A Genome-Wide Characterization of the Xyloglucan Endotransglucosylase/Hydrolase Family Genes and Their Functions in the Shell Formation of Pecan
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Identification and Characterization of XTH Family Genes in Pecan
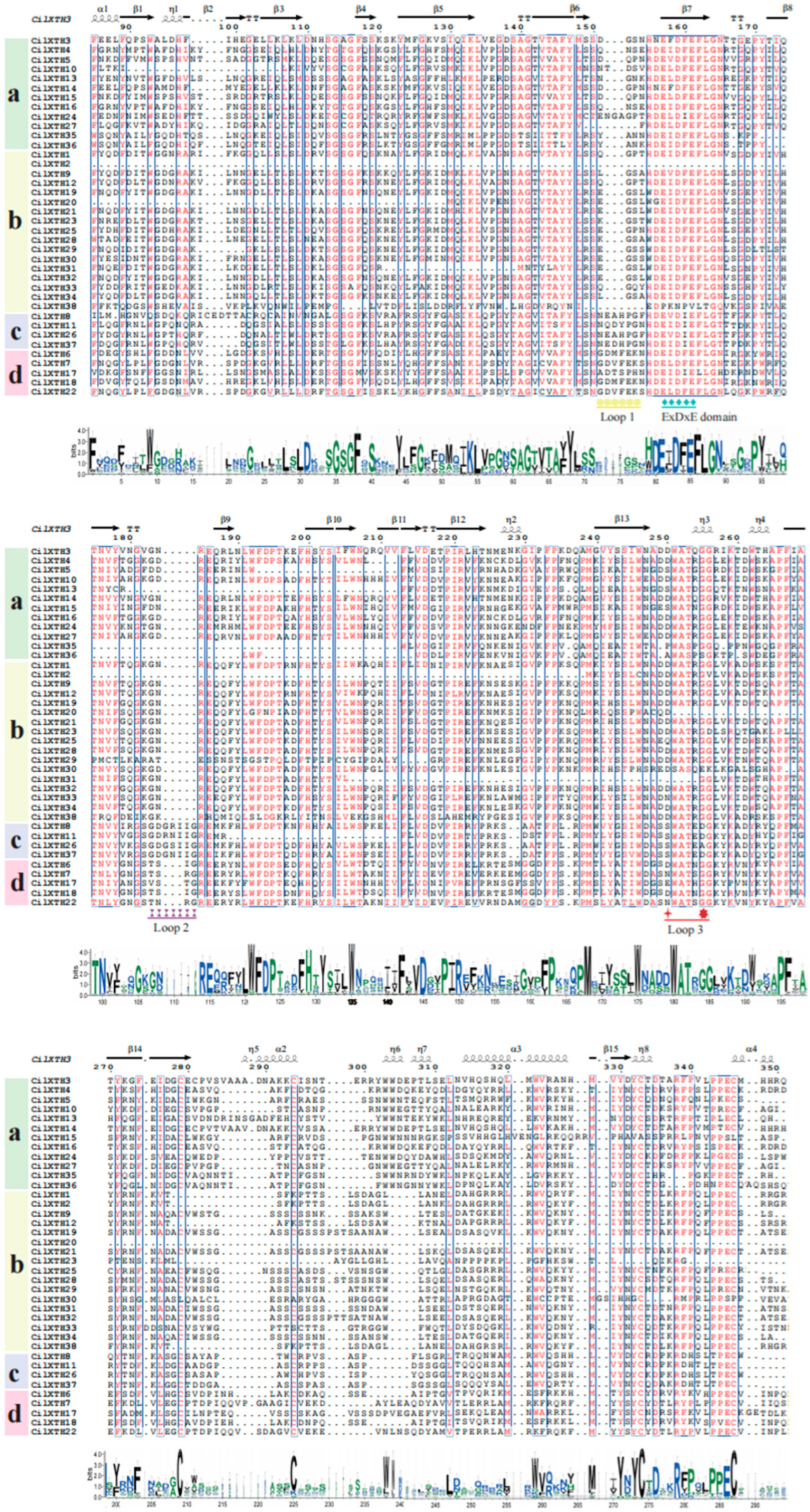
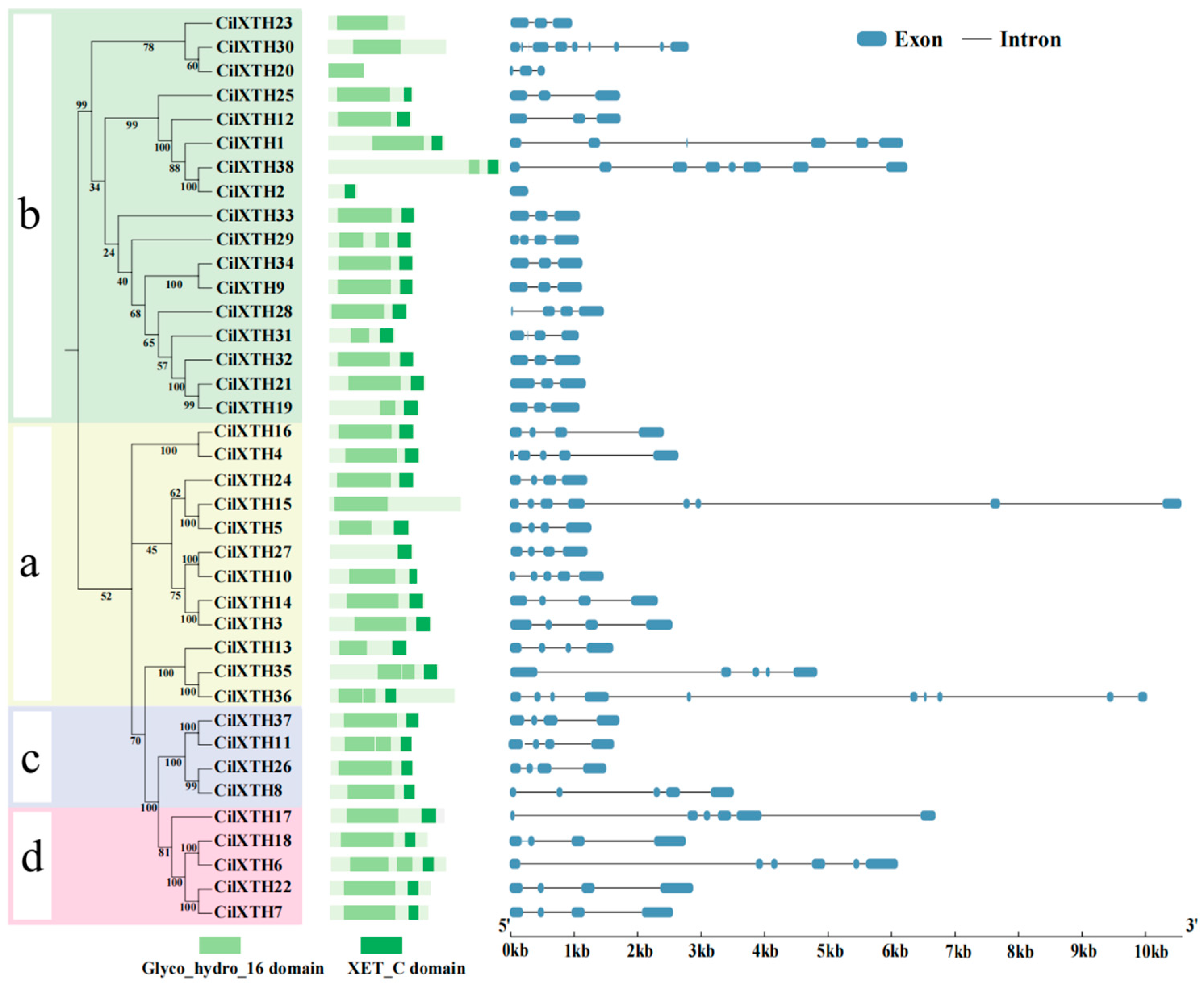
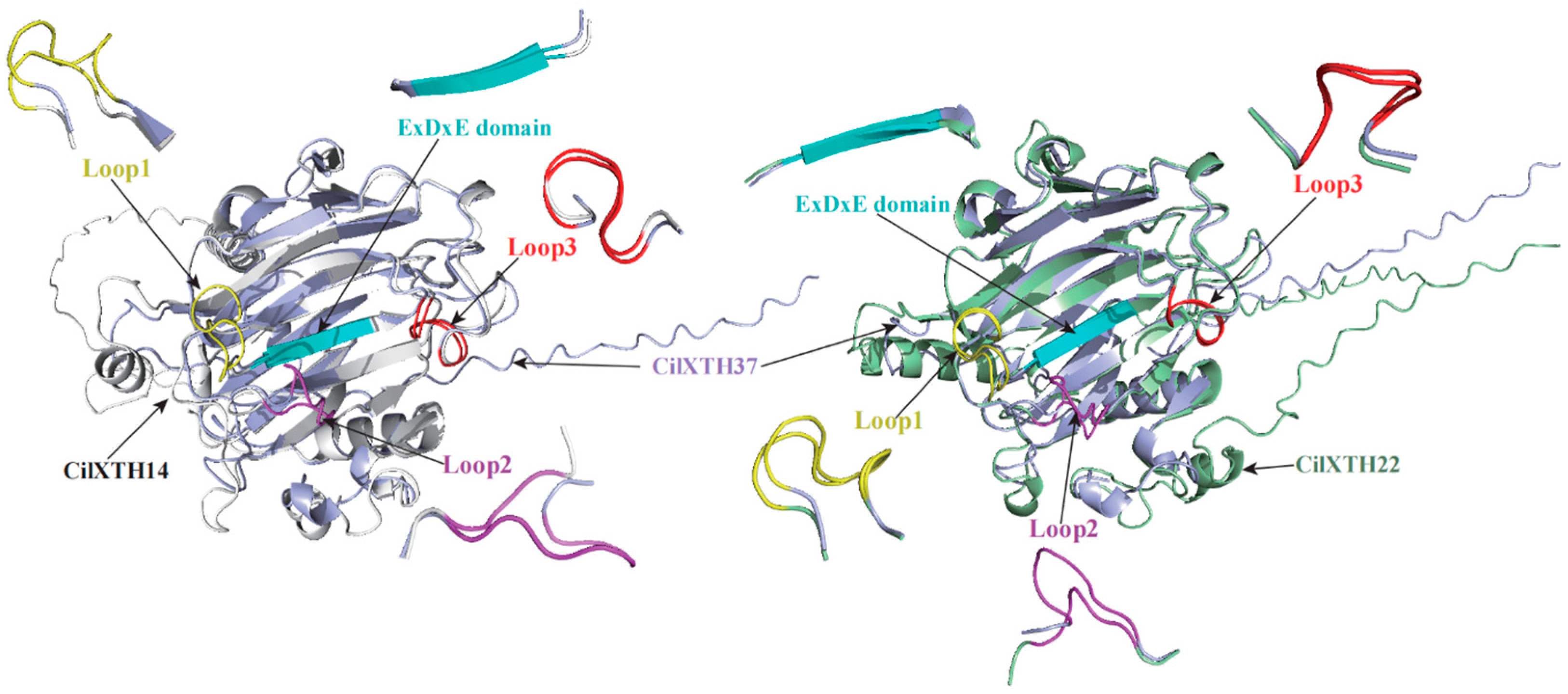
2.3. Multiple Sequence Alignment, Conserved Domain, and Gene Structure Analysis
2.4. Evolutionary Relationships of XTH Genes in Pecan
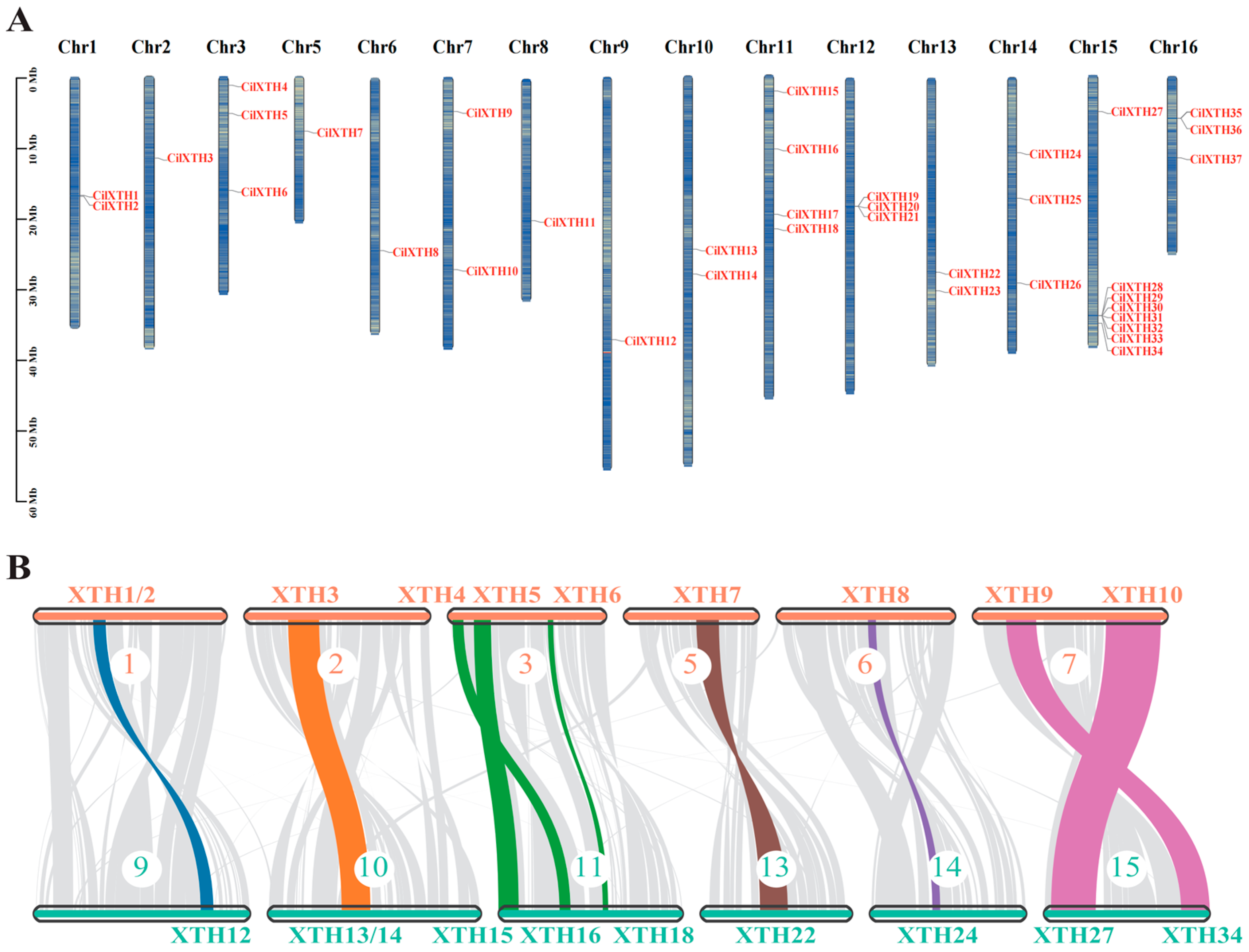
2.5. Chromosomal Distribution, Collinearity, and Evolutionary Analysis
2.6. RNA Extraction, Library Construction, Expression Profiling, and Gene Co-Expression Analysis
2.7. Cis-Regulatory Elements Analysis of CilXTH Genes
3. Results
3.1. The Identification and Structural Features of CiXTH Genes from the Pecan Genome
3.2. Phylogenetic Analysis of XTH Genes in Pecan
3.3. Chromosome Distribution and Collinearity Analysis of CilXTH Genes
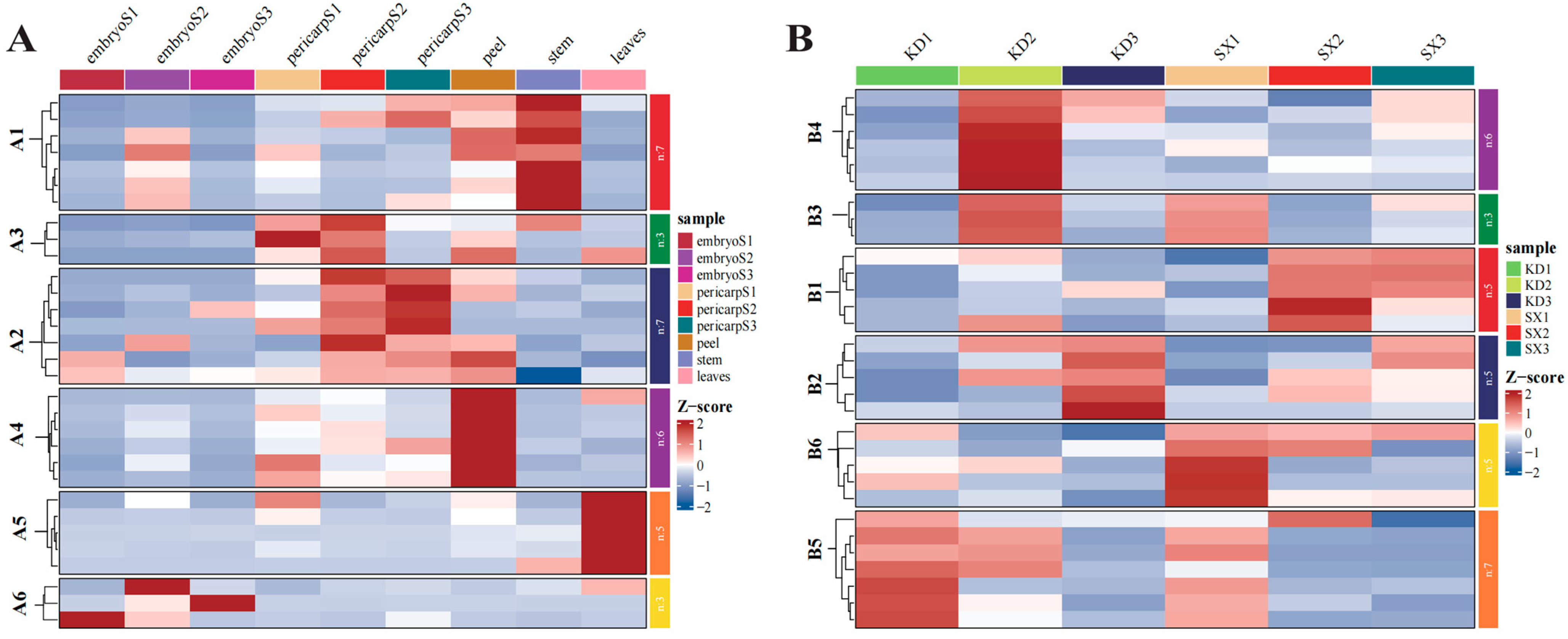
3.4. Expression Profiles of CilXTH Genes in Different Tissues and Pecan Varieties
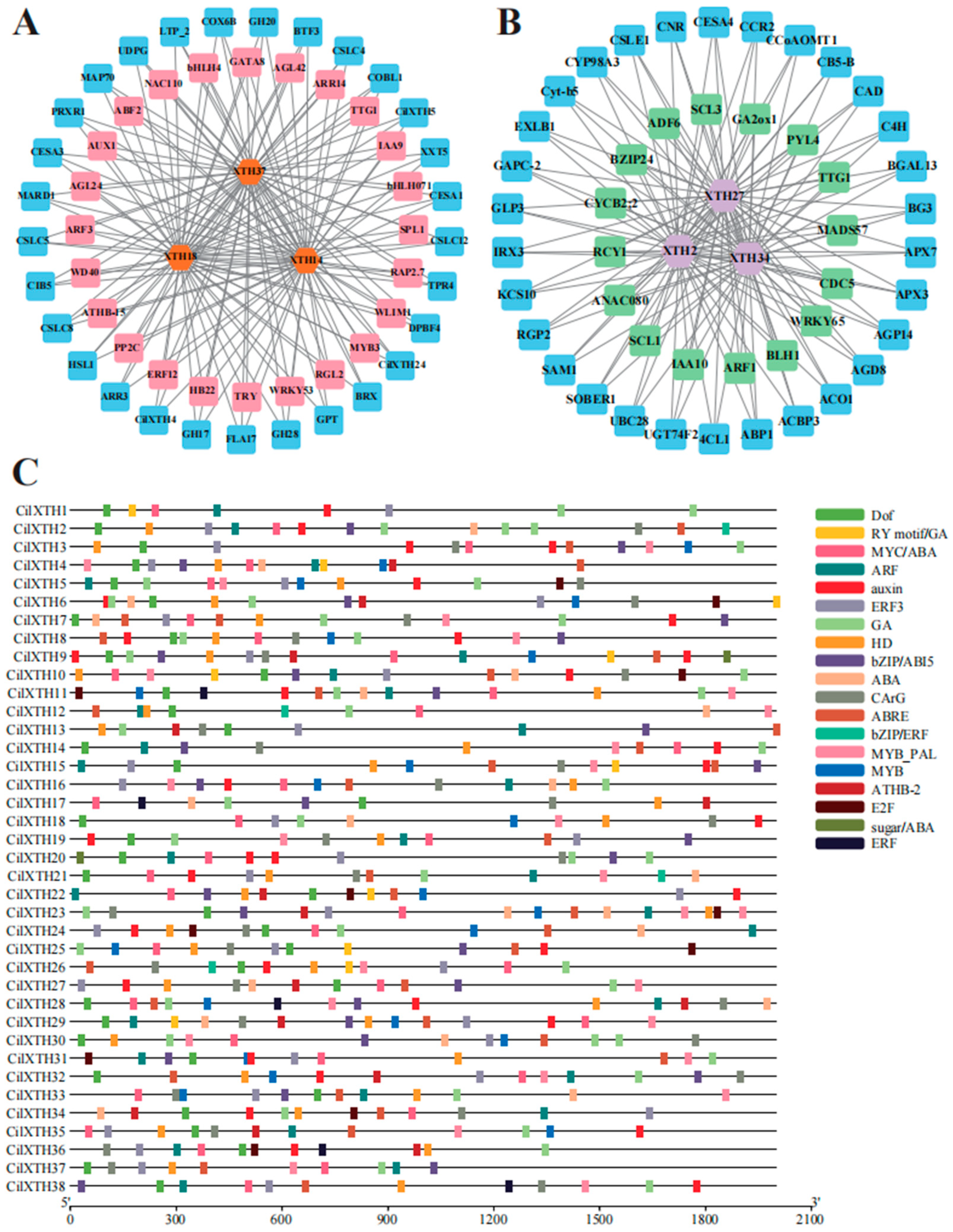
3.5. Gene Regulatory Network of CilXTH Genes in Pecan Shell Development
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| XTH | Xyloglucan endotransglucosylases/hydrolases |
| SCW | Secondary cell wall |
| XyG | Xyloglucan |
| XET | Xyloglucan endotransglucosylase |
| XEH | Xyloglucan hydrolase |
| GH16 | Glycoside hydrolase family 16 |
| KD | Caddo |
| SX | Shaoxing |
| DAP | Days after pollination |
| MW | Molecular weight |
| pI | Isoelectric point |
| ML | Maximum Likelihood |
| kDa | kiloDalton |
| aa | Amino acids |
| CDS | Coding DNA Sequence |
| CW | Cell wall |
| C | Cytoplasm |
| CP | Chloroplast |
| SBP | Selenium-binding protein |
| Mb | Megabases |
| GA | Gibberellic Acid |
| ABA | Abscisic Acid |
References
- Le Gall, H.; Philippe, F.; Domon, J.M.; Gillet, F.; Pelloux, J.; Rayon, C. Cell Wall Metabolism in Response to Abiotic Stress. Plants 2015, 4, 112–166. [Google Scholar] [CrossRef] [PubMed]
- Cosgrove, D.J. Growth of the Plant Cell Wall. Nat. Rev. Mol. Cell Biol. 2005, 6, 850–861. [Google Scholar] [CrossRef]
- Xu, H.; Giannetti, A.; Sugiyama, Y.; Zheng, W.; Schneider, R.; Watanabe, Y.; Oda, Y.; Persson, S. Secondary Cell Wall Patterning-Connecting the Dots, Pits and Helices. Open Biol. 2022, 12, 210208. [Google Scholar] [CrossRef]
- Cosgrove, D.J.; Jarvis, M.C. Comparative Structure and Biomechanics of Plant Primary and Secondary Cell Walls. Front. Plant Sci. 2012, 3, 204. [Google Scholar] [CrossRef]
- von Schantz, L.; Gullfot, F.; Scheer, S.; Filonova, L.; Cicortas Gunnarsson, L.; Flint, J.E.; Daniel, G.; Nordberg-Karlsson, E.; Brumer, H.; Ohlin, M. Affinity Maturation Generates Greatly Improved Xyloglucan-Specific Carbohydrate Binding Modules. BMC Biotechnol. 2009, 9, 92. [Google Scholar] [CrossRef]
- Takahashi, D.; Johnson, K.L.; Hao, P.; Tuong, T.; Erban, A.; Sampathkumar, A.; Bacic, A.; Livingston, D.P., 3rd; Kopka, J.; Kuroha, T.; et al. Cell Wall Modification by the Xyloglucan Endotransglucosylase/Hydrolase XTH19 Influences Freezing Tolerance after Cold and Sub-Zero Acclimation. Plant Cell Environ. 2021, 44, 915–930. [Google Scholar] [CrossRef]
- Li, Y.; Hua, J.; Hou, X.; Qi, N.; Li, C.; Wang, C.; Yao, Y.; Huang, D.; Zhang, H.; Liao, W. Brassinosteroids Is Involved in Methane-Induced Adventitious Root Formation Via Inducing Cell Wall Relaxation in Marigold. BMC Plant Biol. 2023, 23, 2. [Google Scholar] [CrossRef] [PubMed]
- Hrmova, M.; Stratilová, B.; Stratilová, E. Broad Specific Xyloglucan:Xyloglucosyl Transferases Are Formidable Players in the Re-Modelling of Plant Cell Wall Structures. Int. J. Mol. Sci. 2022, 23, 1656. [Google Scholar] [CrossRef]
- Holland, C.; Simmons, T.J.; Meulewaeter, F.; Hudson, A.; Fry, S.C. Three Highly Acidic Equisetum XTHs Differ from Hetero-Trans-β-Glucanase in Donor Substrate Specificity and Are Predominantly Xyloglucan Homo-Transglucosylases. J. Plant Physiol. 2020, 251, 153210. [Google Scholar] [CrossRef] [PubMed]
- Eklöf, J.M.; Brumer, H. The Xth Gene Family: An Update on Enzyme Structure, Function, and Phylogeny in Xyloglucan Remodeling. Plant Physiol. 2010, 153, 456–466. [Google Scholar] [CrossRef]
- Viborg, A.H.; Terrapon, N.; Lombard, V.; Michel, G.; Czjzek, M.; Henrissat, B.; Brumer, H. A Subfamily Roadmap of the Evolutionarily Diverse Glycoside Hydrolase Family 16 (GH16). J. Biol. Chem. 2019, 294, 15973–15986. [Google Scholar] [CrossRef] [PubMed]
- Yokoyama, R.; Nishitani, K. A Comprehensive Expression Analysis of All Members of a Gene Family Encoding Cell-Wall Enzymes Allowed Us to Predict Cis-Regulatory Regions Involved in Cell-Wall Construction in Specific Organs of Arabidopsis. Plant Cell Physiol. 2001, 42, 1025–1033. [Google Scholar] [CrossRef] [PubMed]
- Uozu, S.; Tanaka-Ueguchi, M.; Kitano, H.; Hattori, K.; Matsuoka, M. Characterization of XET-Related Genes of Rice. Plant Physiol. 2000, 122, 853–859. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Z.; Zhang, X.; Yao, W.; Gao, Y.; Zhao, K.; Guo, Q.; Zhou, B.; Jiang, T. Genome-Wide Identification and Expression Analysis of the Xyloglucan Endotransglucosylase/Hydrolase Gene Family in Poplar. BMC Genomics 2021, 22, 804. [Google Scholar] [CrossRef]
- Dhar, S.; Kim, J.; Yoon, E.K.; Jang, S.; Ko, K.; Lim, J. Short-Root Controls Cell Elongation in the Etiolated Arabidopsis hypocotyl. Mol. Cells 2022, 45, 243–256. [Google Scholar] [CrossRef]
- Hyodo, H.; Yamakawa, S.; Takeda, Y.; Tsuduki, M.; Yokota, A.; Nishitani, K.; Kohchi, T. Active Gene Expression of a Xyloglucan Endotransglucosylase/Hydrolase Gene, XTH9, in Inflorescence Apices Is Related to Cell Elongation in Arabidopsis thaliana. Plant Mol. Biol. 2003, 52, 473–482. [Google Scholar] [CrossRef]
- Li, Y.; Zheng, X.; Wang, C.; Hou, D.; Li, T.; Li, D.; Ma, C.; Sun, Z.; Tian, Y. Pear Xyloglucan Endotransglucosylase/Hydrolases Pcbru1 Promotes Stem Growth through Regulating Cell Wall Elongation. Plant Sci. 2021, 312, 111026. [Google Scholar] [CrossRef]
- Kushwah, S.; Banasiak, A.; Nishikubo, N.; Derba-Maceluch, M.; Majda, M.; Endo, S.; Kumar, V.; Gomez, L.; Gorzsas, A.; McQueen-Mason, S.; et al. Arabidopsis XTH4 and XTH9 Contribute to Wood Cell Expansion and Secondary Wall Formation. Plant Physiol. 2020, 182, 1946–1965. [Google Scholar] [CrossRef]
- Vissenberg, K.; Oyama, M.; Osato, Y.; Yokoyama, R.; Verbelen, J.P.; Nishitani, K. Differential Expression of AtXTH17, AtXTH18, AtXTH19 and AtXTH20 Genes in Arabidopsis Roots. Physiological Roles in Specification in Cell Wall Construction. Plant Cell Physiol. 2005, 46, 192–200. [Google Scholar] [CrossRef]
- Matsui, A.; Yokoyama, R.; Seki, M.; Ito, T.; Shinozaki, K.; Takahashi, T.; Komeda, Y.; Nishitani, K. AtXTH27 Plays an Essential Role in Cell Wall Modification During the Development of Tracheary Elements. Plant J. 2005, 42, 525–534. [Google Scholar] [CrossRef]
- Nishikubo, N.; Takahashi, J.; Roos, A.A.; Derba-Maceluch, M.; Piens, K.; Brumer, H.; Teeri, T.T.; Stålbrand, H.; Mellerowicz, E.J. Xyloglucan Endo-Transglycosylase-Mediated Xyloglucan Rearrangements in Developing Wood of Hybrid Aspen. Plant Physiol. 2011, 155, 399–413. [Google Scholar] [CrossRef] [PubMed]
- Nakaune, M.; Hanada, A.; Yin, Y.G.; Matsukura, C.; Yamaguchi, S.; Ezura, H. Molecular and Physiological Dissection of Enhanced Seed Germination Using Short-Term Low-Concentration Salt Seed Priming in Tomato. Plant Physiol. Biochem. 2012, 52, 28–37. [Google Scholar] [CrossRef] [PubMed]
- Fu, M.M.; Cao, F.; Qiu, C.W.; Liu, C.; Tong, T.; Feng, X.; Cai, S.; Chen, Z.H.; Wu, F. Xyloglucan Endotransglucosylase-Hydrolase 1 Is a Negative Regulator of Drought Tolerance in Barley Via Modulating Lignin Biosynthesis and Stomatal Closure. Plant Physiol. Biochem. 2024, 216, 109171. [Google Scholar] [CrossRef]
- Badenes, M.L.; Byrne, D.H. Fruit Breeding; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2012; Volume 8. [Google Scholar]
- Toro-Vazquez, J.F.; Charó-Alonso, M.A.; Pérez-Briceño, F. Fatty Acid Composition and Its Relationship with Physicochemical Properties of Pecan (Carya illinoensis) Oil. J. Am. Oil Chem. Soc. 1999, 76, 957–965. [Google Scholar] [CrossRef]
- Liu, J.; Xue, T.; Ren, L.; Cui, M.; Jiang, T.; Yang, X. Study on Pecan Seed Germination Influenced by Seed Endocarp. Open Life Sci. 2022, 17, 851–855. [Google Scholar] [CrossRef]
- Xiao, L.; Yu, M.; Zhang, Y.; Hu, J.; Zhang, R.; Wang, J.; Guo, H.; Zhang, H.; Guo, X.; Deng, T.; et al. Chromosome-Scale Assembly Reveals Asymmetric Paleo-Subgenome Evolution and Targets for the Acceleration of Fungal Resistance Breeding in the Nut Crop, Pecan. Plant Commun. 2021, 2, 100247. [Google Scholar] [CrossRef] [PubMed]
- Potter, S.C.; Luciani, A.; Eddy, S.R.; Park, Y.; Lopez, R.; Finn, R.D. Hmmer Web Server: 2018 Update. Nucleic Acids Res. 2018, 46, W200–W204. [Google Scholar] [CrossRef]
- Gasteiger, E.; Gattiker, A.; Hoogland, C.; Ivanyi, I.; Appel, R.D.; Bairoch, A. Expasy: The Proteomics Server for in-Depth Protein Knowledge and Analysis. Nucleic Acids Res. 2003, 31, 3784–3788. [Google Scholar] [CrossRef]
- Chou, K.C.; Shen, H.B. Plant-Mploc: A Top-Down Strategy to Augment the Power for Predicting Plant Protein Subcellular Localization. PLoS ONE 2010, 5, e11335. [Google Scholar] [CrossRef]
- Høie, M.H.; Kiehl, E.N.; Petersen, B.; Nielsen, M.; Winther, O.; Nielsen, H.; Hallgren, J.; Marcatili, P. Netsurfp-3.0: Accurate and Fast Prediction of Protein Structural Features by Protein Language Models and Deep Learning. Nucleic Acids Res. 2022, 50, W510–W515. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly Accurate Protein Structure Prediction with Alphafold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive Tree of Life (Itol) V5: An Online Tool for Phylogenetic Tree Display and Annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y.; et al. Tbtools-Ii: A “One for All, All for One” Bioinformatics Platform for Biological Big-Data Mining. Mol. Plant 2023, 16, 1733–1742. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Tang, H.; Debarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.H.; Jin, H.; Marler, B.; Guo, H.; et al. Mcscanx: A Toolkit for Detection and Evolutionary Analysis of Gene Synteny and Collinearity. Nucleic Acids Res. 2012, 7, e49. [Google Scholar] [CrossRef]
- Sleator, R.D. Jcvi-Syn3.0—A Synthetic Genome Stripped Bare! Bioengineered 2016, 7, 53–56. [Google Scholar] [CrossRef]
- Sun, P.; Jiao, B.; Yang, Y.; Shan, L.; Li, T.; Li, X.; Xi, Z.; Wang, X.; Liu, J. Wgdi: A User-Friendly Toolkit for Evolutionary Analyses of Whole-Genome Duplications and Ancestral Karyotypes. Mol. Plant 2022, 15, 1841–1851. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Xiao, L.; Zhang, Z.; Zhang, R.; Wang, Z.; Huang, C.; Huang, R.; Luan, Y.; Fan, T.; Wang, J.; et al. The Genomes of Pecan and Chinese Hickory Provide Insights into Carya Evolution and Nut Nutrition. Gigascience 2019, 8, giz036. [Google Scholar] [CrossRef]
- Xanthopoulou, A.; Moysiadis, T.; Bazakos, C.; Karagiannis, E.; Karamichali, I.; Stamatakis, G.; Samiotaki, M.; Manioudaki, M.; Michailidis, M.; Madesis, P.; et al. The Perennial Fruit Tree Proteogenomics Atlas: A Spatial Map of the Sweet Cherry Proteome and Transcriptome. Plant J. 2022, 109, 1319–1336. [Google Scholar] [CrossRef]
- Brandine, G.S.; Smith, A.D. Falco: High-Speed Fastqc Emulation for Quality Control of Sequencing Data. F1000Research 2019, 8, 1874. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-Based Genome Alignment and Genotyping with Hisat2 and Hisat-Genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef] [PubMed]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. Stringtie Enables Improved Reconstruction of a Transcriptome from Rna-Seq Reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef] [PubMed]
- Huynh-Thu, V.A.; Irrthum, A.; Wehenkel, L.; Geurts, P. Inferring Regulatory Networks from Expression Data Using Tree-Based Methods. PLoS ONE 2010, 5, e12776. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Stratilová, B.; Šesták, S.; Stratilová, E.; Vadinová, K.; Kozmon, S.; Hrmova, M. Engineering of Substrate Specificity in a Plant Cell-Wall Modifying Enzyme through Alterations of Carboxyl-Terminal Amino Acid Residues. Plant J. 2023, 116, 1529–1544. [Google Scholar] [CrossRef] [PubMed]
- Blanc, G.; Wolfe, K.H. Widespread Paleopolyploidy in Model Plant Species Inferred from Age Distributions of Duplicate Genes. Plant Cell 2004, 16, 1667–1678. [Google Scholar] [CrossRef]
- Lynch, M.; Conery, J.S. The Evolutionary Fate and Consequences of Duplicate Genes. Science 2000, 290, 1151–1155. [Google Scholar] [CrossRef]
- Cao, J.; Lv, Y.; Li, X. Interspaced Repeat Sequences Confer the Regulatory Functions of AtXTH10, Important for Root Growth in Arabidopsis. Plants 2019, 8, 130. [Google Scholar] [CrossRef]
- Gao, Y.; Wang, L.; Li, D.; Qi, D.; Fang, F.; Luo, Y.; Zhang, H.; Zhang, S. Genome-Wide Characterization of the Xyloglucan Endotransglucosylase/Hydrolase Family Genes and Their Response to Plant Hormone in Sugar Beet. Plant Physiol. Biochem. 2024, 206, 108239. [Google Scholar] [CrossRef]
- Zhang, Z.; Wang, N.; Jiang, S.; Xu, H.; Wang, Y.; Wang, C.; Li, M.; Liu, J.; Qu, C.; Liu, W.; et al. Analysis of the Xyloglucan Endotransglucosylase/Hydrolase Gene Family During Apple Fruit Ripening and Softening. J. Agric. Food Chem. 2017, 65, 429–434. [Google Scholar] [CrossRef]
- Van Sandt, V.S.; Suslov, D.; Verbelen, J.P.; Vissenberg, K. Xyloglucan Endotransglucosylase Activity Loosens a Plant Cell Wall. Ann. Bot. 2007, 100, 1467–1473. [Google Scholar] [CrossRef]
- Derba-Maceluch, M.; Awano, T.; Takahashi, J.; Lucenius, J.; Ratke, C.; Kontro, I.; Busse-Wicher, M.; Kosik, O.; Tanaka, R.; Winzéll, A.; et al. Suppression of Xylan Endotransglycosylase PtxtXyn10A Affects Cellulose Microfibril Angle in Secondary Wall in Aspen Wood. New Phytol. 2015, 205, 666–681. [Google Scholar] [CrossRef] [PubMed]
- Yokoyama, R.; Uwagaki, Y.; Sasaki, H.; Harada, T.; Hiwatashi, Y.; Hasebe, M.; Nishitani, K. Biological Implications of the Occurrence of 32 Members of the XTH (Xyloglucan Endotransglucosylase/Hydrolase) Family of Proteins in the Bryophyte Physcomitrella patens. Plant J. 2010, 64, 645–656. [Google Scholar] [CrossRef] [PubMed]
- Bhatti, M.H.T.; Han, S.; Sami, A.; Haider, M.Z.; Shafiq, M.; Ali, M.; Ahmad, S.; Ali, Q.; Sabir, I.A.; Manzoor, M.A. Genome-Wide Analysis of XTH Gene Family in Cucumber (Cucumis sativus) against Different Insecticides to Enhance Defense Mechanism. Plant Stress 2024, 13, 100538. [Google Scholar] [CrossRef]
- Witasari, L.D.; Huang, F.C.; Hoffmann, T.; Rozhon, W.; Fry, S.C.; Schwab, W. Higher Expression of the Strawberry Xyloglucan Endotransglucosylase/Hydrolase Genes FvXTH9 and FvXTH6 Accelerates Fruit Ripening. Plant J. 2019, 100, 1237–1253. [Google Scholar] [CrossRef]
- Zhao, S.; Kang, Y.; Lin, Y.; Zheng, X.; Wu, Y.; Yang, Z. A Genome-Wide Identification and Expression Analysis of the Xyloglucan Endotransglucosylase/Hydrolase Gene Family in Melon (Cucumis melo L.). Horticulturae 2024, 10, 1017. [Google Scholar] [CrossRef]
- Van Sandt, V.S.; Guisez, Y.; Verbelen, J.P.; Vissenberg, K. Analysis of a Xyloglucan Endotransglycosylase/Hydrolase (XTH) from the Lycopodiophyte Selaginella Kraussiana Suggests That XTH Sequence Characteristics and Function Are Highly Conserved During the Evolution of Vascular Plants. J. Exp. Bot. 2006, 57, 2909–2922. [Google Scholar] [CrossRef]
- Bourquin, V.; Nishikubo, N.; Abe, H.; Brumer, H.; Denman, S.; Eklund, M.; Christiernin, M.; Teeri, T.T.; Sundberg, B.; Mellerowicz, E.J. Xyloglucan Endotransglycosylases Have a Function During the Formation of Secondary Cell Walls of Vascular Tissues. Plant Cell 2002, 14, 3073–3088. [Google Scholar] [CrossRef]
- Seale, M. Cell Wall Remodeling During Wood Development. Plant Physiol. 2020, 182, 1800–1801. [Google Scholar] [CrossRef]
- Baumann, M.J.; Eklöf, J.M.; Michel, G.; Kallas, A.M.; Teeri, T.T.; Czjzek, M.; Brumer, H., 3rd. Structural Evidence for the Evolution of Xyloglucanase Activity from Xyloglucan Endo-Transglycosylases: Biological Implications for Cell Wall Metabolism. Plant Cell 2007, 19, 1947–1963. [Google Scholar] [CrossRef]
- Park, Y.B.; Cosgrove, D.J. Xyloglucan and its Interactions with Other Components of the Growing Cell Wall. Plant Cell Physiol. 2015, 56, 180–194. [Google Scholar] [CrossRef] [PubMed]
- Cosgrove, D.J. Wall structure and wall loosening. A look backwards and forwards. Plant Physiol. 2001, 125, 131–134. [Google Scholar] [CrossRef] [PubMed]
- Takahisa, H.; Rumi, K. Functions of Xyloglucan in Plant Cells. Mol. Plant 2011, 4, 17–24. [Google Scholar]
- Genovesi, V.; Fornalé, S.; Fry, S.C.; Ruel, K.; Ferrer, P.; Encina, A.; Sonbol, F.M.; Bosch, J.; Puigdomènech, P.; Rigau, J.; et al. Zmxth1, a New Xyloglucan Endotransglucosylase/Hydrolase in Maize, Affects Cell Wall Structure and Composition in Arabidopsis thaliana. J. Exp. Bot. 2008, 59, 875–889. [Google Scholar] [CrossRef]
- Oh, M.-H.; Romanow, W.G.; Smith, R.C.; Zamski, E.; Sasse, J.; Clouse, S.D. Soybean Bru1 Encodes a Functional Xyloglucan Endotransglycosylase That Is Highly Expressed in Inner Epicotyl Tissues During Brassinosteroid-Promoted Elongation. Plant Cell Physiol. 1998, 39, 124–130. [Google Scholar] [CrossRef]
- Kaewthai, N.; Gendre, D.; Eklöf, J.M.; Ibatullin, F.M.; Ezcurra, I.; Bhalerao, R.P.; Brumer, H. Group III-a XTH Genes of Arabidopsis Encode Predominant Xyloglucan Endohydrolases That Are Dispensable for Normal Growth. Plant Physiol. 2013, 161, 440–454. [Google Scholar] [CrossRef]

| Gene Name | Gene ID | CDS (bp) | Protein (aa) | Exon | pI | Basic Proteins (%) | MW (Da) | Sub-Loc |
|---|---|---|---|---|---|---|---|---|
| CilXTH1 | Cil_01G_00632V2 | 1197 | 398 | 6 | 9.26 | 13.57 | 45,652.15 | CW/C |
| CilXTH2 | Cil_01G_00640V2 | 297 | 98 | 1 | 10.02 | 19.39 | 11,506.32 | CW |
| CilXTH3 | Cil_02G_00706V2 | 1056 | 351 | 4 | 5.68 | 12.54 | 40,021.45 | CW |
| CilXTH4 | Cil_03G_00077V2 | 939 | 312 | 5 | 8.92 | 14.10 | 36,247.32 | CW/C |
| CilXTH5 | Cil_03G_00411V2 | 828 | 275 | 4 | 9.07 | 13.45 | 31,919.27 | CW |
| CilXTH6 | Cil_03G_01067V2 | 1191 | 395 | 6 | 6.18 | 14.65 | 45,167.80 | CW |
| CilXTH7 | Cil_05G_00828V2 | 1011 | 336 | 4 | 8.98 | 13.99 | 38,764.06 | CW |
| CilXTH8 | Cil_06G_01204V2 | 882 | 293 | 5 | 9.36 | 15.70 | 33,792.43 | CW |
| CilXTH9 | Cil_07G_00376V2 | 873 | 290 | 3 | 8.80 | 11.38 | 32,412.66 | CW/C |
| CilXTH10 | Cil_07G_01585V2 | 912 | 303 | 5 | 8.62 | 14.52 | 34,514.17 | CW |
| CilXTH11 | Cil_08G_01020V2 | 834 | 277 | 4 | 6.29 | 10.47 | 30,791.31 | CW |
| CilXTH12 | Cil_09G_02459V2 | 858 | 285 | 3 | 9.36 | 12.28 | 32,268.65 | CW/C |
| CilXTH13 | Cil_10G_01144V2 | 795 | 264 | 4 | 5.16 | 11.74 | 30,541.09 | CW |
| CilXTH14 | Cil_10G_01312V2 | 981 | 326 | 4 | 6.00 | 12.27 | 37,277.34 | CW |
| CilXTH15 | Cil_11G_00212V2 | 1353 | 450 | 8 | 9.78 | 15.11 | 50,254.36 | CW |
| CilXTH16 | Cil_11G_00890V2 | 855 | 293 | 4 | 8.16 | 12.97 | 34,115.55 | CW/C |
| CilXTH17 | Cil_11G_01428V2 | 1176 | 391 | 6 | 8.90 | 14.83 | 44,026.61 | CW |
| CilXTH18 | Cil_11G_01492V2 | 1005 | 334 | 4 | 6.86 | 14.67 | 38,381.49 | CW |
| CilXTH19 | Cil_12G_00855V2 | 930 | 309 | 3 | 5.09 | 8.41 | 34,160.98 | CW/C |
| CilXTH20 | Cil_12G_00856V2 | 366 | 121 | 3 | 6.06 | 9.92 | 13,971.90 | CW/C |
| CilXTH21 | Cil_12G_00857V2 | 993 | 330 | 3 | 6.07 | 9.39 | 36,838.15 | CW/C |
| CilXTH22 | Cil_13G_01077V2 | 1041 | 346 | 4 | 9.25 | 14.74 | 39,987.48 | CW |
| CilXTH23 | Cil_13G_01231V2 | 786 | 261 | 3 | 7.74 | 10.34 | 28,932.94 | CW/C |
| CilXTH24 | Cil_14G_00900V2 | 879 | 292 | 4 | 4.67 | 10.27 | 34,105.96 | CW |
| CilXTH25 | Cil_14G_01287V2 | 876 | 291 | 3 | 7.11 | 12.71 | 33,539.68 | CW/C |
| CilXTH26 | Cil_14G_01719V2 | 852 | 283 | 4 | 9.24 | 14.13 | 32,416.48 | CW |
| CilXTH27 | Cil_15G_00375V2 | 858 | 285 | 4 | 6.70 | 12.28 | 32,546.05 | CW |
| CilXTH28 | Cil_15G_01868V2 | 804 | 267 | 4 | 4.66 | 7.87 | 30,032.17 | CW/C |
| CilXTH29 | Cil_15G_01869V2 | 864 | 287 | 4 | 8.76 | 9.41 | 31,723.85 | CW |
| CilXTH30 | Cil_15G_01870V2 | 1218 | 345 | 10 | 8.90 | 12.84 | 44,668.86 | CW |
| CilXTH31 | Cil_15G_01871V2 | 681 | 226 | 4 | 4.60 | 8.53 | 25,207.82 | CW |
| CilXTH32 | Cil_15G_01872V2 | 882 | 293 | 3 | 5.01 | 9.43 | 32,463.03 | CW/C |
| CilXTH33 | Cil_15G_01873V2 | 893 | 297 | 3 | 5.94 | 8.97 | 34,074.27 | CW/C |
| CilXTH34 | Cil_15G_01963V2 | 873 | 290 | 3 | 5.93 | 10.56 | 32,377.34 | CW/C |
| CilXTH35 | Cil_16G_00439V2 | 1128 | 375 | 5 | 9.38 | 10.33 | 42,085.19 | CW/CP |
| CilXTH36 | Cil_16G_00440V2 | 1281 | 426 | 10 | 5.41 | 7.08 | 48,044.13 | CW/CP |
| CilXTH37 | Cil_16G_00824V2 | 901 | 300 | 4 | 5.71 | 13.87 | 33,734.39 | CW |
| CilXTH38 | Cil_73S_00004V2 | 1779 | 592 | 8 | 7.03 | 14.19 | 66,148.33 | CW |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wen, M.; Zhou, Z.; Sun, J.; Meng, F.; Xi, X.; Liu, A.; Yu, A. A Genome-Wide Characterization of the Xyloglucan Endotransglucosylase/Hydrolase Family Genes and Their Functions in the Shell Formation of Pecan. Horticulturae 2025, 11, 609. https://doi.org/10.3390/horticulturae11060609
Wen M, Zhou Z, Sun J, Meng F, Xi X, Liu A, Yu A. A Genome-Wide Characterization of the Xyloglucan Endotransglucosylase/Hydrolase Family Genes and Their Functions in the Shell Formation of Pecan. Horticulturae. 2025; 11(6):609. https://doi.org/10.3390/horticulturae11060609
Chicago/Turabian StyleWen, Mengyun, Zekun Zhou, Jing Sun, Fanqing Meng, Xueliang Xi, Aizhong Liu, and Anmin Yu. 2025. "A Genome-Wide Characterization of the Xyloglucan Endotransglucosylase/Hydrolase Family Genes and Their Functions in the Shell Formation of Pecan" Horticulturae 11, no. 6: 609. https://doi.org/10.3390/horticulturae11060609
APA StyleWen, M., Zhou, Z., Sun, J., Meng, F., Xi, X., Liu, A., & Yu, A. (2025). A Genome-Wide Characterization of the Xyloglucan Endotransglucosylase/Hydrolase Family Genes and Their Functions in the Shell Formation of Pecan. Horticulturae, 11(6), 609. https://doi.org/10.3390/horticulturae11060609







