Abstract
Papilionanthe Schltr. (Vandeae, Epidendroideae, Orchidaceae) is distinguished by its terete leaves and showy flowers, with significant horticultural and medicinal values. However, its systematic position in Aeridinae has been controversial and not been paid attention to or clarified. This study is focused on the complete chloroplast (cp) genomic data of P. biswasiana and P. teres in order to provide some genomic data for its phylogenetic relationship. The cp genomes of two Papilionanthe were 148,183 bp (P. biswasiana) and 148,145 bp (P. teres) in length, with similar GC content (36.5–36.6%). Comparative cp genomes of two Papilionanthe species and the other eight related taxa revealed differences in sequence analysis and statistics. A phylogenetic analysis based on CDS from complete cp genomes supported the notion that Papilionanthe is a monophyletic clade and closer to Luisia than to Paraphalaenopsis. It was obvious that there are four well-supported clades in Aeridinae, which could hold a significant implication for the phylogenetic relationship between Papilionanthe and other alliances in Aeridinae. Furthermore, the taxonomic positions of V. flabellata and H. himalacia were also reconfirmed herein by phylogenetic analysis of the cp genomes.
1. Introduction
Papilionanthe Schltr. (Aeridinae, Vandeae, Epidendroideae, Orchidaceae) is a small genus, consisting of about 12 species, that is distributed in India, China, Southeast Asia, and the Malay Archipelago [1]. There are four species recorded in China, including one endemic species [2] and the recently described P. motuoensis J.D.Ya & D.Z.Li. [3]. Some of them in China and SE Asia are perennial herbaceous orchids and epiphytic on tree trunks in forests [2], and others in Malaysia and Borneo often grow with shrubs and tall grasses for support in peat swamps [1]. Members of Papilionanthe can be easily distinguished from their Aeridinae alliance by the elongated stem with flesh and terete leaves and by the flat and large flowers. Most Papilionanthe species possess highly ornamental and medicinal values but have become scarce and endangered because of their habitat fragmentation and the over-exploration of wild resources [1,4].
However, the phylogenetic relationship of Papilionanthe in Aeridinae has been unclear since it was described based on Vanda teres (Roxburgh) Lindley [1,2]. It has been noted that the intergeneric delimitation and systematic positioning of Aeridinae are greatly problematic and also regarded as “a black pit” [5,6,7], since the group comprises about 1350 species in approximately 90 genera [8]. Previous research has been focused on Vanda Jones ex R.Br. [9], Aerides Lour. [10], and Gastrochilus D.Don [11], but the leafless orchid of Papilionanthe has been neglected. Papilonanthe was presumed to be related to Vanda and Aerides based on morphological features [5,12] and then to Holcoglossum Schltr. [13], Luisia Gaudich., and Paraphalaenopsis A.D.Hawkes. [8,14] based on molecular evidence. Taxonomic uncertainty has arisen due to the inconsistent treatment of Vanda species with terete leaves. Some of these Vanda species were transferred to Papilionanthe based on their morphological characteristics [5,12], while two others were reassigned to Papilionanthe following analyses using random amplified polymorphic DNA (RAPD) markers [15]. This inconsistency has contributed to confusion regarding the delimitation of these genera. Meanwhile, Papilionanthe is expected to be closer to Aerides than to Vanda, since A. cylindrica Lindl. and P. teres (Roxb.) Schltr. share a similar floral morphology [5]. In contrast, Papilionanthe was also presumed to be near to Holcoglossum, since P. teres and P. biswasiana (Ghose & Mukerjee) Garay were found to be sisters to the latter based on ITS, trnL–F, and matK analysis [13]. Thereafter, these two species of Papilionanthe were grouped into the “Vanda–Aerides alliance” [14,16], together with its three related genera (Vanda, Aerides, and Holcoglossum) and another three genera (Ascocentrum Schltr., Neofinetia Hu., and Rhynchostylis Bl.) based on DNA sequence data [17]. And, later, it was shown that Papilionanthe should be related to Luisia and Paraphalaenopsis, since they clustered into a clade with a strong support, a sister to Holcoglossum–Vanda, based on trnL–F, ITS, and cp DNA markers, but insufficient attention was paid to clarifying the confusing relationship of Papilionanthe [8,14].
The chloroplast genome, characterized by significantly reduced nucleotide substitutions and gene rearrangements relative to the nuclear genome, has emerged as a premier model for plant phylogenetics [8,11,13]. The chloroplast (cp) genome has been widely used in the phylogenetics of Orchidaceae, including Dendrobium Sw. [18], Phalaenopsis Blume [19], and Phaius Lour. [20]. And a thorough understanding of the phylogeny of some taxa in the “Vanda–Aerides alliance”, such as Vanda [9], Aerides [10], Holcoglossum [21], and Luisia [22,23], has been gained via cp genomic evidence, but, to date, no such research has concentrated specifically on Papilionanthe. Recently, the partial cp genome of Papilionanthe motuoensis (accession number: OR772949) and the complete cp genome of P. teres (accession number: OR772950) were sequenced and some plastid genes were selected as molecular markers for the identification of the former as a new species [3]. In fact, the phylogenetic relationship of Papilionanthe in the “Aerides–Vanda alliance” has not been resolved by molecular data, which presents an obstacle to understanding its phylogeny and conservation [1]. To address this, herein, the cp genomes of P. teres and P. biswasiana were characterized in detail and compared with those of eight related species from Luisia, Paraphalaenopsis, and Holcoglossum. Therefore, this study aimed to achieve the following: (1) characterize complete cp genome structures of two Papilionanthe species; (2) select some distinct genomic features for phylogenetic implications based on a cp genome comparison between Papilionanthe and its related taxon; and (3) reconstruct a phylogenetic tree based on cp genomic data to clarify the phylogenetic relationship of Papilionanthe in Aeridinae.
2. Materials and Methods
2.1. Plant Materials, Chloroplast Genome Sequencing, Assembly, and Annotation
Plants of Papilionanthe biswasiana and P. teres were cultivated in Xishuangbanna Tropical Botanical Garden, Chinese Academy of Sciences, Yunnan, China. Leaf samples were collected from cultivated plants for DNA extraction, and their voucher specimens were stored in the Herbarium of Southwest Forestry University (HSFU, Lilu20200720 and Lilu20200919, lilu@swfu.edu.cn). The Tiangen DNA kit (TIANGEN, Beijing, China) was employed to extract genomic DNA via the modified CTAB method [24]. Paired-end libraries with an average insert size of approximately 400 bp were prepared using a TruSeq DNA Sample Prep Kit (Illumina, Inc., San Diego, CA, USA) according to the manufacturer’s instructions [25]. The libraries were sequenced on the Illumina HiSeq 2500 platform at Personalbio (two times 150 bp; Illumina, Shanghai, China). Fastp v0.23.1 was employed to filter the raw data and generate high-quality reads [25]. The clean reads were assembled by the GetOrganelle version 1.7.7.1 [26] and the new sequences were annotated using the Geneious Prime version 2020.0.4 [27]. The complete cp genome of P. biswasiana (accession number: PQ505028) and P. teres (accession number: PQ505027) were submitted to GenBank. The OGDRAW program was used to draw the circular genome maps [28].
2.2. Sequence Analysis and Statistics
REPuter v2.74 was used to distinguish the long repeats [29]. The 50 bp and 30 bp were set to maximum and minimum repeat sizes, while 3 was set as the Hamming distance [23]. To identify SSRs, the MISA–web was utilized. Parameters 10, 5, 4, 3, 3, and 3 were used to identify six types of SSRs [30,31].
MEGA11 was used to analyze the sequences [32] and find the relative synonymous codon usage (RSCU) and amino acid frequencies [33]. To draw the RSCU figure, PhyloSuite version 1.2.2 was employed [34,35]. The EMBOSS program was used to define the GC content of the three positions [36].
2.3. Structure Variations and Highly Variable Regions
The mVISTA tool in Shuffle–LAGAN mode was employed for pairwise alignments and analyses of sequence divergence between Papilionanthe biswasiana, P. teres, and eight related Aeridinae species (Table S1) [37]. Contraction and extension operations of the IR borders between the four major areas in 10 cp genome sequences were implemented through CPJSdraw v1.0.0, an online application platform [38].
2.4. Positive Selection Analysis
PhyloSuite version 1.2.2 enabled the extraction of CDS sequences in two Papilionanthe and another eight related species from Aeridinae (Table S1) [34,35]. MAFFT version 7 alignment was applied to the single-copy CDS sequences [39]. MEGA 11 with Neighbor-Joining (NJ) methods was utilized to construct the phylogenetic tree based on CDS [32]. CodeML algorithm implemented in EasyCodeML calculated the non-synonymous (dN) and synonymous (dS) substitution rates [40] and discovered the M8 mode for selection suites to detect the protein-coding genes in the two Papilionanthe species and eight related species.
2.5. Phylogenetic Analysis
A total of 110 cp genomes of orchid species were utilized in the phylogenetic analysis process (Table S1). The ingroup comprises genomic data from 103 Aeridinae species, of which 101 species were sourced from the NCBI database. Seven species from Dendrobium were selected as the outgroup. Single-CDS sequences from cp genomes were employed in the phylogenetic analysis. These single-CDS sequences were extracted using PhyloSuite version 1.2.2 [34,35], aligned using MAFFT version 7 [39], trimmed using Gblocks [41], and concatenated using plugins in PhyloSuite version 1.2.2 [34,35]. The phylogenetic relationships were reconstructed through Maximum Likelihood (ML) analysis in IQTREE 2, employing the GTR + F + R2 model on CDS sequences with 5000 ultrafast bootstrap (UFBoot) [42,43,44].
3. Results
3.1. General Features of Two Chloroplast Genomes
The two Papilionanthe species exhibit highly similar structural organization in their cp genomes. The cp genomes of P. biswasiana and P. teres measure 148,183 bp and 148,145 bp in total length, respectively (Figure 1, Table S2). Their cp genomes are organized into a large single-copy (LSC) region (85,910 bp and 85,701 bp), a small single-copy (SSC) region (11,591 bp and 11,600 bp), and two inverted repeats (IR) regions (25,341 bp and 25,422 bp). Both cp genomes reflect a total GC content range from 36.5 to 36.6%, with inverted IR regions displaying the highest GC content (43.2%), surpassing that of the SSC (27.6–27.7%) and LSC (33.8%) regions (Table S2).
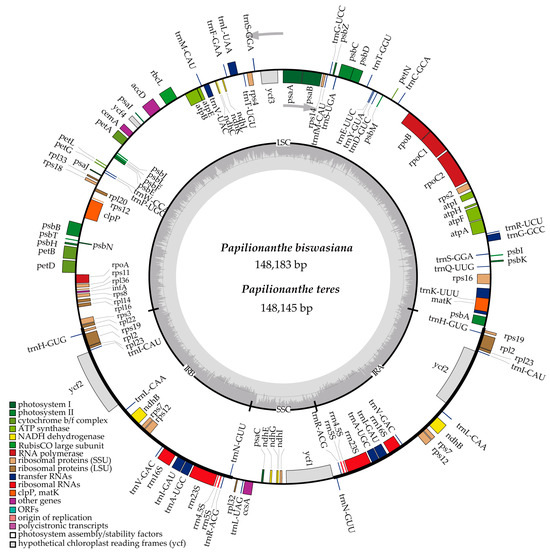
Figure 1.
Circular chloroplast genome map of Papilionanthe biswasiana and P. teres. Genes drawn inside and outside of the circle are transcribed in clockwise and counterclockwise directions separately. Darker and lighter gray area in the inner circle corresponds to the GC and AT content, respectively. Genes belonging to different functional groups are color-coded in the outmost circle.
Both contain 128 genes in two cp genomes, including 5 pseudogenes, 77 CDSs, 38 tRNAs, and 8 rRNAs (Table S3). A total of 110 unique genes are identified in each cp genome. Then, 62 CDS genes and 21 tRNA genes are found within the LSC regions of the two cp genomes. In addition, the SSC region contained a single tRNA gene, while discovering five CDS genes in P. biswasiana and six CDS genes in P. teres. In the IR regions, four rRNA genes, six CDS genes, and eight tRNA genes are duplicated (Table S3). In addition, ndh B/C/E/G/I/J/K genes are identified in the cp genome of two Papilionanthe species (Figure 1, Table S3).
3.2. Repeat Analysis
A comparison of the cp genome of two Papilionanthe species is conducted in order to elucidate their inter-species variations. There are 73 SSRs detected in each of them, which showed some difference between them. Six types of SSRs are observed in P. biswasiana, including mononucleotide (52), dinucleotide (10), trinucleotide (6), tetranucleotide (3), pentanucleotide (1), and hexanucleotide (1). In contrast, five types of SSRs are identified in P. teres, containing mononucleotide (49), dinucleotide (11), trinucleotide (5), tetranucleotide (7), and pentanucleotide (1) (Table S4). And what is more, four types of long repeats (range from 30 to 39 bp) are identified, including complement (C), forward (F), palindromic (P), and reverse (R), and two types of long repeats (over 40 bp) are also observed, including forward (F) and palindromic (P) (Table S5). However, the long repeats ranging from 20 to 29 bp in length are not detected in both cp genomes (Table S5).
3.3. Codon Usage Analysis
Codon usage frequency and relative synonymous codon usage (RSCU) are analyzed and compared for varied genomic features between two cp genomes here and eight cp genomes of the related taxa, downloaded from NCBI (Table S6). The CDS in 10 cp genomes is composed of codons varying between 49,021 and 49,746, which encode 20 amino acids (Figure S1, Table S6). Among the six synonymous codons encoding Leucine, CUU exhibits the highest RSCU value of 1.319, while CUG codon has the lowest at 0.551. Then, 30 codons (RSCU > 1.00) and 29 codons (RSCU < 1.00) are observed in all amino acids except for Methionine and Tryptophan, with no preference (RSCU = 1.00) (Figure S1, Table S6).
3.4. Positive Selection Analysis
The Bayes Empirical Bayes (BEB) method identifies 52 genes under positive selection, containing only one substantial positive selection site in 50 genes but two positive selection sites in atpA (one subunit of ATP synthase gene in the LSC region) and rpl23 genes (genes for large subunit of ribosomein in the IR regions) among 10 cp genomes. And LSC has the highest number of positive selections (Tables S7 and S8).
3.5. Comparative Genome Analysis
Structurally, both the two Papilionanthe cp genomes and those of the eight related species exhibit high conservation. In the 10 cp genomes, the rpl22 gene spans the JLB junctions (Figure 2). The rpl32 gene is occurred in the SSC region and the trnN gene is found in the IR regions (Figure 2). Notably, ycf1 gene extends the JSA junctions in three Paraphalaenopsis species and two Holcoglossum species, but this situation does not happen in the two Papilionanthe species and three Luisia species (Figure 2).
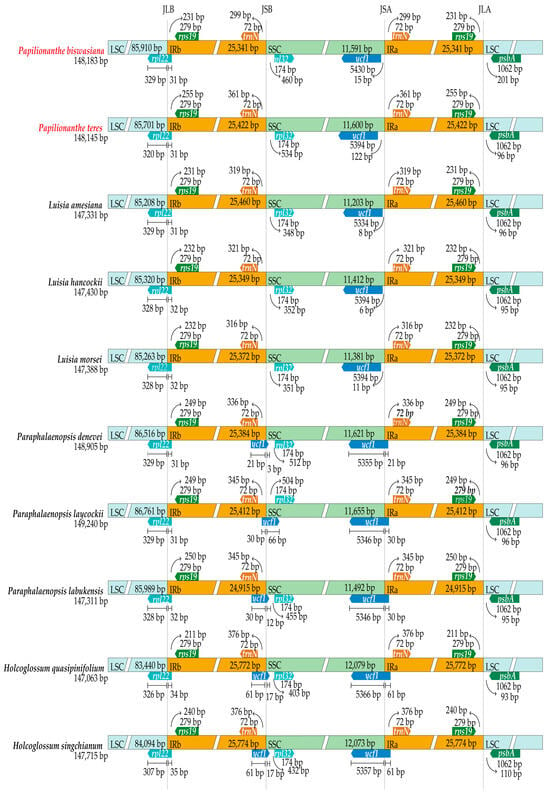
Figure 2.
Comparison of the borders of LSC, SSC, and IR regions among two Papilionanthe species and another eight related species.
3.6. Structure Variations and Highly Variable Regions
Based on the annotation of the cp genome of Papilionanthe teres as the reference, the cp genome sequences of P. biswasiana and the other eight species from Aeridinae are compared by mVISTA (Figure 3). The nucleotide sequences in non-coding regions emerged with greater divergence than those of coding regions among the 10 cp genomes (Figure 3). Three CDSs exhibit significantly higher Pi values, including matK (0.011675347), ycf1 (0.017386189), and psbJ (0.013550136) (Figure 4A, Table S9). Three locations are found to have high Pi values (>0.05) for the intergenic spacer (IGS), including atpH–atpI (0.15658502), psbE–petL (0.087026207), and clpP–psbB (0.056823555) (Figure 4B, Table S10). CDSs (0.00–0.01, average 0.005004492) exhibit a lower Pi value compared to IGS locations (0.01–0.16, average 0.024585135) (Figure 4, Tables S9 and S10).
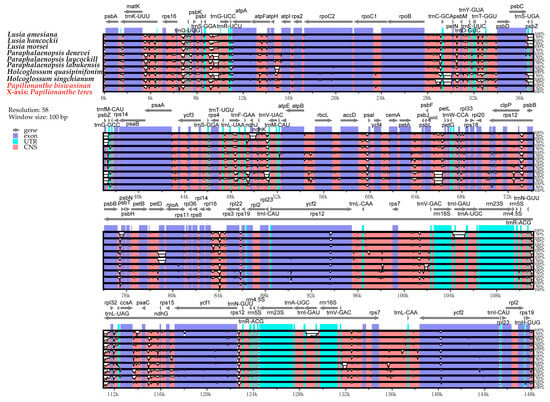
Figure 3.
Sequence comparison of chloroplast genomes of two Papilionanthe species and eight related species using mVISTA. Genome regions are color-coded as exon, rRNA gene, and conserved non-coding sequences (CNS).
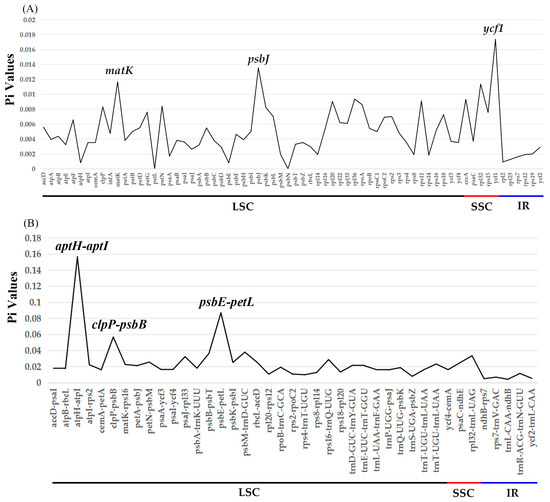
Figure 4.
Comparative analysis of nucleotide diversity (Pi) among two Papilionanthe species and eight related species: (A): CDS regions; (B): IGS regions.
3.7. Phylogenetic Analysis
The phylogenetic tree obtained here indicates that all 103 Aeridinae species sampled are clustered into a monophyletic group with a strong support (UFBoot: 100). Among them, four clades well-supported as the monophytic groups are recognized clearly with strong supports (Figure 5), including (I) Holcoglossum clade, (II) Vanda–Neofinetia clade, (III) Luisia–Papilionanthe–Paraphalaenopsis clade, and (IV) Aerides–Renanthera clade.
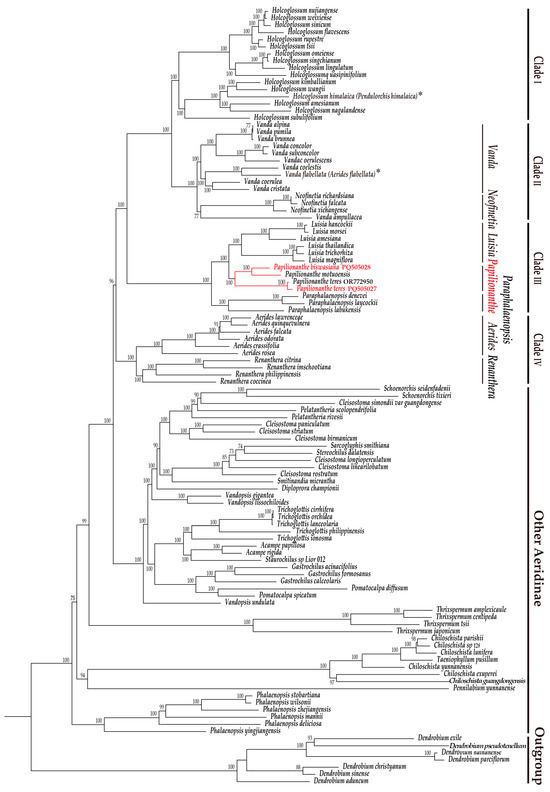
Figure 5.
A phylogenetic tree based on complete chloroplast genome data of four individuals of 3 Papilionanthe species and another 99 related species from Aeridinae, with 7 Dendrobium species as outgroup. Papilionanthe species (P. biswasiana and P. teres) highlighted in red were sequenced and assembled here. An asterisk (*) indicates the uncertain species.
- (I.)
- The Holcoglossum clade comprises 16 Holcoglossum species, a sister to Vanda–Neofinetia clade, with strong supports (UFBoot: 100), containing three distinct subclades, as well as the uncertain species of H. himalaica (Deb, Sengupta & Malick) Aver. The Holcoglossum clade seemed to be related to the Vanda–Neofinetia clade.
- (II.)
- The Vanda–Neofinetia clade is composed of two monophyletic subclades with a strong support (UFBoot: 100), including Vanda subclade (11 species) and Neofinetia subclade (3 species). In Vanda subclade, V. ampullacea (Roxb.) L.M.Gardiner. appears to be a sister to the other 10 species, including the uncertain species of V. flabellata (Rolfe ex Downie) Christenson. And here, V. flabellata seems to be close to V. coelestis (Rchb. f.) Motes.
- (III.)
- The Luisia–Papilionanthe–Paraphalaenopsis clade contains four individuals of three Papilionanthe species, six Luisia species, and three Paraphalaenopsis species with a strong support (UFBoot: 100). Analytical results show that Papilionanthe species grouped into a well-supported clade (UFBoot: 100), being close to Luisia while distant from Paraphalaenopsis.
- (IV.)
- The Aerides–Renanthera clade is composed of six Aerides species and four Renanthera Lour. species with a strong support (UFBoot: 100). It is a sister to the other three clades above, namely, Holcoglossum clade, Vanda–Neofinetia clade, and Luisia–Papilionanthe–Paraphalaenopsis clade.
4. Discussion
There are some similar genomic features of the two Papilionanthe species, including the GC content, gene content, and order of cp genome. And GC content analysis revealed that the IR region consistently surpassed the LSC and SSC regions, which was consistent with other orchids, such as in Phaius [20], Paphiopedilum Pfitzer. [45], Goodyera R.Br. [46], and Bulbophyllum Thouars. [47]. Seven ndh (B/C/E/G/I/J/K) genes were exhibited in the cp genome of two Papilionanthe species. The ndh gene pseudogenization and functional loss have been widely reported in multiple orchids [48,49], prominently observed in some epiphytic lineages [50,51,52,53]. Meanwhile, there are other distinctly genomic features between two Papilionanthe species, including the number of SSRs, the length and number of long repeat sequences, IR boundary, highly variable sequences, and positive selection sites. And some features are distinguished from its three related genera, which could be further discussed for some taxonomic significance, as below.
4.1. Comparative Chloroplast Genome Between Papilionanthe biswasiana and P. teres
Some distinct cp genomic features between the two Papilionanthe species are selected, including the number and type of simple sequence repeats (SSRs) and the length of long repeats. There were 73 SSRs detected in each of two cp genomes of Papilionanthe. It was shown that five types of SSRs were commonly identified in P. biswasiana and P. teres, including mononucleotide, dinucleotide, trinucleotide, tetranucleotide, and pentanucleotide. However, hexanucleotide was present in P. biswasiana but absent in P. teres. Due to their high variability, plastome markers like SSRs and large repeats were useful in resolving taxonomic ambiguities among related species [54,55]. For example, the state of hexanucleotide was varied in species of Paraphalaenopsis, present in P. labukensis and P. denevel but absent in P. laycokii [56]. This was also found in Aerides species, being observed in A. flabellata but not detected in A. rosea Lodd. ex Lindl. & Paxton. [10]. In addition, hexanucleotide was not identified in two Phalaenopsis species [19] and six Luisia species [23]. And what is more, there were 49 long repeats detected in the two cp genomes. However, the long repeats were varied in length between two Papionanthe species and its related species. The long repeats ranged from 30 bp to more than 40 bp, and no repeat was detected above 20–29 bp in length in Papilionanthe biswasiana and P. teres. Almost all repeats were in the range of more than 30 bp in length, with an exception of a range of 20–29 bp in three Paraphalaenopsis species [56]. Therefore, SSRs and long repeats could be used as the important genomic data for phylogenetic analysis.
4.2. Comparative Analysis of Chloroplast Genome Between Papilionanthe and Other Eight Species from the Related Genera
The cp genomes of two Papilionanthe species investigated here were compared with those of eight species from the three related genera downloaded from the NCBI database in order to provide new molecular evidence for the phylogenetic relationship of Papilionanthe. A comparative analysis revealed that the two cp genomes shared some genomic features with the other eight cp genomes of the three related genera, including Luisia, Paraphalaenopsis, and Holcoglossum, with the highest similarity with those of Luisia (97–98%).
The IR region exhibited the most conservative section in the 10 cp genomes from Aeridinae, but its boundaries demonstrate frequent contractions and expansions. The feature was considered to underpin the evolution of cp genome, acting as the primary driver for variation in cp genome length [57]. Significant differences at the JSB and JSA junction areas were detected, which possessed phylogenetic insights. The ycf1 gene extended the JSA junctions in three Paraphalaenopsis species and two Holcoglossum species, but this situation did not happen in the two Papilionanthe species and three Luisia species. Therefore, it is suggested that the expansion or contraction of the IR boundary in Aeridinae species might be attributed to the variations in ycf1 genes.
Divergent regions, when used as molecular markers, demonstrated significant potential in enhancing DNA barcoding accuracy and resolving phylogenetic relationships [58]. Comparative analysis of the 10 chloroplast genomes revealed significantly higher sequence divergence in non-coding regions relative to coding regions, a pattern consistent with observations in other orchids including Coelogyne Lindl. [59], Polystachya Hook. [60], and Platanthera [61]. It was shown that genes matK, ycf1, and psbJ had significantly higher Pi values. Similar to matK, ycf1 serves as a DNA marker in phylogenetic analysis [62]. Here, atpH–atpI, psbE–petL, and clpP–psbB also possessed a higher degree of variability in the 10 cp genomes. It was verified that cp genomes in most of Orchidaceae members exhibited a diversity array of highly variable sequences, such as in Goodyera [46], Coelogyne [59], Polystachya [60], and Pholidota Lindl. ex Hook. [63].
The dN/dS ratio (ω) has emerged as a critical parameter to define adaptive genetic divergence among taxa and model the selective pressures [64,65,66]. There were 52 genes significantly identified under positive selection, containing only one substantial positive selection site in 50 genes, but two positive selection sites in atpA and rpl23 genes in the 10 cp genomes. Similarly, the rps4, petL, and atpH genes were also found in other orchids, such as in Bulbophyllum [47] and Phalaenopsis [19]. It seemed that these genes might be used for identification and phylogenetic research of Aeridinae.
4.3. The Intergeneric Relationship of Papilionanthe
Papilionanthe was well distinguished from other members in Aeridinae by the terete leaf and showy flower with a spur connated to the lip [2]. However, its phylogenetic relationship in Aeridinae has not been resolved with focus. It was previously presumed to be related to Vanda [5,12] and Aerides [5] by the morphological evidence, but recently to be close to Holcoglossum [13], Luisia, and Paraphalaenopsis [8,14] based on the molecular DNA sequence. However, none of these studies involved have to date focused specifically on Papilionanthe, which seemed to be an obstacle for a better understanding of its phylogeny [1].
Here, a phylogenetic tree was reconstructed based on cp genomes of two Papilionanthe species and those of the other taxa from Aeridinae, as many as available, in order to provide new genomic evidence for its phylogenetic relationship. Some clades of the phylogenetic tree based on the cp genome were similar to those previously obtained by molecular DNA sequences [8]. Here, it was clear that there were four clades well-supported as four monotypic groups, representative of eight related genera involved, including (I) Holcoglossum clade, (II) Vanda–Neofinetia clade, (III) Luisia–Papilionanthe–Paraphalaenopsis clade, and (IV) Aerides–Renanthera clade. The four monophyletic clades obtained here could provide significant implications for the phylogenetic relationship of Papilionanthe in Aeridinae as below.
Papilionanthe should be a monophyletic genus and related to Luisia and Paraphalaenopsis, since members sampled from the three genera were clustered into a clade of Luisia–Papilionanthe–Paraphalaenopsis. This phylogenetic relationship among these three genera might be attributed to their shared characteristic of terete leaves [1]. Among this clade, species from each genus formed an independent subclade, indicating that each of them could be monophyletic taxa. Four individuals of three Papilionanthe species were grouped into a subclade, being a sister to Luisia. It was also indicated that Papilionanthe should be closer to Luisia than to Paraphalaenopsis in this clade. And, in this subclade Papilionanthe, the newly described P. motuoensis was nearer to P. biswasiana than to P. teres. However, P. motuoensis appeared to be closer to P. teres and P. subulata, while P. biswasiana seemed to be more related to P. hookeriana based on seven combined DNA sequences [3]. Therefore, the infrageneric phylogeny of Papilionanthe should be further studied by more evidence with samples expanded.
Papilionanthe was supposed to be close to Vanda and Aerides [5,12], but the latter two genera were more closely related to other genera, which were grouped into, respectively, Vanda–Neofinetia clade and Aerides–Renanthera clade. It was strongly supported that Vanda seemed to be closer to Neofinetia than to Papilionanthe and that Aerides appeared to be more related to Renanthera based on cp genomic data here. Simultaneously, the monophyletic Vanda s.l., consisting of Vanda s.s. and Neofinetia, should be reconsidered, since an uncertain species of V. ampullacea was distant from the other 10 Vanda species but a sister to a subclade of Neofinetia with a moderate support. In addition, it was reconfirmed that A. flabellata should be treated as a synonym of V. flabellata here, as it was embedded into other members of Vanda but not within Aerides. This species was placed in Aerides because it was distinguished from other Vanda members by a flower with a long column foot and motile lip [6,17,67] and supported by a molecular phylogeny on a plastid matK gene [7]. However, it was moved from Aerides into Vanda by the combined molecular data (nrITS and matK, trnL, and trnL–F) [8] and Aerides cp genomic data [10].
It was also presumed that Papilionanthe should be close to Holcoglossum [13], but here, 16 Holcoglossum species from the latter were grouped into a separated clade, being a sister to clade Vanda–Neofinetia. It might be attributed to the samples limited without any members of Luisia nor Paraphalaenopsis selected [13]. Meanwhile, H. himalaicum has been known as a problematic species in Aeridinae, and its taxonomic position in Holcoglossum was reconfirmed here. And, obviously, H. himalaicum could not be placed in several genera proposed previously, such as Vanda [68], Ascocentrum [69], and Pendulorchis Z.J.Liu, K.Wei Liu & G.Q.Zhang. [70]. In addition, H. himalaicum shares evidently common features with other Holcoglossum members such as stems enclosed by a persistent leaf sheath, terete and fleshy leaf, a cylindric and curved spur, and a stout column with foot [2,70].
5. Conclusions
Through comprehensive sequencing and characterization of complete cp genomes, Papilionanthe biswasiana and P. teres were analyzed alongside eight other species from three genera within Aeridinae. To derive phylogenetic insights, several cp genome features were examined, such as genome feature, codon utilization patterns, repetitive sequences, and DNA polymorphisms. And a phylogenetic tree based on 103 cp genomes from Aeridinae taxa was reconstructed in order to clarify its phylogenetic relationship. It shows that Papilionanthe is monophyletic and closer to Luisia and Paraphalaenopsis than to Vanda, Aerides, and Holcoglossum based on cp genomic data here, while Vanda is close to Neofinetia and Aerides is near to Renanthera. And then, members of Papilionanthe, Luisia, and Paraphalaenopsis are clustered into a clade as a sister to Holcoglossum–Vanda–Neofinetia clade. It shows that there are four clades well-supported as four monotypic groups, which could provide a significant implication for the phylogenetic relationship of Papilionanthe in Aeridinae. Furthermore, the taxonomic position of two problematic species, including V. flabellata and H. himalacia, was also re-clarified.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/horticulturae11060641/s1, Figure S1. Maps of relative synonymous codon usage (RSCU) of 10 chloroplast genomes from two Papilionanthe species and eight species related in Aeridinae; Table S1. List of 110 species used in the phylogenetic analysis; Table S2. Basic features of two cp genomes; Table S3. Gene annotation for the chloroplast genome of two Papilionanthe species; Table S4. Number of different SSR units in two cp genomes; Table S5. The detailed information of long repeats in the two Papilionanthe species; Table S6. The relative synonymous codon usage of the two Papilionanthe species and eight species related; Table S7. The positive selection sites of 10 cp genomes; Table S8. Positive selection sites were detected in the cp genome of the two Papilionanthe species and eight species related; Table S9. Comparison of the nucleotide variability (PI) among CDS regions of the two Papilionanthe species and eight species related; Table S10. Comparison of the nucleotide variability among IGS regions of the two Papilionanthe species and eight species related.
Author Contributions
Conceptualization, L.L.; Data curation, Y.X. and K.T.; Funding acquisition, L.L.; Methodology, L.L.; Project administration, L.L.; Software, Y.X., K.T., D.M., Y.W. and J.X.; Writing—original draft, Y.X.; Writing—review and editing, Y.X., Y.L. and L.L. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by National Nature Science Foundation of China (NSFC 32060049 and NSFC 32270225) and Yunnan Fundamental Research Projects (Grant No. 202501BD07000–018).
Data Availability Statement
The datasets generated and analyzed in this research are available in the NCBI BioProject (PRJNA1172001 and PRJNA1172398, SRA: SRR30963468 and SRR30978210).
Acknowledgments
We appreciate Yuxiao Zhang (Southwest Forestry University) for providing computer server.
Conflicts of Interest
The authors declare no competing interests.
Abbreviations
The following abbreviations are used in this manuscript:
| GC | Guanine–cytosine |
| bp | Base pairs |
| CDS | Coding sequences |
| cp | Chloroplast |
| dN | Non synonymous |
| dS | Synonymous |
| IRa | Inverted repeat A |
| IRb | Inverted repeat B |
| LSC | Large single copy |
| ML | Maximum likelihood |
| NCBI | National Center for Biotechnology Information |
| Pi | Nucleotide diversity |
| rRNA | Ribosomal RNA |
| RSCU | Relative synonymous codon usage |
| SSC | Small single copy region |
| SSRs | Simple sequence repeats |
| tRNA | Transfer RNA |
| IGS | Intergenic spacer |
| mRNA | Messenger RNA |
References
- Pridgeon, A.M.; Cribb, P.J.; Chase, M.W.; Rasmussen, F.N. Genera Orchidacearum Volume 6: Epidendroideae (Part Three); Oxford University Press: Oxford, UK, 2014; pp. 218–220. [Google Scholar]
- Chen, X.Q.; Wood, J.J. Papilionanthe Schltr. In Flora of China: Orchidaceae; Science Press: Beijing, China, 2009; Volume 25, pp. 477–478. [Google Scholar]
- Ya, J.D.; Wang, W.T.; Liu, Y.L.; Jiang, H.; Han, Z.D.; Zhang, T.; Huang, H.; Cai, J.; Li, D.Z. Five new and noteworthy species of Epidendroideae (Orchidaceae) from southwestern China based on morphological and phylogenetic evidence. PhytoKeys 2023, 235, 211–236. [Google Scholar] [CrossRef]
- Ahmed, A.M.; Rahman, M.A. Wild epiphytic Bangladeshi orchids Cymbidium aloifolium (L.) Sw. and Papilionanthe teres (Roxb.) Lindl. Potentially modulates the immune functions in Swiss albino mice. J. Adv. Vet. Anim. Res. 2021, 8, 479–488. [Google Scholar] [CrossRef]
- Garay, L.A. On the systematics of the monopodial orchids II. In Botanical Museum Leaflets; Harvard University: Cambridge, MA, USA, 1974; pp. 369–376. [Google Scholar]
- Seidenfaden, G. Orchid Genera in Thailand XIV: Fifty–Nine Vandoid Genera; Council for Nordic Publications in Botany: Copenhagen, Denmark, 1988. [Google Scholar]
- Topik, H.; Yukawa, T.; Ito, M. Molecular phylogenetics of subtribe Aeridinae (Orchidaceae): Insights from plastid matK and nuclear ribosomal ITS sequences. J. Plant Res. 2005, 118, 271–284. [Google Scholar] [CrossRef] [PubMed]
- Zou, L.H.; Huang, J.X.; Zhang, G.Q.; Liu, Z.J.; Zhuang, X.Y. A molecular phylogeny of Aeridinae (Orchidaceae: Epidendroideae) inferred from multiple nuclear and chloroplast regions. Mol. Phylogenetics Evol. 2015, 85, 247–254. [Google Scholar] [CrossRef]
- Wang, X.T.; Liu, D.K.; Zhu, W.Y.; Zheng, S.Z.; Yu, X.Y.; Ai, Y.; Zhang, Q.H. The complete chloroplast genome of an ornamental orchid, Vanda coerulescens (Orchidaceae). Mitochondrial DNA Part B 2020, 10, 465–467. [Google Scholar] [CrossRef] [PubMed]
- Tao, K.; Tao, L.; Huang, J.; Duan, H.; Luo, Y.; Li, L. Complete chloroplast genome structural characterization of two Aerides (Orchidaceae) species with a focus on phylogenetic position of Aerides flabellata. BMC Genom. 2024, 25, 552. [Google Scholar] [CrossRef] [PubMed]
- Zhou, P.; Lei, W.S.; Shi, Y.K.; Liu, Y.Z.; Luo, Y.; Li, J.H.; Xiang, X.G. Plastome Evolution, Phylogenomics, and DNA Barcoding Investigation of Gastrochilus (Aeridinae, Orchidaceae), with a Focus on the Systematic Position of Haraella retrocalla. Int. J. Mol. Sci. 2024, 25, 8500. [Google Scholar] [CrossRef]
- Christenson, E.A. Taxonomy of the Aeridinae with an infrageneric classification of Vanda Jones ex R. Br. In Proceedings of the 14th World Orchid Conference; HMSO Publications: Edinburgh, UK, 1994; pp. 206–216. [Google Scholar]
- Liu, Z.J.; Chen, L.J.; Chen, S.C.; Cai, J.; Tsai, W.C.; Hsiao, Y.Y.; Rao, W.H. Paraholcoglossum and Tsiorchis, Two New Orchid Genera Established by Molecular and Morphological Analyses of the Holcoglossum Alliance. PLoS ONE 2011, 6, e24864. [Google Scholar] [CrossRef]
- Gardiner, L.M.; Kocyan, A.; Motes, M.; Roberts, D.L.; Emerson, B.C. Molecular phylogenetics of Vanda and related genera (Orchidaceae). Bot. J. Linn. Soc. 2013, 173, 549–572. [Google Scholar] [CrossRef]
- Lim, S.H.; Teng, P.P.; Lee, Y.H.; Goh, C.J. RAPD Analysis of Some Species in the Genus Vanda (Orchidaceae). Ann. Bot. 1999, 83, 193–196. [Google Scholar] [CrossRef]
- Christenson, E.A.; Saito, K.; Tanaka, R. The Taxonomy of Aerides and Related Genera. In Proceedings of the 12th World Orchid Conference; 12th World Orchid Conference Organizing Committee: Tokyo, Japan, 1987; pp. 35–40. [Google Scholar]
- Kocyan, A.; Vogel, E.F.; Conti, E.; Gravendeel, B. Molecular phylogeny of Aerides (Orchidaceae) based on one nuclear and two plastid markers: A step forward in understanding the evolution of the Aeridinae. Mol. Phylogenetics Evol. 2008, 48, 422–443. [Google Scholar] [CrossRef]
- Wu, X.Y.; Li, T.Z.; Chen, G.Z.; Xu, Q.; Pan, Y.Y. The complete chloroplast genome of Dendrobium longicornu (Orchidaceae). Mitochondrial DNA Part B 2019, 4, 3776–3777. [Google Scholar] [CrossRef]
- Tao, L.; Duan, H.; Tao, K.; Luo, Y.; Li, Q.; Li, L. Complete chloroplast genome structural characterization of two Phalaenopsis (Orchidaceae) species and comparative analysis with their alliance. BMC Genom. 2023, 24, 359. [Google Scholar] [CrossRef] [PubMed]
- Tao, K.; Tang, L.; Luo, Y.; Li, L. Complete chloroplast genome of eight Phaius (Orchidaceae) species from China: Comparative analysis and phylogenetic relationship. BMC Plant Biol. 2025, 25, 37. [Google Scholar] [CrossRef]
- Li, Z.H.; Ma, X.; Wang, D.Y.; Li, Y.X.; Wang, C.W.; Jin, X.H. Evolution of plastid genomes of Holcoglossum (Orchidaceae) with recent radiation. BMC Evol. Biol. 2019, 19, 63. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Lu, Z.; Wang, J.; Zhang, H.; Jiang, M. Complete chloroplast genome of a traditional medicinal plant Luisia hancockii Rolfe 1896: Genomic features and phylogenetic relationship within subtribe Aeridinae (Orchidaceae). Mitochondrial DNA Part B 2023, 30, 1149–1153. [Google Scholar] [CrossRef]
- Ma, L.; Zhou, C.Y.; Chen, J.L.; Liu, D.K.; Lan, S.; Liu, Z.J. Comparative Analysis of Luisia (Aeridinae, Orchidaceae) Plastomes Shed Light on Plastomes Evolution and Barcodes Investigation. Genes 2024, 15, 20. [Google Scholar] [CrossRef] [PubMed]
- Healey, A.; Furtado, A.; Cooper, T.; Henry, R. Protocol: A simple method for extracting next-generation sequencing quality genomic DNA from recalcitrant plant species. Plant Methods 2014, 10, 21. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Jin, J.J.; Yu, W.B.; Yang, J.B.; Song, Y.; DePamphilis, C.W.; Yi, T.S. GetOrganelle: A fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 2020, 21, 241. [Google Scholar] [CrossRef]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef] [PubMed]
- Shaw, J.; Shafer, H.L.; Leonard, O.R.; Kovach, M.J.; Schorr, M.; Morris, A.B. Chloroplast DNA sequence utility for the lowest phylogenetic and phylogeographic inferences in angiosperms: The tortoise and the hare IV. Am. J. Bot. 2014, 101, 1987–2004. [Google Scholar] [CrossRef] [PubMed]
- Kurtz, S.; Choudhuri, J.V.; Ohlebusch, E.; Schleiermacher, C.; Stoye, J.; Giegerich, R. REPuter: The manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res. 2001, 29, 4633–4642. [Google Scholar] [CrossRef]
- Beier, S.; Thiel, T.; Münch, T.; Scholz, U.; Mascher, M. MISA–web: A web server for microsatellite prediction. Bioinformatics 2017, 33, 2583–2585. [Google Scholar] [CrossRef]
- Thiel, T.; Michalek, W.; Varshney, R.; Graner, A. Exploiting EST databases for the development and characterization of gene-derived SSR–markers in barley (Hordeum vulgare L.). Theor. Appl. Genet. 2003, 106, 411–422. [Google Scholar] [CrossRef]
- Kumar, S.; Nei, M.; Dudley, J.; Tamura, K. MEGA: A biologist–centric software for evolutionary analysis of DNA and protein sequences. Brief. Bioinformatic 2008, 9, 299–306. [Google Scholar] [CrossRef]
- Bylaiah, S.; Shedole, S.; Suresh, K.P.; Gowda, L.; Patil, S.S.; Indrabalan, U.B. Analysis of Codon Usage Bias in Cya, Lef, and Pag Genes Exists in px01 Plasmid of Bacillus anthracis. In ICT Analysis and Applications; Springer Nature: Singapore, 2022; pp. 1–9. [Google Scholar]
- Xiang, C.Y.; Gao, F.; Jakovlić, I.; Lei, H.P.; Hu, Y.; Zhang, H.; Zou, H.; Wang, G.; Zhang, D. Using PhyloSuite for molecular phylogeny and tree–based analyses. iMeta 2023, 2, e87. [Google Scholar] [CrossRef]
- Zhang, D.; Gao, F.; Jakovlić, I.; Zou, H.; Zhang, J.; Li, W.X.; Wang, G.T. PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour. 2020, 20, 348–355. [Google Scholar] [CrossRef] [PubMed]
- Rice, P.; Longden, I.; Bleasby, A. EMBOSS: The European Molecular Biology Open Software Suite. Trends Genet. 2000, 16, 276–277. [Google Scholar] [CrossRef]
- Brudno, M.; Malde, S.; Poliakov, A.; Do, C.B.; Couronne, O.; Dubchak, I.; Batzoglou, S. Glocal alignment: Finding rearrangements during alignment. Bioinformatics 2003, 19, i54–i62. [Google Scholar] [CrossRef]
- Li, H.; Guo, Q.; Xu, L.; Gao, H.; Liu, L.; Zhou, X. CPJSdraw: Analysis and visualization of junction sites of chloroplast genomes. PeerJ 2023, 11, e15326. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Gao, F.; Chen, C.; Arab, D.A.; Du, Z.; He, Y.; Ho, S.Y.W. EasyCodeML: A visual tool for analysis of selection using CodeML. Mol. Biol. Evol. 2019, 9, 3891–3898. [Google Scholar] [CrossRef]
- Talavera, G.; Castresana, J. Improvement of Phylogenies after Removing Divergent and Ambiguously Aligned Blocks from Protein Sequence Alignments. Syst. Biol. 2007, 56, 564–577. [Google Scholar] [CrossRef]
- Hoang, D.T.; Chernomor, O.; von Haeseler, A.; Minh, B.Q.; Vinh, L.S. UFBoot2: Improving the Ultrafast Bootstrap Approximation. Mol. Biol. Evol. 2018, 35, 518–522. [Google Scholar] [CrossRef]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; Haeseler, A. IQ-TREE 2: New Models and Efficient Methods for Phylogenetic Inference in the Genomic Era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.D.; Peng, D.H.; Lan, S.R.; Chen, J.; Fu, W.Q. The complete chloroplast genome sequence of Paphiopedilum purpuratum (Orchidaceae). Mitochondrial DNA Part B 2019, 4, 3910–3911. [Google Scholar] [CrossRef]
- Kim, T.H.; Kim, J.H. Molecular Phylogeny and Historical Biogeography of Goodyera R. Br. (Orchidaceae): A Case of the Vicariance Between East Asia and North America. Front. Plant Sci. 2022, 13, 850170. [Google Scholar] [CrossRef]
- Wu, Y.; Zeng, M.Y.; Wang, H.X.; Lan, S.; Liu, Z.J.; Zhang, S. The Complete Chloroplast Genomes of Bulbophyllum (Orchidaceae) Species: Insight into Genome Structure Divergence and Phylogenetic Analysis. Int. J. Mol. Sci. 2024, 25, 2665. [Google Scholar] [CrossRef]
- Feng, Y.L.; Wicke, S.; Li, J.W.; Han, Y.; Lin, C.S.; Li, D.Z. Lineage–specific reductions of plastid genomes in an orchid tribe with partially and fully mycoheterotrophic species. Genome Biol. Evol. 2016, 8, 2164–2175. [Google Scholar] [CrossRef]
- Kim, Y.K.; Jo, S.; Cheon, S.H.; Joo, M.J.; Hong, J.R.; Kwak, M. Plastome Evolution and Phylogeny of Orchidaceae, with 24 New Sequences. Front. Plant Sci. 2020, 11, 22. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.K.; Tu, X.D.; Zhao, Z.; Zeng, M.Y.; Zhang, S.; Ma, L. Plastid phylogenomic data yield new and robust insights into the phylogeny of Cleisostoma–Gastrochilus clades (Orchidaceae, Aeridinae). Mol. Phylogenetics Evol. 2020, 145, 106729. [Google Scholar] [CrossRef]
- Niu, Z.; Zhu, S.; Pan, J.; Li, L.; Sun, J.; Ding, X. Comparative analysis of Dendrobium plastomes and utility of plastomic mutational hotspots. Sci. Rep. 2017, 7, 2073. [Google Scholar]
- Zavala-Páez, M.; Vieira, L.D.N.; Baura, V.A.D.; Balsanelli, E.; Souza, E.M.D.; Cevallos, M.C.; Smidt, E.D.C. Comparative plastid genomics of neotropical Bulbophyllum (Orchidaceae; Epidendroideae). Front. Plant Sci. 2020, 11, 799. [Google Scholar] [CrossRef]
- Kim, Y.K.; Jo, S.; Cheon, S.H.; Kwak, M.; Kim, Y.D.; Kim, K.J. Plastome evolution and phylogeny of subtribe Aeridinae (Vandeae, Orchidaceae). Mol. Phylogenetics Evol. 2020, 144, 106721. [Google Scholar] [CrossRef] [PubMed]
- Tauta, D. Hypervariablity of simple sequences as a general source for polymorphic DNA marks. Nucleic Acids Res. 1989, 17, 6463–6471. [Google Scholar] [CrossRef]
- Cato, S.A.; Richardson, T.E. Inter- and intraspecific polymorphism at chloroplast SSR loci and the inheritance of plastids in Pinus radiata D. Don. Theor. Appl. Genet. 1996, 93, 587–592. [Google Scholar] [CrossRef]
- Chen, J.; Wang, F.; Zhao, Z.; Li, M.; Liu, Z.; Peng, D. Complete Chloroplast Genomes and Comparative Analyses of Three Paraphalaenopsis (Aeridinae, Orchidaceae) Species. Int. J. Mol. Sci. 2023, 24, 11167. [Google Scholar] [CrossRef]
- Dugas, D.V.; Hernandez, D.; Koenen, E.J.M.; Schwarz, E.; Straub, S.; Hughes, C.E. Mimosoid legume plastome evolution: IR expansion, tandem repeat expansions and accelerated rate of evolution in clpP. Sci. Rep. 2015, 5, 16958. [Google Scholar] [CrossRef]
- Menezes, A.P.A.; Resende-Moreira, L.C.; Buzatti, R.S.O.; Nazareno, A.G.; Carlsen, M.; Lobo, F.P. Chloroplast genomes of Byrsonima species (Malpighiaceae): Comparative analysis and screening of high divergence sequences. Sci. Rep. 2018, 8, 2210. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.; Li, R.; Tang, S.; Chen, Y.; Yan, Y.; Gao, X. Plastomes of Seven Coelogyne s.l. (Arethuseae, Orchidaceae) Species: Comparative Analysis and Phylogenetic Relationships. Horticulturae 2025, 11, 144. [Google Scholar] [CrossRef]
- Jiang, H.; Tian, J.; Yang, J.; Dong, X.; Zhong, Z.; Mwachala, G. Comparative and phylogenetic analyses of six Kenya Polystachya (Orchidaceae) species based on the complete chloroplast genome sequences. BMC Plant Biol. 2022, 22, 177. [Google Scholar] [CrossRef]
- Han, C.; Ding, R.; Zong, X.; Zhang, L.; Chen, X.; Bo, Q. Structural characterization of Platanthera ussuriensis chloroplast genome and comparative analyses with other species of Orchidaceae. BMC Genom. 2022, 23, 84. [Google Scholar] [CrossRef] [PubMed]
- Dong, W.; Xu, C.; Li, C.; Sun, J.H.; Zuo, Y.J.; Shi, S. ycf1, the most promising plastid DNA barcode of land plants. Sci. Rep. 2015, 5, 8348. [Google Scholar] [CrossRef]
- Li, L.; Wang, W.; Zhang, G.; Wu, K.L.; Fang, L.; Li, M.Z. Comparative analyses and phylogenetic relationships of thirteen Pholidota species (Orchidaceae) inferred from complete chloroplast genomes. BMC Plant Biol. 2023, 23, 269. [Google Scholar] [CrossRef]
- Kryazhimskiy, S.; Plotkin, J.B. The population genetics of dN/dS. PLoS Genet. 2008, 4, e1000304. [Google Scholar] [CrossRef]
- Zuo, L.H.; Shang, A.Q.; Zhang, S.; Yu, X.Y.; Ren, Y.C.; Yang, M.S. The first complete chloroplast genome sequences of Ulmus species by de novo sequencing: Genome comparative and taxonomic position analysis. PLoS ONE 2017, 12, e0171264. [Google Scholar] [CrossRef]
- Williams, M.J.; Zapata, L.; Werner, B.; Barnes, C.P.; Sottoriva, A.; Graham, T.A. Measuring the distribution of fitness effects in somatic evolution by combining clonal dynamics with dN/dS ratios. eLife 2020, 9, e48714. [Google Scholar] [CrossRef]
- Garay, L.A. On the systematics of the monopodial orchids I. In Botanical Museum Leaflets; Harvard University: Cambridge, MA, USA, 1972; Volume 23, pp. 149–212. [Google Scholar]
- Gardiner, L.M. New combinations in the genus Vanda (Orchidaceae). Phytotaxa 2012, 61, 47–54. [Google Scholar] [CrossRef]
- Jiang, H. A new variety of Ascocentrum from Yunnan, China. Acta Bot. Yunnanica 2006, 28, 259–260. [Google Scholar]
- Zhang, G.Q.; Liu, K.W.; Chen, L.J.; Xiao, X.J.; Zhai, J.W.; Li, L.Q. A New Molecular Phylogeny and a New Genus, Pendulorchis, of the Aerides–Vanda Alliance (Orchidaceae: Epidendroideae). PLoS ONE 2013, 8, e60097. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).