Detection of Cassava Mosaic Disease and Assessment of Selected Agronomic Traits of Cassava (Manihot esculenta)
Abstract
1. Introduction
2. Materials and Methods
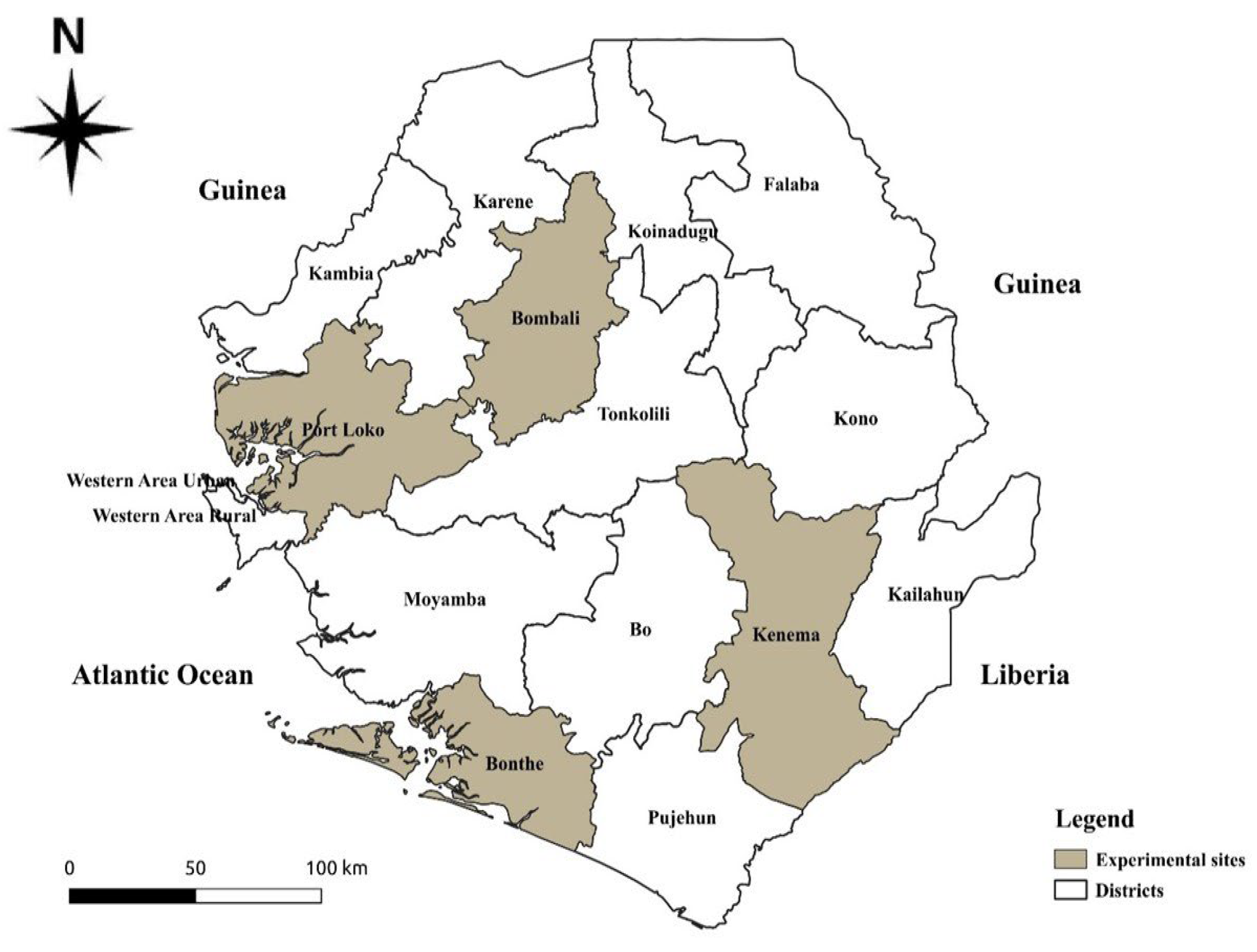
2.1. Identification and Collection of Cassava Planting Material
2.2. Experimental Material, Layout, Design, and Management
2.3. Phenotypic Data Collection and Analysis
2.4. Genotyping, Virus Indexing, and Analysis
3. Results
3.1. Phenotypic Trait Assessment
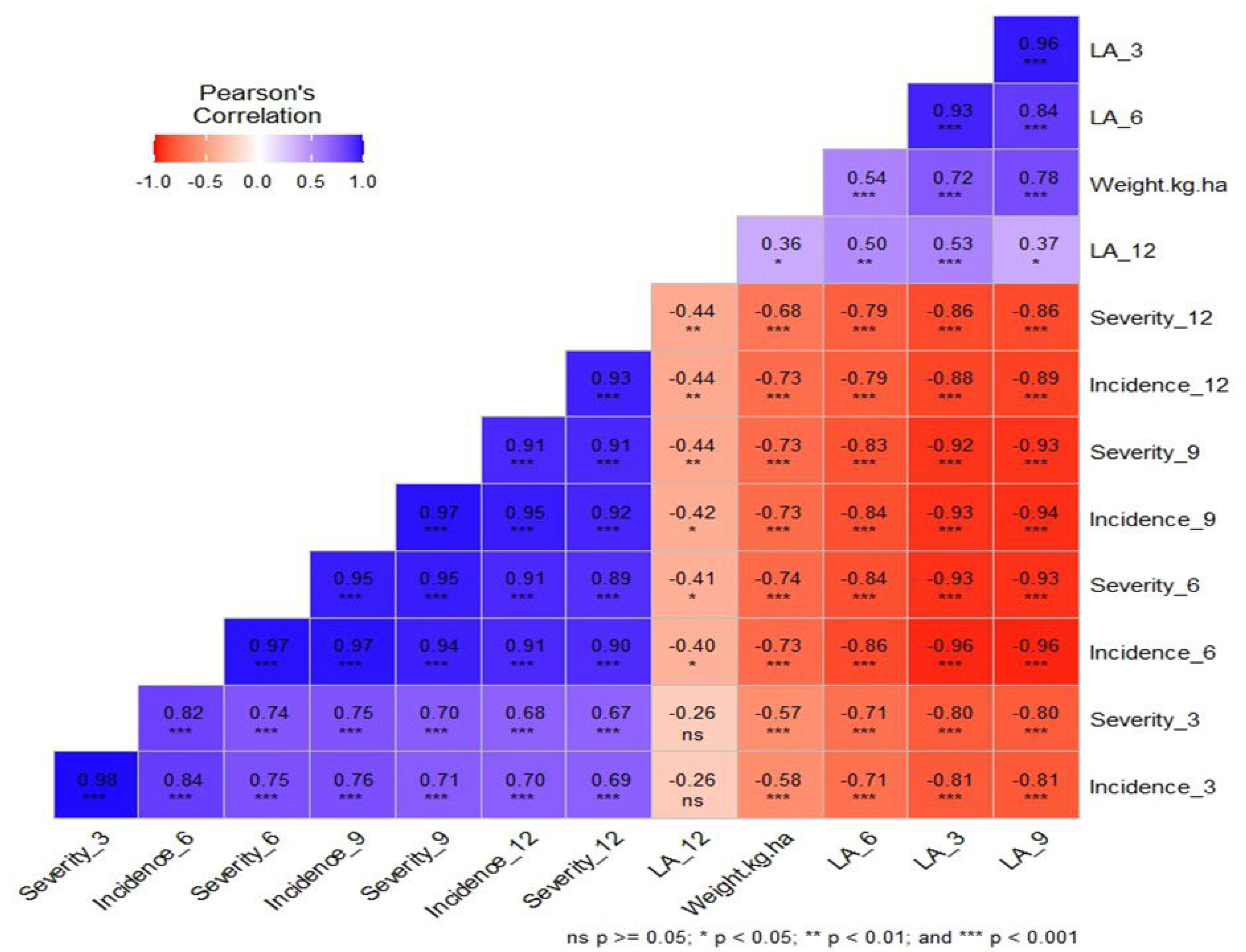
3.2. Correlation Analysis for Trait Associations
3.3. Molecular Detection of Cassava Mosaic Virus Strains Through Virus Indexing
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ACMV | African Cassava Mosaic Virus |
| CTAB | Cetyltrimethylammonium bromide |
| DNA | Deoxyribonucleic acid |
| EACMV | East African Cassava Mosaic Virus |
| CMD | Cassava Mosaic Disease |
| IITA | International Institute of Tropical Agriculture |
| MAP | Months After Planting |
| SEACMV | South East African Cassava Mosaic Virus |
| SLICASS | Sierra Leone Improved Cassava |
| WAVE | Central and West African Virus Epidemiology |
Appendix A
| Variant of the Experiment | Location | Sampling Regime | |||
|---|---|---|---|---|---|
| 3 MAP | 6 MAP | 9 MAP | 12 MAP | ||
| Incidence of CMD | |||||
| Asymptomatic | Bombali | 0.0 f | 27.7 e | 36.3 d | 36.3 d |
| Bonthe | 0.0 f | 30.0 e | 41.7 d | 44.0 d | |
| Kenema | 0.0 f | 28.0 de | 28.0 de | 37.7 d | |
| Port Loko | 0.0 f | 30.3 e | 58.3 c | 86.7 ab | |
| Improved | Bombali | 0.0 f | 0.0 f | 0.0 f | 0.0 f |
| Bonthe | 0.0 f | 0.0 f | 0.0 f | 0.0 f | |
| Kenema | 0.0 f | 0.0 f | 0.0 f | 0.0 f | |
| Port Loko | 0.0 f | 0.0 f | 0.0 f | 0.0 f | |
| Symptomatic | Bombali | 38.3 d | 100.0 a | 100.0 a | 95.7 a |
| Bonthe | 41.7 d | 80.0 b | 93.3 a | 81.7 b | |
| Kenema | 65.0 c | 100.0 a | 100.0 a | 100.0 a | |
| Port Loko | 100.0 a | 100.0 a | 100.0 a | 96.7 a | |
| Severity of CMD | |||||
| Asymptomatic | Bombali | 1.0 f | 2.0 e | 2.8 cd | 2.8 cd |
| Bonthe | 1.0 f | 2.2 e | 2.7 d | 2.7 d | |
| Kenema | 1.0 f | 2.3 e | 2.4 cde | 2.7 d | |
| Port Loko | 1.0 f | 2.3 e | 3.0 c | 3.3 c | |
| Improved | Bombali | 1.0 f | 1.0 f | 1.0 f | 1.0 f |
| Bonthe | 1.0 f | 1.0 f | 1.0 f | 1.0 f | |
| Kenema | 1.0 f | 1.0 f | 1.0 f | 1.0 f | |
| Port Loko | 1.0 f | 1.0 f | 1.0 f | 1.0 f | |
| Symptomatic | Bombali | 2.0 e | 4.7 a | 4.7 a | 4.3 a |
| Bonthe | 2.7 d | 3.8 bc | 4.7 a | 3.3 c | |
| Kenema | 3.4 c | 4.0 b | 4.7 a | 4.7 a | |
| Port Loko | 4.0 b | 4.0 b | 4.3 ab | 4.3 ab | |
| Location | ||||
|---|---|---|---|---|
| Variant of the Experiment | Bombali | Bonthe | Kenema | Port Loko |
| CMD Incidence | ||||
| Asymptomatic | 25.1 f | 28.9 f | 23.4 f | 43.8 e |
| Improved | 0.0 g | 0.0 g | 0.0 g | 0.0 g |
| Symptomatic | 83.5 c | 74.2 d | 91.3 b | 99.2 a |
| CMD severity | ||||
| Asymptomatic | 2.2 c | 2.1 c | 2.1 c | 2.4 c |
| Improved | 1.0 d | 1.0 d | 1.0 d | 1.0 d |
| Symptomatic | 3.9 ab | 3.6 b | 4.2 a | 4.2 a |
| Whitefly abundance | ||||
| Asymptomatic | 70.2 f | 80.2 e | 71.1 f | 59.8 g |
| Improved | 51.3 gh | 40.4 h | 50.2 gh | 38.6 h |
| Symptomatic | 142.9 c | 120.0 d | 155.3 b | 160.8 a |
| Sampling Regime | |||||
|---|---|---|---|---|---|
| Variant of the Experiment | Location | 3 MAP | 6 MAP | 9 MAP | 12 MAP |
| Asymptomatic | Bombali | 79.3 f | 76.3 fg | 72.7 f g | 52.3 gh |
| Bonthe | 82.0 f | 96.7 e | 81.0 f | 61.0 g | |
| Kenema | 74.7 fg | 65.0 g | 74.7 fg | 70.0 fg | |
| Port Loko | 68.3 fg | 56.7 g | 62.7 gh | 51.3 gh | |
| Improved | Bombali | 51.3 gh | 53.7 gh | 53.3 gh | 47.0 h |
| Bonthe | 75.3 fg | 34.3 i | 28.3 i | 23.7 i | |
| Kenema | 60.0 gh | 51.0 h | 43.7 h | 46.0 h | |
| Port Loko | 47.7 h | 34.3 i | 39.0 hi | 33.3 hi | |
| Symptomatic | Bombali | 136.7 d | 176.7 b | 180.0 b | 78.3 f |
| Bonthe | 100.0 ef | 133.3 d | 156.7 c | 90.0 f | |
| Kenema | 130.3 d | 195.0 a | 189.3 b | 106.7 e | |
| Port Loko | 157.0 c | 207.0 a | 180.0 b | 99.3 | |
Appendix B
References
- Fei, S.; Mahama, A.A.; Singh, A.K.; Singh, A. Cassava Breeding. In Crop Improvement; Suza, W.P., Lamkey, K.R., Eds.; Iowa State University Digital Press: Ames, ID, USA, 2023; Available online: https://iastate.pressbooks.pub/cropimprovement/chapter/cassava-breeding/ (accessed on 28 January 2022).
- Owolade, O.F.; Dixon, A.G.O.; Adeoti, A.Y.A. Diallel analysis of cassava genotypes to anthracnose disease. World J. Agric. Sci. 2006, 2, 98–104. [Google Scholar]
- Sesay, J.V.; Lebbie, A.; Wadsworth, R.; Nuwamanya, E.; Bado, S.; Norman, P.E. Genetic structure and diversity study of cassava (Mannihot esculenta) germplasm for African cassava mosaic disease and fresh storage root yield. Open J. Gen. 2023, 13, 23–47. [Google Scholar] [CrossRef]
- Esuma, W.; Nanyonjo, A.R.; Miiro, R.; Angudubo, S.; Kawuki, R.S. Men and women’s perception of yellow-root cassava among rural farmers in eastern Uganda. Agric. Food Secur. 2019, 8, 10. [Google Scholar] [CrossRef]
- Chávez, A.L.; Sánchez, T.; Jaramillo, G.; Bedoya, J.M.; Echeverry, J.; Bolaños, E.A.; Ceballos, H.; Iglesias, C.A. Variation of quality traits in cassava roots evaluated in landraces and improved clones. Euphytica 2005, 143, 125–133. [Google Scholar] [CrossRef]
- Kintché, K.; Hauser, S.; Mahungu, M.; Ndonda, A.; Lukombo, S.; Nhamo, N.; Uzokwe, V.N.E.; Yomeni, M.; Ngamitshara, J.; Ekoko, B.; et al. Cassava yield loss in farmer fields was mainly caused by low soil fertility and suboptimal management practices in two provinces of the Democratic Republic of Congo. Eur. J. Agron. 2017, 89, 107–123. [Google Scholar] [CrossRef]
- Legg, J.P.; Owor, B.; Sseruwagi, P.; Ndunguru, J. Cassava mosaic virus disease in East and Central Africa: Epidemiology and management of a regional pandemic. Adv. Virol. Res. 2006, 67, 355–418. [Google Scholar] [CrossRef]
- Crespo-Bellido, A.; Hoyer, J.S.; Dubey, D.; Jeannot, R.B.; Duffy, S. Interspecies recombination has driven the macroevolution of cassava mosaic begomoviruses. J. Virol. 2021, 95, e00541-21. [Google Scholar] [CrossRef]
- Maruthi, M.N.; Seal, S.; Colvin, J.; Briddon, R.W.; Bull, S.E. East African cassava mosaic Zanzibar virus—A recombinant begomovirus species with a mild phenotype. Arch. Virol. 2004, 149, 2365–2377. [Google Scholar] [CrossRef] [PubMed]
- Legg, J.P.; Fauquet, C.M. Cassava mosaic geminiviruses in Africa. Plant Mol. Biol. 2004, 56, 585–599. [Google Scholar] [CrossRef]
- Fondong, V.N.; Pita, J.S.; Rey, C.; Beachy, R.N.; Fauquet, C.M. Evidence of synergism between African cassava mosaic virus and the new double recombinant Geminivirus infecting cassava in Cameroon. J. Gen. Virol. 2000, 81, 287–297. [Google Scholar] [CrossRef]
- Zhou, X.; Robinson, D.; Harrison, D.B. Types of variation in DNA-A among isolates of East African cassava mosaic viruses from Kenya, Malawi and Tanzania. J. Gen. Virol. 1998, 79, 2835–2840. [Google Scholar] [CrossRef] [PubMed]
- Berrie, L.C.; Rybicki, E.P.; Rey, M.E.C. Complete nucleotide sequence and host range of South African cassava mosaic virus: Further evidence for recombination amongst begomoviruses. J. Gen. Virol. 2001, 82, 53–58. [Google Scholar] [CrossRef] [PubMed]
- Adjata, K.D.; Muller, E.; Aziadekey, M.; Gumedzoe, Y.M.D.; Peterschmitt, M. Incidence of cassava viral diseases and first identification of East African cassava mosaic virus and Indian cassava mosaic virus by PCR in Cassava (Manihot esculenta Crantz) fields in Togo. Am. J. Plant Physiol. 2008, 3, 73–80. [Google Scholar] [CrossRef]
- Harimalala, M.A.; Villemot, J.; Hoareau, M.; Zinga, I.; Ranomenjanahary, S.; Reynaud, B.; Lefeuvre, P.; Lett, J.M. Phylogénie et phylogéographie des begomovirus associés à la maladie de la mosaïque du manioc à Madagascar. In Proceedings of the 13 Èmes Rencontres de Virologie Végétale (RVV 2011), Aussois, France, 16–20 January 2011. [Google Scholar]
- IITA (International Institute of Tropical Agriculture). Cassava in Tropical Africa, A Reference Manual; IITA: Ibadan, Nigeria, 1990; p. 176. [Google Scholar]
- Zhou, X.; Liu, Y.; Calvert, L.; Munoz, C.; Otim-Nap, G.W.; Robinson, D.J.; Harrison, B.D. Evidence that DNA-A of Geminivirus associated with severe cassava mosaic disease in Uganda has arisen by interspecific recombination. J. Gen. Virol. 1997, 78, 2101–2111. [Google Scholar] [CrossRef]
- Neuenschwander, P.; Hughes, J.; Ogbe, F.; Ngatse, J.M.; Legg, J.P. Occurrence of the Uganda variant of east African cassava mosaic virus (EACMVUG) in Western Democratic Republic of Congo and the Congo Republic defines the westernmost extent of the CMD pandemic in East/Central Africa. Plant Pathol. 2002, 51, 385. [Google Scholar] [CrossRef]
- Monde, G.; Walangululu, J.; Winter, S.; Bragard, C. Dual infection by cassava begomoviruses in two leguminous species (Fabaceae) in Yangambi, North-Eastern Democartic Republic of Congo. Arch. Virol. 2010, 155, 1865–1869. [Google Scholar] [CrossRef]
- Legg, J.P.; Abele, S.; Obiero, H.; Jeremiah, S.; Bigirimana, S.; Ntawuruhunga, P. The cassava mosaic virus disease pandemic and its impact on people’s livelihoods in East and Central Africa. Phytopathology 2005, 95, 129–130. [Google Scholar]
- Owor, B.; Legg, J.P.; OkaoOkuja, G.; Obonyo, R.; Ogenga-Latigo, M.W. The effect of cassava mosaic geminiviruses on symptoms severity, growth and root yield of a cassava mosaic virus disease- susceptible cultivar in Uganda. Ann. Appl. Biol. 2004, 145, 331–337. [Google Scholar] [CrossRef]
- Mallowa, S.O.; Isutsa, D.K.; Kamau, A.W.; Obonyo, R.; Legg, J.P. Current characteristics of cassava mosaic disease in postepidemic areas increase the range of possible management options. Ann. Appl. Biol. 2006, 149, 137–144. [Google Scholar] [CrossRef]
- Kamilaris, A.; Prenafeta-Boldú, F.X. Deep learning in agriculture: A survey. Comp. Electron. Agric. 2018, 147, 70–90. [Google Scholar] [CrossRef]
- Mrisho, L.M.; Mbilinyi, N.A.; Ndalahwa, M.; Ramcharan, A.M.; Kehs, A.K.; McCloskey, P.C.; Murithi, H.; Hughes, D.P.; Legg, J.P. Accuracy of a smartphone-based object detection model, PlantVillage Nuru, in identifying the foliar symptoms of the viral diseases of cassava–CMD and CBSD. Front. Plant Sci. 2020, 11, 590889. [Google Scholar] [CrossRef]
- Ramcharan, A.; Baranowski, K.; McCloskey, P.; Ahmed, B.; Legg, J.; Hughes, D.P. Deep learning for image-based cassava disease detection. Front. Plant Sci. 2017, 8, 1852. [Google Scholar] [CrossRef] [PubMed]
- Ramcharan, A.; McCloskey, P.; Baranowski, K.; Mbilinyi, N.; Mrisho, L.; Ndalahwa, M.; Legg, J.; Hughes, D.P. A mobile-based deep learning model for cassava disease diagnosis. Front. Plant Sci. 2019, 10, 272. [Google Scholar] [CrossRef] [PubMed]
- Cuellar, W.; Mwanzia, L.; Lourido, D.; Garcia, C.; Martínez, A.; Cruz, P.; Pino, L.; Tohme, J. PestDisPlace: Monitoring the distribution of pests and diseases. Version 2.0 International Center for Tropical Agriculture (CIAT). 2018. Available online: https://pestdisplace.org (accessed on 1 January 2019).
- Siriwan, W.; Jimenez, J.; Hemniam, N.; Saokham, K.; Lopez-Alvarez, D.; Leiva, A.M.; Martinez, A.; Mwanzia, L.; Lopez-Lavalle, L.A.B.; Cueller, W.J. Surveillance and diagnostics of the emergent Sri Lankan cassava mosaic virus (Fam. Geminiviridae) in Southeast Asia. Virus Res. 2020, 285, 197959. [Google Scholar] [CrossRef] [PubMed]
- Sseruwagi, P.; Sserubombwe, W.S.; Legg, J.P.; Ndunguru, J.; Thresh, J.M. Methods of surveying the incidence and severity of cassava mosaic disease and whitefly vector populations on cassava in Africa: A review. Virus Res. 2004, 100, 129–142. [Google Scholar] [CrossRef]
- Ariyo, O.A.; Dixon, A.G.; Atiri, G.I. Whitefly Bemisia tabaci (Homoptera: Aleyrodidae) infestation on cassava genotypes grown at different ecozones in Nigeria. J. Econ. Entomol. 2005, 98, 611–617. [Google Scholar] [CrossRef]
- Permingeat, H.R.; Romagnoli, M.V.; Sesma, J.I.; Vallejos, R.H. A simple method for isolating DNA of high yield and quality from cotton (shape Gossypium hirsutum L.) leaves. Plant Mol. Biol. Rep. 1998, 16, 89. [Google Scholar] [CrossRef]
- Aljanabi, S.M.; Martinez, I. Universal and rapid salt-extraction of high-quality genomic DNA for PCR-based techniques. Nucleic Acids Res. 1997, 25, 4692–4693. [Google Scholar] [CrossRef]
- Pita, J.S.; Fondong, V.N.; Sangare, A.; Otim-Nape, G.W.; Ogwal, S.; Fauquet, C.M. Recombination, pseudorecombination and synergism of geminiviruses are determinant keys to the epidemic of severe cassava mosaic disease in Uganda. J. Gen. Virol. 2001, 82, 655–665. [Google Scholar] [CrossRef]
- Matic, S.; Pais da Cunha, A.T.; Thompson, J.R.; Tepfer, M. An analysis of viruses associated with cassava mosaic disease in three Angolan provinces. J. Plant Pathol. 2012, 94, 443–450. [Google Scholar]
- Alabi, O.J.; Kumar, P.L.; Naidu, R.A. Multiplex PCR method for detecting Africa cassava mosaic virus and east Africa cassava mosaic virus in cassava. J. Virol. Methods 2008, 154, 111–120. [Google Scholar] [CrossRef]
- Bos, L.; Parlevliet, J.E. Concepts and terminology on plant/pest relationships: Toward consensus in plant pathology and crop protection. Ann. Rev. Phytopathol. 1995, 33, 69–102. [Google Scholar] [CrossRef] [PubMed]
- Paudel, D.B.; Sanfaçon, H. Exploring the diversity of mechanisms associated with plant tolerance to virus infection. Front. Plant Sci. 2018, 9, 1575. [Google Scholar] [CrossRef]
- Fauquet, C.; Fargette, D. African cassava mosaic virus: Etiology, epidemiology and control. Plant Dis. 1990, 74, 404–411. [Google Scholar] [CrossRef]
- Amoakon, W.J.-L.; Combala, M.; Pita, J.S.; Mutuku, J.M.; N’Zué, B.; Otron, D.H.; Yéo, E.F.; Kouassi, N.K.; Sié, R. Phenotypic screening and molecular characterization of cassava mosaic disease resistance in Côte d’Ivoire cassava germplasm. Front Sust. Food Syst. 2023, 6, 1052437. [Google Scholar] [CrossRef]
- Soko, D.F.; Ambroise, L.; Siene, C.; Kotchi, V.; Gogbeu, J.; Sere, Y.; Ake, S. Évaluation du niveau de sensibilité et de résistance des variétés de riz différentielles d’ AfricaRice à huit isolats de la panachure jaune du riz (RYMV) de Gagnoa (Côte d’ Ivoire). J. Animal Plant Sci. 2015, 26, 4138–4149. [Google Scholar]
- Rodríguez, M.; Taleisnik, E.; Lenardon, S.; Lascano, R. Are sunflower chlorotic mottle virus infection symptoms modulated by early increases in leaf sugar concentration? J. Plant Physiol. 2010, 167, 1137–1144. [Google Scholar] [CrossRef]
- Rodríguez, M.; Muñoz, N.; Lenardon, S.; Lascano, R. The chlorotic symptom induced by Sunflower chlorotic mottle virus is associated with changes in redox-related gene expression and metabolites. Plant Sci. 2012, 196, 107–116. [Google Scholar] [CrossRef]
- Souza, P.F.N.; Carvalho, F.E.L. Killing two birds with one stone: How do plant viruses break down plant defenses and manipulate cellular processes to replicate themselves? J. Plant Biol. 2019, 62, 170–180. [Google Scholar] [CrossRef]
- Sade, D.; Brotman, Y.; Eybishtz, A.; Cuadros-Inostroza, A.; Fernie, A.R.; Willmitzer, L.; Czosnek, H. Involvement of the hexose transporter gene LeHT1 and of sugars in resistance of tomato to tomato yellow leaf curl virus. Mol. Plant 2013, 6, 1707–1710. [Google Scholar] [CrossRef]
- Nuwamanya, E.; Baguma, Y.; Atwijukire, E.; Acheng, S.; Abidrabo, P.; Omongo, C.A.; Alicai, T. Cassava brown streak disease effects on leaf metabolites and pigment accumulation. Afr. Crop Sci. J. 2017, 25, 33–45. [Google Scholar] [CrossRef]
- Andreola, S.; Rodriguez, M.; Parola, R.; Alemano, S.; Lascano, R. Interactions between soybean, Bradyrhizobium japonicum and Soybean mosaic virus: The effects depend on the interaction sequence. Functional Plant Biol. 2019, 46, 1036. [Google Scholar] [CrossRef] [PubMed]
- Técsi, L.I.; Smith, A.M.; Maule, A.J.; Leegood, R.C. A spatial analysis of physiological changes associated with infection of cotyledons of marrow plants with cucumber mosaic virus. Plant Physiol. 1996, 111, 975–985. [Google Scholar] [CrossRef]
- Wydra, K.; Msikita, W. An overview of the present situation of cassava diseases in West Africa. In Proceedings of the Sixth Triennial Symposium of the International Society of Tropical Root Crops for Poverty Alleviation. In Proceedings of the 6th Triennial Symposium, International Society for Tropical Root Crops-Africa Branch, Lilongwe, Malawi, 22–28 October 1995; Akoroda, M.O., Ekanayake, I.O., Eds.; pp. 198–206. [Google Scholar]
- Alabi, O.J.; Kumar, P.L.; Naidu, R.A. Cassava mosaic disease: A curse to food security in Sub-Saharan Africa. APSnet Features. Am. Phytopathol. Soc. 2011, 1–16. [Google Scholar]
- Ratner, B. The correlation coefficient: Its values range between +1/−1, or do they? J. Target Measure Anal. Market. 2009, 17, 139–142. [Google Scholar] [CrossRef]
- Biabani, A.R.; Pakniyat, H. Evaluation of seed yield-related characters in sesame (Sesamum indicum L.) using factor and path analysis. Pakistan J. Biol. Sci. 2008, 11, 1157–1160. [Google Scholar] [CrossRef]
- Johansen, K.; Sohlbach, M.; Sullivan, B.; Stringer, S.; Peasley, D.; Phinn, S. Mapping banana plants from high spatial resolution orthophotos to facilitate plant health assessment. Remote Sens. 2014, 6, 8261–8286. [Google Scholar] [CrossRef]
- Arakpogun, E.O.; Wanjiru, R.; Whalley, J. Impediments to the implementation of universal service funds in Africa—A cross-country comparative analysis. Telecom. Policy 2017, 41, 617–630. [Google Scholar] [CrossRef]
| Primer | Sequence (5′-3′) | Target Region | Expected Size (bp) | Virus Species | Reference |
|---|---|---|---|---|---|
| JSP001 JSP002 | ATGTCGAAGCGACCAGGAGAT TGTTTATTAATTGCCAATACT | DNA-A (CP) | 783 | ACMV | [33] |
| ACMBVF ACMBVR | TCGGGAGTGATACATGCGAAGGC GGCTACACCAGCTACCTGAAGCT | DNA-B (BV1/BC1) | 628 | ACMV | [34] |
| JSP001 JSP003 | ATGTCGAAGCGACCAGGAGAT CCTTTATTAATTTGTCACTGC | DNA-A (CP) | 780 | EACMV | [33] |
| CMBRepF EACMVRepR | CRTCAATGACGTTGTACCA GGTTTGCAGAGAACTACATC | DNA-A(AC1) | 650 | EACMV | [35] |
| VNF031F VNF032R | GGATACAGATAGGGTTCCCAC GACGAGGACAAGAATTCCAAT | AC2/AC3 | ≈560 | EACMV-CM | [12] |
| Variant of Experiment | Sampling Regime | |||
|---|---|---|---|---|
| 3 MAP | 6 MAP | 9 MAP | 12 MAP | |
| Percent Incidence of CMD | ||||
| Improved | 0 ± 0.00 b | 0.0 ± 0.00 c | 0.00 ± 0.00 c | 0.00 ± 0.00 c |
| Asymptomatic | 0 ± 0.00 b | 28.9 ± 10.87 b | 41.08 ± 20.98 b | 51.17 ± 25.11 b |
| Symptomatic | 61.25 ± 29.63 a | 95.0 ± 11.60 a | 98.33 ± 5.77 a | 93.50 ± 8.67 a |
| p-value | <0.001 | <0.001 | <0.001 | <0.001 |
| Significance | *** | *** | *** | *** |
| Severity of CMD | ||||
| Improved | 1.00 ± 0.00 b | 1.0 ± 0.00 b | 1.00 ± 0.00 c | 1.00 ± 0.00 c |
| Asymptomatic | 1.00 ± 0.00 b | 2.2 ± 0.00 b | 2.73 ± 0.55 b | 2.87 ± 0.50 b |
| Symptomatic | 3.01 ± 1.00 a | 4.1 ± 0.60 a | 4.58 ± 0.51 a | 4.17 ± 0.83 a |
| p-value | <0.001 | <0.001 | <0.001 | <0.001 |
| Significance | *** | *** | *** | *** |
| Whitefly abundance | ||||
| Improved | 51.3 gh | 40.4 h | 50.2 gh | 38.6 h |
| Asymptomatic | 70.2 f | 80.2 e | 71.1 f | 59.8 g |
| Symptomatic | 142.9 c | 120.0 d | 155.3 b | 160.8 a |
| p-value | <0.001 | <0.001 | <0.001 | <0.001 |
| Significance | *** | *** | *** | *** |
| Variant of Experiment | CMD Incidence | CMD Severity | Whitefly Abundance | Number of Nodes per Stem | Storage Root Diameter (cm) | Storage Root Length (cm) | Fresh Storage Root Yield (t ha−1) |
|---|---|---|---|---|---|---|---|
| Improved | 0.0 c | 1.0 c | 45.1 d | 20.7 a | 5.2 a | 87.34 a | 54.9 a |
| Asymptomatic | 30.3 b | 2.2 b | 70.3 c | 14.8 b | 4.0 b | 54.13 b | 32.5 b |
| Symptomatic | 87.0 a | 4.0 a | 144.8 a | 11.9 c | 2.7 c | 36.06 c | 10.5 c |
| Location | |||||||
| Bombali | 36.2 b | 2.4 a | 88.1 b | 15.6 a | 3.8 ab | 57.9 b | 30.0 c |
| Bonthe | 34.4 b | 2.3 ab | 80.2 c | 16.2 a | 4.1 a | 55.5 b | 38.9 a |
| Kenema | 38.2 b | 2.4 a | 92.2 a | 15.3 ab | 4.0 a | 61.6 a | 33.7 b |
| Port Loko | 47.7 a | 2.5 a | 86.4 b | 16.0 a | 4.0 a | 61.6 a | 27.8 c |
| Variant of the Experiment | Leaf Area at 3 Months After Planting | Leaf Area at 6 Months After Planting | Leaf Area at 9 Months After Planting | Leaf Area at 12 Months After Planting |
|---|---|---|---|---|
| Improved | 3.68 ± 0.28 a | 4.52 ± 0.64 a | 9.45 ± 0.49 a | 10.65 ± 0.22 a |
| Asymptomatic | 3.10 ± 0.09 b | 4.05 ± 0.18 b | 7.30 ± 0.23 b | 7.63 ± 0.32 b |
| Symptomatic | 1.99 ± 0.10 c | 3.08 ± 0.22 c | 3.20 ± 0.52 c | 3.26 ± 0.49 c |
| DMRT | 0.15 | 0.33 | 0.36 | 0.30 |
| p-value | <0.001 | <0.001 | <0.001 | <0.001 |
| Significance | *** | *** | *** | *** |
| Variant of Experiment | Location | |||
|---|---|---|---|---|
| Bombali | Bonthe | Kenema | Port Loko | |
| Number of nodes per stem | ||||
| Improved | 20.7 a | 20.7 a | 19.0 b | 22.0 a |
| Asymptomatic | 14.3 c | 15.5 c | 14.0 c | 15.0 c |
| Symptomatic | 11.7 d | 12.4 d | 12.0 d | 11.0 d |
| Storage root diameter (cm) | ||||
| Improved | 5.1 a | 5.1 a | 5.4 a | 5.3 a |
| Asymptomatic | 3.8 c | 3.8 c | 4.1 bc | 4.3 b |
| Symptomatic | 2.5 e | 3.4 d | 2.5 e | 2.5 e |
| Storage root length (cm) | ||||
| Improved | 82.27 b | 80.23 b | 93.43 a | 93.43 a |
| Asymptomatic | 54.9 c | 51.83 c | 54.9 c | 54.9 c |
| Symptomatic | 36.57 d | 34.53 d | 36.57 d | 36.57 d |
| Fresh storage root yield (t ha−1) | ||||
| Improved | 53.1 b | 66.4 a | 46.7 c | 46.7 c |
| Asymptomatic | 31.7 d | 35.7 d | 27.3 e | 27.3 e |
| Symptomatic | 5.2 h | 14.7 f | 9.5 g | 9.5 g |
| Fresh storage root yield loss (%) | ||||
| Asymptomatic | 40.3 a | 46.2 a | 41.5 a | 41.5 a |
| Symptomatic | 90.2 c | 77.9 b | 79.7 b | 79.7 b |
| Variant of Experiment | Status of Samples for CMD Infection | Detection of ACMV Single Infection | Detection of EACMV Single Infection | Detection of ACMV/EACMV Mix Infection | Negative Samples |
|---|---|---|---|---|---|
| Improved | 40 | 0 | 0 | 0 | 40 |
| Asymptomatic | 40 | 26 | 4 | 10 | 0 |
| Symptomatic | 40 | 10 | 10 | 20 | 0 |
| Total | 120 | 36 (30.0%) | 14 (11.7%) | 30 (25.0%) | 40 (33.3%) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Saffa, M.D.; Samura, A.E.; Bah, M.A.; Eni, A.O.; Tibiri, E.B.; Sagnon, A.; Tiendrébéogo, F.; Pita, J.S.; Norman, P.E.; Johnson, R.A.B. Detection of Cassava Mosaic Disease and Assessment of Selected Agronomic Traits of Cassava (Manihot esculenta). Horticulturae 2025, 11, 618. https://doi.org/10.3390/horticulturae11060618
Saffa MD, Samura AE, Bah MA, Eni AO, Tibiri EB, Sagnon A, Tiendrébéogo F, Pita JS, Norman PE, Johnson RAB. Detection of Cassava Mosaic Disease and Assessment of Selected Agronomic Traits of Cassava (Manihot esculenta). Horticulturae. 2025; 11(6):618. https://doi.org/10.3390/horticulturae11060618
Chicago/Turabian StyleSaffa, Musa Decius, Alusaine Edward Samura, Mohamed Alieu Bah, Angela Obiageli Eni, Ezechiel Bionimian Tibiri, Adama Sagnon, Fidèle Tiendrébéogo, Justin Simon Pita, Prince Emmanuel Norman, and Raymonda Adeline Bernardette Johnson. 2025. "Detection of Cassava Mosaic Disease and Assessment of Selected Agronomic Traits of Cassava (Manihot esculenta)" Horticulturae 11, no. 6: 618. https://doi.org/10.3390/horticulturae11060618
APA StyleSaffa, M. D., Samura, A. E., Bah, M. A., Eni, A. O., Tibiri, E. B., Sagnon, A., Tiendrébéogo, F., Pita, J. S., Norman, P. E., & Johnson, R. A. B. (2025). Detection of Cassava Mosaic Disease and Assessment of Selected Agronomic Traits of Cassava (Manihot esculenta). Horticulturae, 11(6), 618. https://doi.org/10.3390/horticulturae11060618






