen, J.; Biggers, E.; et al. The pineapple genome and the evolution of CAM photosynthesis. Nat. Genet. 2015, 47, 1435–1442. [Google Scholar] [CrossRef] [PubMed]
- Goodstein, D.M.; Shu, S.; Howson, R.; Neupane, R.; Hayes, R.D.; Fazo, J.; Mitros, T.; Dirks, W.; Hellsten, U.; Putnam, N.; et al. Phytozome: A comparative platform for green plant genomics. Nucleic Acids Res. 2012, 40, D1178–D1186. [Google Scholar] [CrossRef]
- Ouyang, S.; Zhu, W.; Hamilton, J.; Lin, H.; Campbell, M.; Childs, K.; Thibaud-Nissen, F.; Malek, R.L.; Lee, Y.; Zheng, L.; et al. The TIGR Rice Genome Annotation Resource: Improvements and new features. Nucleic Acids Res. 2007, 35, D883–D887. [Google Scholar] [CrossRef]
- Tuskan, G.A.; Difazio, S.; Jansson, S.; Bohlmann, J.; Grigoriev, I.; Hellsten, U.; Putnam, N.; Ralph, S.; Rombauts, S.; Salamov, A.; et al. The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 2006, 313, 1596–1604. [Google Scholar] [CrossRef]
- Liao, Z.; Zhang, T.; Lei, W.; Wang, Y.; Yu, J.; Wang, Y.; Chai, K.; Wang, G.; Zhang, H.; Zhang, X. A telomere-to-telomere reference genome of ficus (Ficus hispida) provides new insights into sex determination. Hortic. Res. 2024, 11, uhad257. [Google Scholar] [CrossRef]
- Stiehler, F.; Steinborn, M.; Scholz, S.; Dey, D.; Weber, A.P.M.; Denton, A.K. Helixer: Cross-species gene annotation of large eukaryotic genomes using deep learning. Bioinformatics 2021, 36, 5291–5298. [Google Scholar] [CrossRef]
- Blum, M.; Andreeva, A.; Florentino, L.C.; Chuguransky, S.R.; Grego, T.; Hobbs, E.; Pinto, B.L.; Orr, A.; Paysan-Lafosse, T.; Ponamareva, I.; et al. InterPro: The protein sequence classification resource in 2025. Nucleic Acids Res. 2025, 53, D444–D456. [Google Scholar] [CrossRef]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y.; et al. TBtools-II: A “one for all, all for one” bioinformatics platform for biological big-data mining. Mol. Plant 2023, 16, 1733–1742. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Tang, H.; Debarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef]
- Lescot, M.; Dehais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouze, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Yang, L.; Hou, Z.; Liu, C.; Zhu, C.; Qin, Y.; Wang, X. Exogenous γ-aminobutyric acid enhanced salt-alkaline tolerance in mulberry trees through transcriptomic sequencing analysis. Plant Stress. 2024, 14, 100595. [Google Scholar] [CrossRef]
- Anders, S.; Huber, W. Differential expression analysis for sequence count data. Genome Biol. 2010, 11, R106. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Nastou, K.; Koutrouli, M.; Kirsch, R.; Mehryary, F.; Hachilif, R.; Hu, D.; Peluso, M.E.; Huang, Q.; Fang, T.; et al. The STRING database in 2025: Protein networks with directionality of regulation. Nucleic Acids Res. 2025, 53, D730–D737. [Google Scholar] [CrossRef]
- Hou, Z.; Xu, D.; Deng, N.; Li, Y.; Yang, L.; Li, S.; Zhou, H.; Huang, Q.; Wang, X. Comparative proteomics of mulberry leaves at different developmental stages identify novel proteins function related to photosynthesis. Front. Plant Sci. 2021, 12, 797631. [Google Scholar] [CrossRef]
- Hou, Z.; Pang, X.; Hedtke, B.; Grimm, B. In vivo functional analysis of the structural domains of FLUORESCENT (FLU). Plant J. 2021, 107, 360–376. [Google Scholar] [CrossRef]
- Wu, S.; Han, B.; Jiao, Y. Genetic Contribution of Paleopolyploidy to Adaptive Evolution in Angiosperms. Mol. Plant 2020, 13, 59–71. [Google Scholar] [CrossRef] [PubMed]
- Panchy, N.; Lehti-Shiu, M.; Shiu, S.H. Evolution of Gene Duplication in Plants. Plant Physiol. 2016, 171, 2294–2316. [Google Scholar] [CrossRef] [PubMed]
- Freeling, M. Bias in plant gene content following different sorts of duplication: Tandem, whole-genome, segmental, or by transposition. Annu. Rev. Plant Biol. 2009, 60, 433–453. [Google Scholar] [CrossRef] [PubMed]
- Maere, S.; De Bodt, S.; Raes, J.; Casneuf, T.; Van Montagu, M.; Kuiper, M.; Van de Peer, Y. Modeling gene and genome duplications in eukaryotes. Proc. Natl. Acad. Sci. USA 2005, 102, 5454–5459. [Google Scholar] [CrossRef]
- Soltis, P.S.; Soltis, D.E. Ancient WGD events as drivers of key innovations in angiosperms. Curr. Opin. Plant Biol. 2016, 30, 159–165. [Google Scholar] [CrossRef]
- Khan, S.A.; Li, M.Z.; Wang, S.M.; Yin, H.J. Revisiting the Role of Plant Transcription Factors in the Battle against Abiotic Stress. Int. J. Mol. Sci. 2018, 19, 1634. [Google Scholar] [CrossRef]
- Jing, Y.; Lin, R. The VQ Motif-Containing Protein Family of Plant-Specific Transcriptional Regulators. Plant Physiol. 2015, 169, 371–378. [Google Scholar] [CrossRef]
- Mikheyeva, I.V.; Grady, P.J.; Tamburini, F.B.; Lorenz, D.R.; Cam, H.P. Multifaceted genome control by Set1 Dependent and Independent of H3K4 methylation and the Set1C/COMPASS complex. PLoS Genet. 2014, 10, e1004740. [Google Scholar] [CrossRef]
- Zirngibl, M.E.; Araguirang, G.E.; Kitashova, A.; Jahnke, K.; Rolka, T.; Kuhn, C.; Nagele, T.; Richter, A.S. Triose phosphate export from chloroplasts and cellular sugar content regulate anthocyanin biosynthesis during high light acclimation. Plant Commun. 2023, 4, 100423. [Google Scholar] [CrossRef]
- Gao, Z.; Sun, Y.; Zhu, Z.; Ni, N.; Sun, S.; Nie, M.; Du, W.; Irfan, M.; Chen, L.; Zhang, L. Transcription factors LvBBX24 and LvbZIP44 coordinated anthocyanin accumulation in response to light in lily petals. Hortic. Res. 2024, 11, uhae211. [Google Scholar] [CrossRef]
- Liu, W.; Mei, Z.; Yu, L.; Gu, T.; Li, Z.; Zou, Q.; Zhang, S.; Fang, H.; Wang, Y.; Zhang, Z.; et al. The ABA-induced NAC transcription factor MdNAC1 interacts with a bZIP-type transcription factor to promote anthocyanin synthesis in red-fleshed apples. Hortic. Res. 2023, 10, uhad049. [Google Scholar] [CrossRef] [PubMed]
- Dai, F.; Zhuo, X.; Luo, G.; Wang, Z.; Xu, Y.; Wang, D.; Zhong, J.; Lin, S.; Chen, L.; Li, Z.; et al. Genomic Resequencing Unravels the Genetic Basis of Domestication, Expansion, and Trait Improvement in Morus Atropurpurea. Adv. Sci. (Weinh) 2023, 10, e2300039. [Google Scholar] [CrossRef]
- Ma, B.; Wang, H.; Liu, J.; Chen, L.; Xia, X.; Wei, W.; Yang, Z.; Yuan, J.; Luo, Y.; He, N. The gap-free genome of mulberry elucidates the architecture and evolution of polycentric chromosomes. Hortic. Res. 2023, 10, uhad111. [Google Scholar] [CrossRef] [PubMed]
- Duan, L.; Mo, Z.; Fan, Y.; Li, K.; Yang, M.; Li, D.; Ke, Y.; Zhang, Q.; Wang, F.; Fan, Y.; et al. Genome-wide identification and expression analysis of the bZIP transcription factor family genes in response to abiotic stress in Nicotiana tabacum L. BMC Genom. 2022, 23, 318. [Google Scholar] [CrossRef] [PubMed]
- Ko, D.K.; Brandizzi, F. Transcriptional competition shapes proteotoxic ER stress resolution. Nat. Plants 2022, 8, 481–490. [Google Scholar] [CrossRef]
- Ko, D.K.; Brandizzi, F. Dynamics of ER stress-induced gene regulation in plants. Nat. Rev. Genet. 2024, 25, 513–525. [Google Scholar] [CrossRef]
- Ruberti, C.; Lai, Y.; Brandizzi, F. Recovery from temporary endoplasmic reticulum stress in plants relies on the tissue-specific and largely independent roles of bZIP28 and bZIP60, as well as an antagonizing function of BAX-Inhibitor 1 upon the pro-adaptive signaling mediated by bZIP28. Plant J. 2018, 93, 155–165. [Google Scholar] [CrossRef]
- Azeem, F.; Tahir, H.; Ijaz, U.; Shaheen, T. A genome-wide comparative analysis of bZIP transcription factors in G. arboreum and G. raimondii (Diploid ancestors of present-day cotton). Physiol. Mol. Biol. Plants 2020, 26, 433–444. [Google Scholar] [CrossRef]
- Yang, Y.; Li, J.; Li, H.; Yang, Y.; Guang, Y.; Zhou, Y. The bZIP gene family in watermelon: Genome-wide identification and expression analysis under cold stress and root-knot nematode infection. PeerJ 2019, 7, e7878. [Google Scholar] [CrossRef]
- Zhijun, Z.; Peiyao, Y.; Bing, H.; Ruifang, M.; Vinod, K.K.; Ramakrishnan, M. Genome-wide identification and expression characterization of the DoG gene family of moso bamboo (Phyllostachys edulis). BMC Genom. 2022, 23, 357. [Google Scholar] [CrossRef]
- Wang, Y.; Salasini, B.C.; Khan, M.; Devi, B.; Bush, M.; Subramaniam, R.; Hepworth, S.R. Clade I TGACG-Motif Binding Basic Leucine Zipper Transcription Factors Mediate BLADE-ON-PETIOLE-Dependent Regulation of Development. Plant Physiol. 2019, 180, 937–951. [Google Scholar] [CrossRef] [PubMed]
- Das, P.; Lakra, N.; Nutan, K.K.; Singla-Pareek, S.L.; Pareek, A. A unique bZIP transcription factor imparting multiple stress tolerance in Rice. Rice 2019, 12, 58. [Google Scholar] [CrossRef] [PubMed]
- Nijhawan, A.; Jain, M.; Tyagi, A.K.; Khurana, J.P. Genomic survey and gene expression analysis of the basic leucine zipper transcription factor family in rice. Plant Physiol. 2008, 146, 333–350. [Google Scholar] [CrossRef] [PubMed]
- Wei, K.; Chen, J.; Wang, Y.; Chen, Y.; Chen, S.; Lin, Y.; Pan, S.; Zhong, X.; Xie, D. Genome-wide analysis of bZIP-encoding genes in maize. DNA Res. 2012, 19, 463–476. [Google Scholar] [CrossRef]
- Wright, S.I.; Bi, I.V.; Schroeder, S.G.; Yamasaki, M.; Doebley, J.F.; McMullen, M.D.; Gaut, B.S. The effects of artificial selection on the maize genome. Science 2005, 308, 1310–1314. [Google Scholar] [CrossRef]
- Xu, D.; Lin, L.; Liu, X.; Wangzha, M.; Pang, X.; Feng, L.; Wan, B.; Wu, G.Z.; Yu, J.; Rochaix, J.D.; et al. Characterization of a tomato chlh mis-sense mutant reveals a new function of ChlH in fruit ripening. Plant Biotechnol. J. 2025, 23, 911–926. [Google Scholar] [CrossRef]
- Lin, R.; Zhang, W.; Tian, R.; Zhang, L.; Hong, J.; Wang, L.; Kang, H.; Yu, J.; Zhou, Y. CPK27 enhances cold tolerance by promoting flavonoid biosynthesis through phosphorylating HY5 in tomato. New Phytol. 2025, 246, 2174–2191. [Google Scholar] [CrossRef]
- Sun, T.; Hazra, A.; Lui, A.; Zeng, S.; Wang, X.; Rao, S.; Owens, L.A.; Fei, Z.; Zhao, Y.; Mazourek, M.; et al. GLKs directly regulate carotenoid biosynthesis via interacting with GBFs in plants. New Phytol. 2025, 246, 645–665. [Google Scholar] [CrossRef]
- Wang, J.; Li, X.; Li, J.; Dong, H.; Hu, Z.; Xia, X.; Yu, J.; Zhou, Y. Manipulating the Light Systemic Signal HY5 Greatly Improve Fruit Quality in Tomato. Adv. Sci. (Weinh) 2025, e2500110. [Google Scholar] [CrossRef]
- Wiese, A.J.; Steinbachova, L.; Timofejeva, L.; Cermak, V.; Klodova, B.; Ganji, R.S.; Limones-Mendez, M.; Bokvaj, P.; Hafidh, S.; Potesil, D.; et al. Arabidopsis bZIP18 and bZIP52 Accumulate in Nuclei Following Heat Stress where They Regulate the Expression of a Similar Set of Genes. Int. J. Mol. Sci. 2021, 22, 530. [Google Scholar] [CrossRef]
- Yang, Y.; Kong, Q.; Tee, W.T.; Li, Y.; Low, P.M.; Patra, B.; Guo, L.; Yuan, L.; Ma, W. Transcription factor bZIP52 modulates Arabidopsis seed oil biosynthesis through interaction with WRINKLED1. Plant Physiol. 2023, 192, 2628–2639. [Google Scholar] [CrossRef] [PubMed]
- Gibalova, A.; Steinbachova, L.; Hafidh, S.; Blahova, V.; Gadiou, Z.; Michailidis, C.; Muller, K.; Pleskot, R.; Duplakova, N.; Honys, D. Characterization of pollen-expressed bZIP protein interactions and the role of ATbZIP18 in the male gametophyte. Plant Reprod. 2017, 30, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Dannfald, A.; Carpentier, M.C.; Merret, R.; Favory, J.J.; Deragon, J.M. Plant response to intermittent heat stress involves modulation of mRNA translation efficiency. Plant Physiol. 2025, 197, kiae648. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.; Yuan, Y.; Tang, Z.; Huang, Y.; Kang, C.; Deng, X.; Xu, Q. Retrotransposon promoter of Ruby1 controls both light- and cold-induced accumulation of anthocyanins in blood orange. Plant Cell Environ. 2019, 42, 3092–3104. [Google Scholar] [CrossRef]
- Li, S.; Yang, Y.; Yu, J.; Zhou, H.; Hou, Z.; Wang, X. Molecular and metabolic insights into purplish leaf coloration through the investigation of two mulberry (Morus alba) genotypes. BMC Plant Biol. 2024, 24, 61. [Google Scholar] [CrossRef]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Q.; Fang, H.; Zhou, H.; Wang, X.; Hou, Z. Identification and Characterization of bZIP Gene Family Combined Transcriptome Analysis Revealed Their Functional Roles on Abiotic Stress and Anthocyanin Biosynthesis in Mulberry (Morus alba). Horticulturae 2025, 11, 694. https://doi.org/10.3390/horticulturae11060694
Liu Q, Fang H, Zhou H, Wang X, Hou Z. Identification and Characterization of bZIP Gene Family Combined Transcriptome Analysis Revealed Their Functional Roles on Abiotic Stress and Anthocyanin Biosynthesis in Mulberry (Morus alba). Horticulturae. 2025; 11(6):694. https://doi.org/10.3390/horticulturae11060694
Chicago/Turabian StyleLiu, Qinghua, Haowen Fang, Hong Zhou, Xiling Wang, and Zhiwei Hou. 2025. "Identification and Characterization of bZIP Gene Family Combined Transcriptome Analysis Revealed Their Functional Roles on Abiotic Stress and Anthocyanin Biosynthesis in Mulberry (Morus alba)" Horticulturae 11, no. 6: 694. https://doi.org/10.3390/horticulturae11060694
APA StyleLiu, Q., Fang, H., Zhou, H., Wang, X., & Hou, Z. (2025). Identification and Characterization of bZIP Gene Family Combined Transcriptome Analysis Revealed Their Functional Roles on Abiotic Stress and Anthocyanin Biosynthesis in Mulberry (Morus alba). Horticulturae, 11(6), 694. https://doi.org/10.3390/horticulturae11060694




