Characterization of SPL Family Genes in Banana Reveals the Critical Role of MaSPL1 in Regulating Fruit Ripening
Abstract
1. Introduction
2. Materials and Methods
2.1. SPL Transcription Factors in M. acuminata DH Pahang
2.2. Gene Structure and Phylogenetic Analysis
2.3. Synteny Analysis
2.4. Cis-Acting Element and Collinearity Analysis
2.5. Expression Profiles of MaSPL Genes in Different Tissues
2.6. Transient Overexpression in Banana Fruits
2.7. RNA Extraction and RT-qPCR Analysis
3. Results
3.1. Identification of MaSPL Gene Family Members in M. acuminata DH Pahang
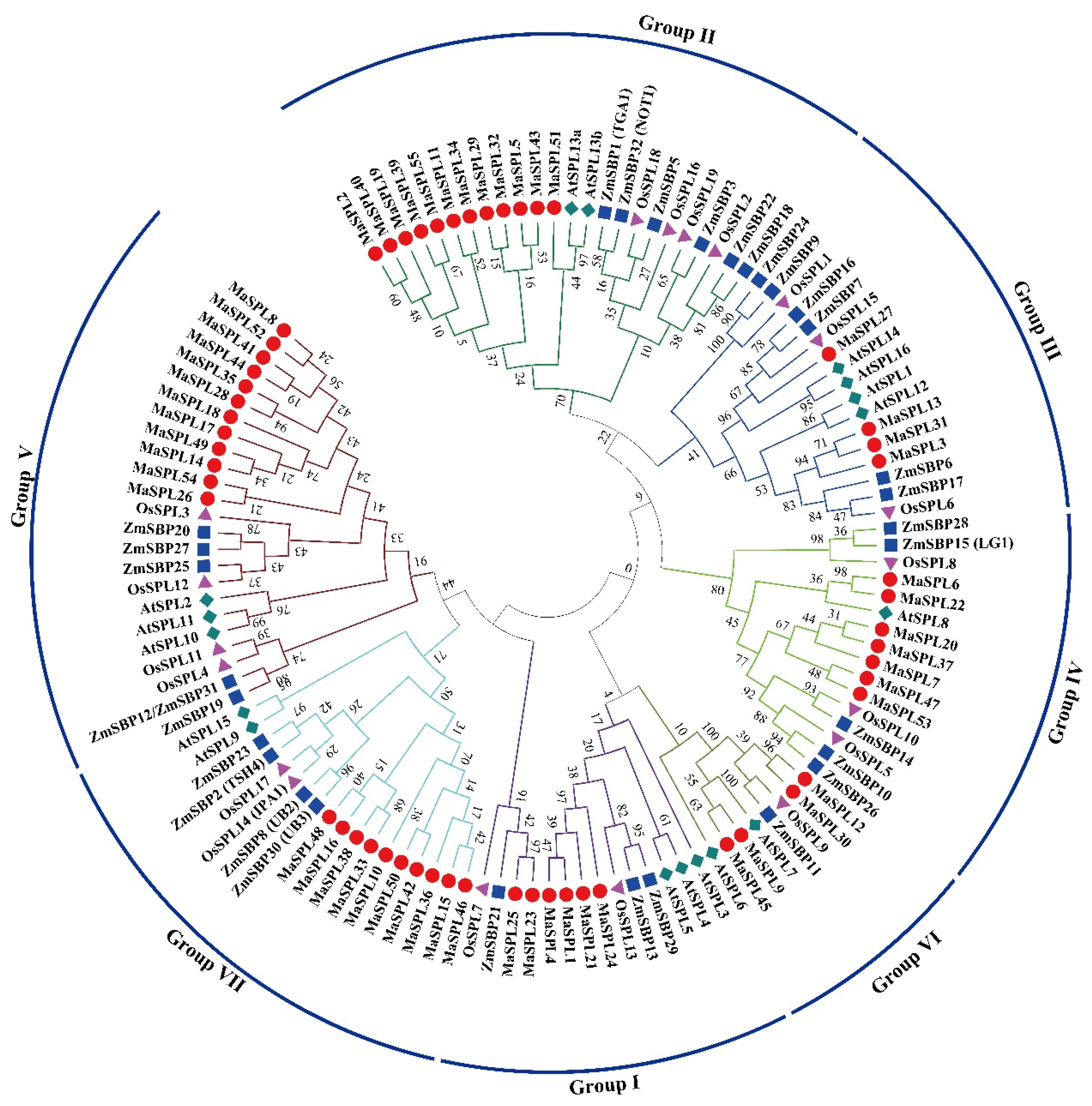
3.2. Phylogenetic Tree of M. acuminata DH Pahang, A. thaliana, O. sativa, and Z. mays SPL Proteins
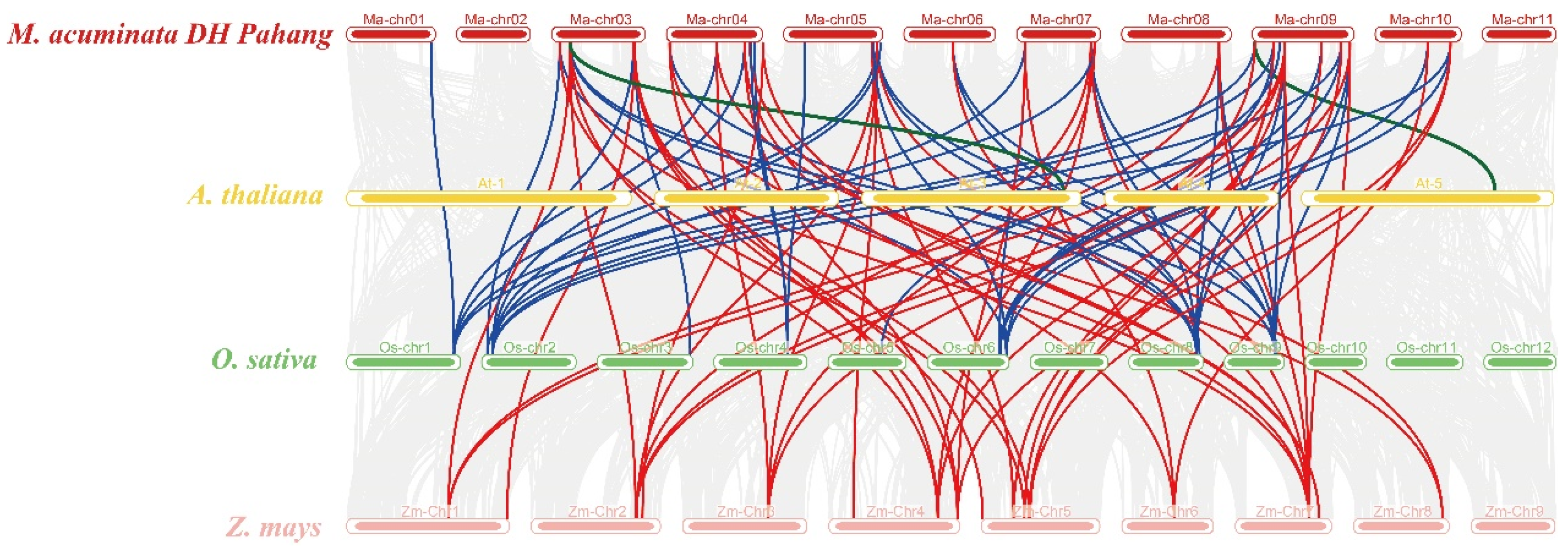
3.3. The Chromosomal Distribution, Duplication Events, and Syntenic Analysis of the Maspl Genes
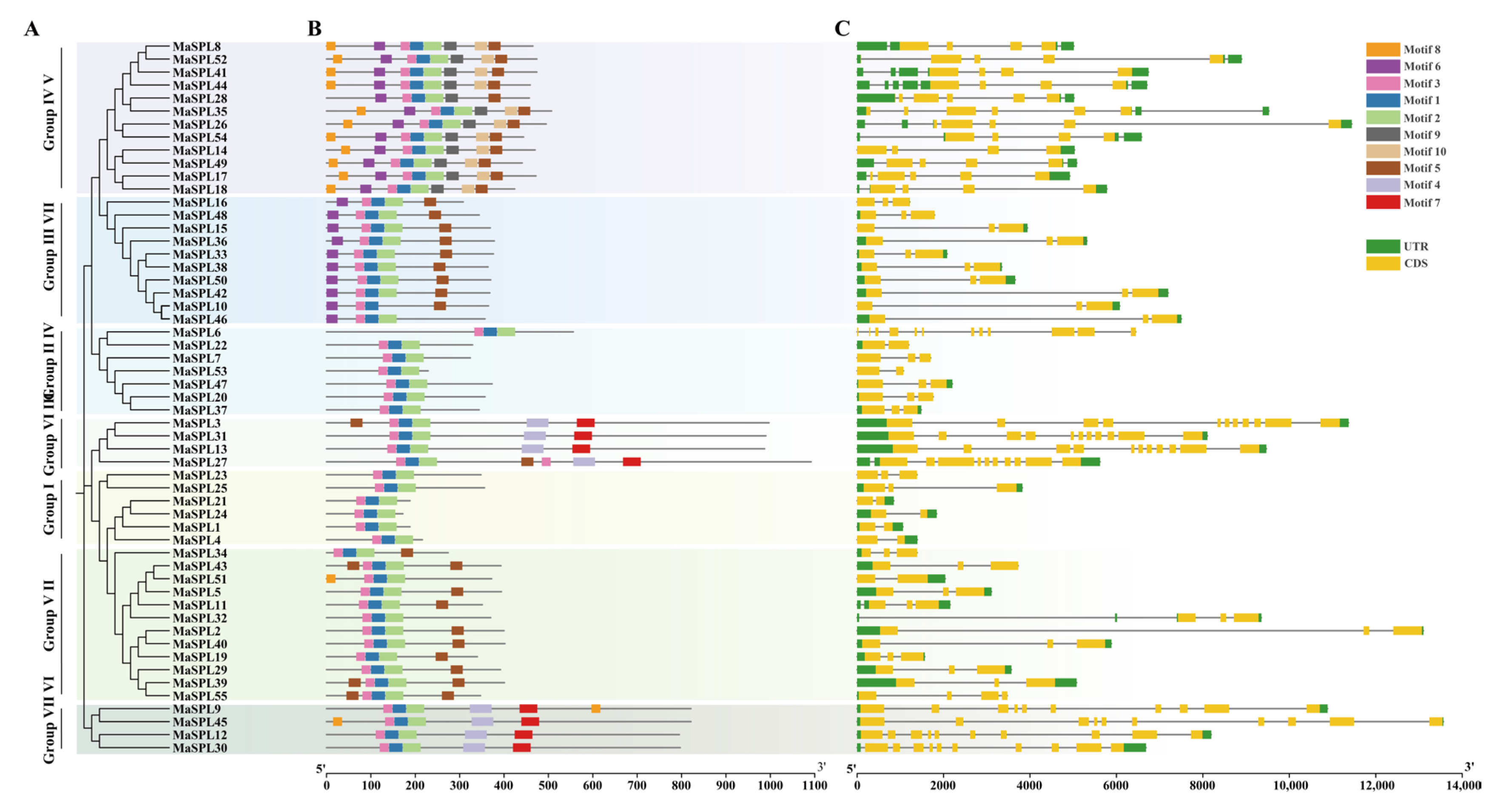
3.4. Phylogenetic Analysis, Conserved Motifs, and Gene Structure Analysis of MaSPLs in M. acuminata DH Pahang
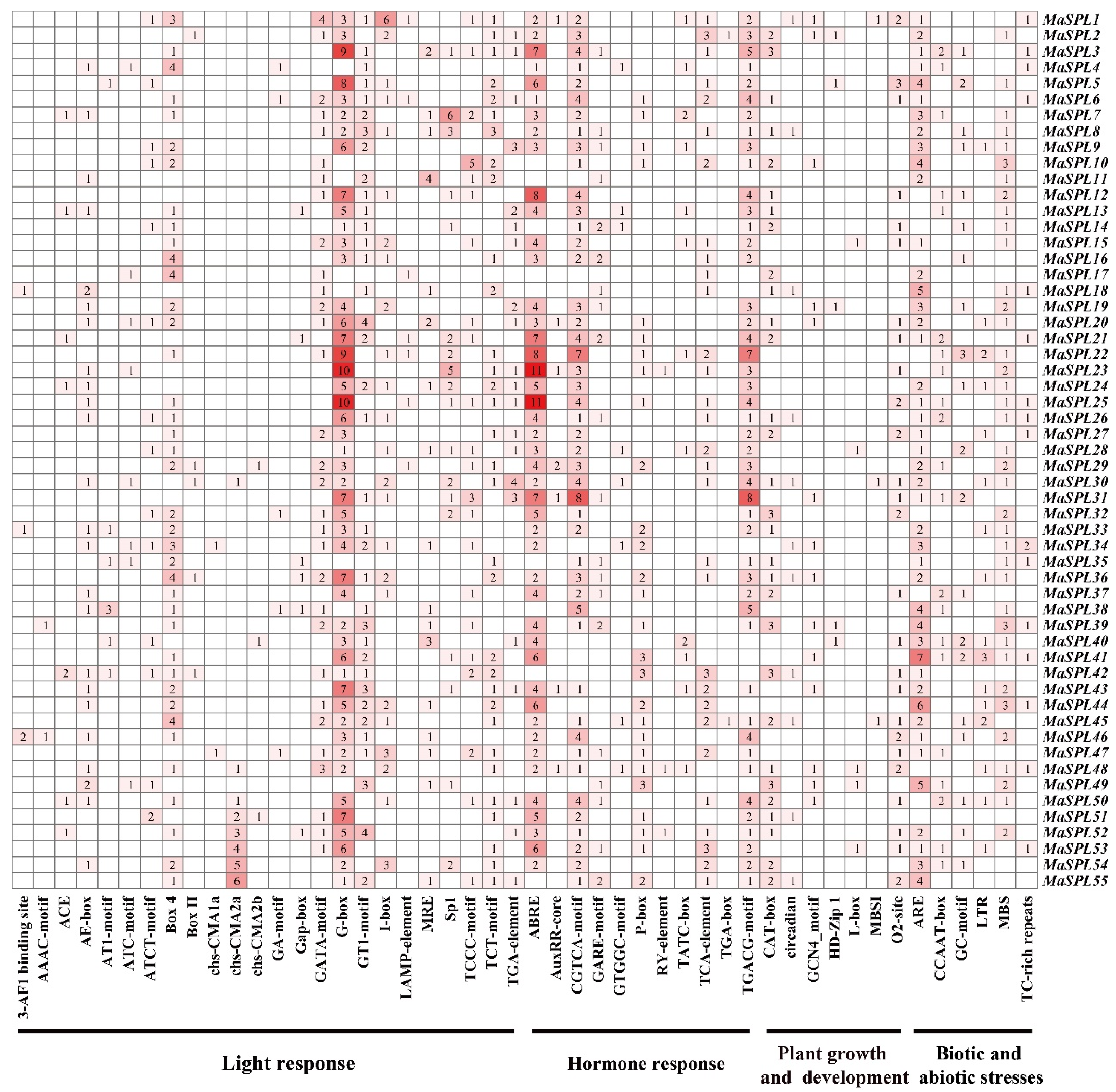
3.5. Analysis of Cis-Acting Elements in the Promoter Regions of MaSPL Genes in M. acuminata DH Pahang
3.6. Expression Profile of MaSPL Genes in Different Tissues
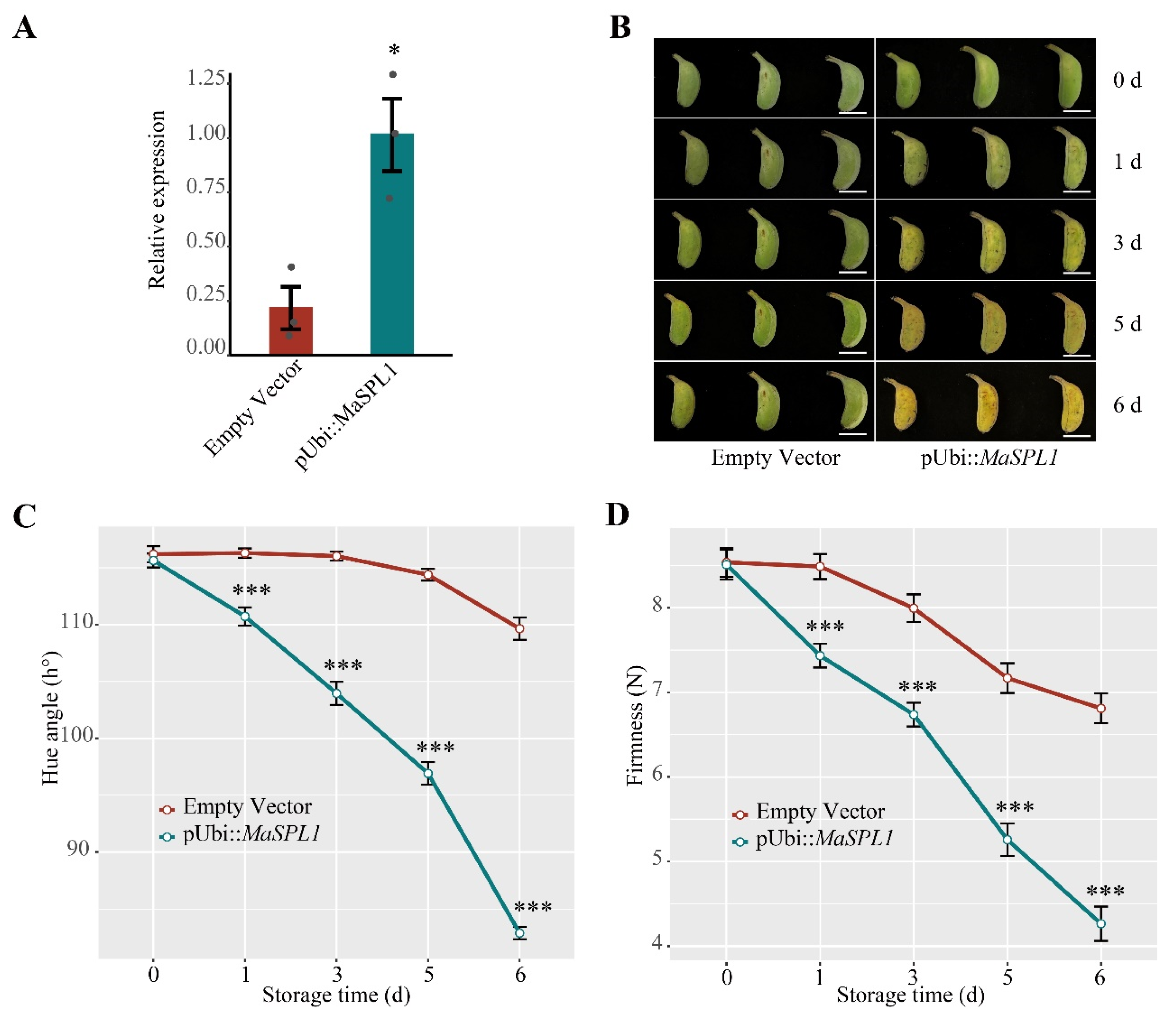
3.7. Transient Overexpression of MaSPL1 in Banana Fruits Accelerates the Fruit Ripening
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Klein, J.; Saedler, H.; Huijser, P. A new family of DNA binding proteins includes putative transcriptional regulators of theAntirrhinum majus floral meristem identity geneSQUAMOSA. Mol. Genet. Genom. 1996, 250, 7–16. [Google Scholar] [CrossRef] [PubMed]
- Yamasaki, K.; Kigawa, T.; Inoue, M.; Tateno, M.; Yamasaki, T.; Yabuki, T.; Aoki, M.; Seki, E.; Matsuda, T.; Nunokawa, E.; et al. A Novel zinc-binding motif revealed by solution structures of DNA-binding domains of arabidopsis SBP-family transcription factors. J. Mol. Biol. 2004, 337, 49–63. [Google Scholar] [CrossRef]
- Birkenbihl, R.P.; Jach, G.; Saedler, H.; Huijser, P. Functional dissection of the plant-specific SBP-domain: Overlap of the DNA-binding and nuclear localization domains. J. Mol. Biol. 2005, 352, 585–596. [Google Scholar] [CrossRef]
- Huijser, P.; Klein, J.; Lönnig, W.; Meijer, H.; Saedler, H.; Sommer, H. Bracteomania, an inflorescence anomaly, is caused by the loss of function of the MADS-box gene squamosa in Antirrhinum majus. EMBO J. 1992, 11, 1239–1249. [Google Scholar] [CrossRef]
- Cardon, G.; Höhmann, S.; Klein, J.; Nettesheim, K.; Saedler, H.; Huijser, P. Molecular characterisation of the Arabidopsis SBP-box genes. Gene 1999, 237, 91–104. [Google Scholar] [CrossRef]
- Cai, C.; Guo, W.; Zhang, B. Genome-wide identification and characterization of SPL transcription factor family and their evolution and expression profiling analysis in cotton. Sci. Rep. 2018, 8, 1–13. [Google Scholar] [CrossRef]
- Tripathi, R.K.; Goel, R.; Kumari, S.; Dahuja, A. Genomic organization, phylogenetic comparison, and expression profiles of the SPL family genes and their regulation in soybean. Dev. Genes Evol. 2017, 227, 101–119. [Google Scholar] [CrossRef]
- Lu, Y.G.; Ouyang, B.; Zhang, J.H.; Wang, T.T.; Lu, C.; Han, Q.Q.; Zhao, S.N.; Ye, Z.B.; Li, H.X. Genomic organization, phy-logenetic comparison and expression profiles of annexin gene family in tomato (Solanum lycopersicum). Gene 2012, 499, 14–24. [Google Scholar] [CrossRef]
- Song, J.; Ma, D.; Yin, J.; Yang, L.; He, Y.; Zhu, Z.; Tong, H.; Chen, L.; Zhu, G.; Liu, Y.; et al. Genome-wide characterization and expression profiling of squamosa promoter binding protein-like (sbp) transcription factors in wheat (Triticum aestivum L.). Agronomy 2019, 9, 527. [Google Scholar] [CrossRef]
- Xie, K.B.; Wu, C.Q.; Xiong, L.Z. Genomic organization, differential expression, and interaction of SQUAMOSA promoter-binding-like transcription factors and microRNA156 in rice. Plant Physiol. 2006, 142, 280–293. [Google Scholar] [CrossRef]
- Wei, H.; Zhao, Y.; Xie, Y.; Wang, H. Exploiting SPL genes to improve maize plant architecture tailored for high-density planting. J. Exp. Bot. 2018, 69, 4675–4688. [Google Scholar] [CrossRef] [PubMed]
- Shao, Y.; Zhou, H.-Z.; Wu, Y.; Zhang, H.; Lin, J.; Jiang, X.; He, Q.; Zhu, J.; Li, Y.; Yu, H.; et al. OsSPL3, an SBP-domain protein, regulates crown root development in rice. Plant Cell 2019, 31, 1257–1275. [Google Scholar] [CrossRef]
- Kong, D.; Pan, X.; Jing, Y.; Zhao, Y.; Duan, Y.; Yang, J.; Wang, B.; Liu, Y.; Shen, R.; Cao, Y.; et al. ZmSPL10/14/26 are required for epidermal hair cell fate specification on maize leaf. New Phytol. 2021, 230, 1533–1549. [Google Scholar] [CrossRef]
- Unte, U.S.; Sorensen, A.-M.; Pesaresi, P.; Gandikota, M.; Leister, D.; Saedler, H.; Huijser, P. SPL8, an SBP-Box gene that affects pollen SAC development in Arabidopsis. Plant Cell 2003, 15, 1009–1019. [Google Scholar] [CrossRef]
- Yang, R.; Li, P.; Mei, H.; Wang, D.; Sun, J.; Yang, C.; Hao, L.; Cao, S.; Chu, C.; Hu, S.; et al. Fine-tuning of MiR528 accumulation modulates flowering time in rice. Mol. Plant 2019, 12, 1103–1113. [Google Scholar] [CrossRef]
- Yang, J.; Wei, H.; Hou, M.; Chen, L.; Zou, T.; Ding, H.; Jing, Y.; Zhang, X.; Zhao, Y.; Liu, Q.; et al. ZmSPL13 and ZmSPL29 act together to promote vegetative and reproductive transition in maize. New Phytol. 2023, 239, 1505–1520. [Google Scholar] [CrossRef]
- Yamaguchi, A.; Wu, M.-F.; Yang, L.; Wu, G.; Poethig, R.S.; Wagner, D. The MicroRNA-regulated SBP-Box transcription factor SPL3 is a direct upstream activator of LEAFY, FRUITFULL, and APETALA1. Dev. Cell 2009, 17, 268–278. [Google Scholar] [CrossRef]
- Wang, L.; Ming, L.C.; Liao, K.Y.; Xia, C.J.; Sun, S.Y.; Chang, Y.; Wang, H.K.; Fu, D.B.; Xu, C.H.; Wang, Z.J.; et al. Bract suppression regulated by the miR156/529-SPLs-NL1-PLA1 module is required for the transition from vegetative to reproductive branching in rice. Mol. Plant. 2021, 14, 1168–1184. [Google Scholar] [CrossRef]
- Ishii, T.; Numaguchi, K.; Miura, K.; Yoshida, K.; Thanh, P.T.; Htun, T.M.; Yamasaki, M.; Komeda, N.; Matsumoto, T.; Terauchi, R.; et al. OsLG1 regulates a closed panicle trait in domesticated rice. Nat. Genet. 2013, 45, 462–465. [Google Scholar] [CrossRef]
- Jiao, Y.; Wang, Y.; Xue, D.; Wang, J.; Yan, M.; Liu, G.; Dong, G.; Zeng, D.; Lu, Z.; Zhu, X.; et al. Regulation of OsSPL14 by OsmiR156 defines ideal plant architecture in rice. Nat. Genet. 2010, 42, 541–544. [Google Scholar] [CrossRef]
- Miura, K.; Ikeda, M.; Matsubara, A.; Song, X.-J.; Ito, M.; Asano, K.; Matsuoka, M.; Kitano, H.; Ashikari, M. OsSPL14 promotes panicle branching and higher grain productivity in rice. Nat. Genet. 2010, 42, 545–549. [Google Scholar] [CrossRef] [PubMed]
- Kong, D.; Li, C.; Xue, W.; Wei, H.; Ding, H.; Hu, G.; Zhang, X.; Zhang, G.; Zou, T.; Xian, Y.; et al. UB2/UB3/TSH4-anchored transcriptional networks regulate early maize inflorescence development in response to simulated shade. Plant Cell 2022, 35, 717–737. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Wu, G.; Zhao, Y.; Wang, H.H.; Dai, Z.; Xue, W.; Yang, J.; Wei, H.; Shen, R.; Wang, H. DWARF53 interacts with transcription factors UB2/UB3/TSH4 to regulate maize tillering and tassel branching. Plant Physiol. 2021, 187, 947–962. [Google Scholar] [CrossRef]
- Zhao, B.; Xu, M.; Zhao, Y.; Li, Y.; Xu, H.; Li, C.; Kong, D.; Xie, Y.; Zheng, Z.; Wang, B.; et al. Overexpression of ZmSPL12 confers enhanced lodging resistance through transcriptional regulation of D1 in maize. Plant Biotechnol. J. 2022, 20, 622–624. [Google Scholar] [CrossRef]
- Wang, J.; Zhou, L.; Shi, H.; Chern, M.; Yu, H.; Yi, H.; He, M.; Yin, J.; Zhu, X.; Li, Y.; et al. A single transcription factor promotes both yield and immunity in rice. Science 2018, 361, 1026–1028. [Google Scholar] [CrossRef]
- Zhang, X.-F.; Yang, C.-Y.; Lin, H.-X.; Wang, J.-W.; Xue, H.-W. Rice SPL12 coevolved with GW5 to determine grain shape. Sci. Bull. 2021, 66, 2353–2357. [Google Scholar] [CrossRef]
- Song, X.; Meng, X.; Guo, H.; Cheng, Q.; Jing, Y.; Chen, M.; Liu, G.; Wang, B.; Wang, Y.; Li, J.; et al. Targeting a gene regulatory element enhances rice grain yield by decoupling panicle number and size. Nat. Biotechnol. 2022, 40, 1403–1411. [Google Scholar] [CrossRef]
- Wang, S.; Li, S.; Liu, Q.; Wu, K.; Zhang, J.; Wang, S.; Wang, Y.; Chen, X.; Zhang, Y.; Gao, C.; et al. The OsSPL16-GW7 regulatory module determines grain shape and simultaneously improves rice yield and grain quality. Nat. Genet. 2015, 47, 949–954. [Google Scholar] [CrossRef]
- Wang, S.; Wu, K.; Yuan, Q.; Liu, X.; Liu, Z.; Lin, X.; Zeng, R.; Zhu, H.; Dong, G.; Qian, Q.; et al. Control of grain size, shape and quality by OsSPL16 in rice. Nat. Genet. 2012, 44, 950–954. [Google Scholar] [CrossRef]
- Wei, W.; Yang, Y.-Y.; Wu, C.-J.; Kuang, J.-F.; Chen, J.-Y.; Shan, W. MaSPL16 positively regulates fruit ripening in bananas via the direct transcriptional induction of MaNAC029. Hortic. Adv. 2023, 1, 1–13. [Google Scholar] [CrossRef]
- Dorweiler, J.; Stec, A.; Kermicle, J.; Doebley, J. Teosinte glume architecture 1: A genetic locus controlling a key step in maize evolution. Science 1993, 262, 233–235. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Gou, Y.; Heng, Y.; Ding, W.; Li, Y.; Zhou, D.; Li, X.; Liang, C.; Wu, C.; Wang, H.; et al. Targeted manipulation of grain shape genes effectively improves outcrossing rate and hybrid seed production in rice. Plant Biotechnol. J. 2023, 21, 381–390. [Google Scholar] [CrossRef]
- Lai, T.; Wang, X.; Ye, B.; Jin, M.; Chen, W.; Wang, Y.; Zhou, Y.; Blanks, A.M.; Gu, M.; Zhang, P.; et al. Molecular and functional characterization of the SBP-box transcription factor SPL-CNR in tomato fruit ripening and cell death. J. Exp. Bot. 2020, 71, 2995–3011. [Google Scholar] [CrossRef]
- Obando-Ulloa, J.M.; Jowkar, M.; Moreno, E.; Souri, M.K.; Martínez, J.A.; Bueso, M.C.; Monforte, A.J.; Fernández-Trujillo, J.P. Discrimination of climacteric and non-climacteric melon fruit at harvest or at the senescence stage by quality traits. J. Sci. Food Agric. 2009, 89, 1743–1753. [Google Scholar] [CrossRef]
- Luo, Q.; Wei, W.; Yang, Y.-Y.; Wu, C.-J.; Chen, J.-Y.; Lu, W.-J.; Kuang, J.-F.; Shan, W. E3 ligase MaNIP1 degradation of NON-YELLOW COLORING1 at high temperature inhibits banana degreening. Plant Physiol. 2023, 192, 1969–1981. [Google Scholar] [CrossRef]
- Si, J.; Fan, Z.Q.; Wu, C.J.; Yang, Y.Y.; Shan, W.; Kuang, J.F.; Lu, W.J.; Wei, W.; Chen, J.Y. MaHsf24, a novel negative modulator, regulates cold tolerance in banana fruits by repressing the expression of HSPs and antioxidant enzyme genes. Plant Biotechnol. J. 2024, 22, 2873–2886. [Google Scholar] [CrossRef]
- Wu, C.; Deng, W.; Shan, W.; Liu, X.; Zhu, L.; Cai, D.; Wei, W.; Yang, Y.; Chen, J.; Lu, W.; et al. Banana MKK1 modulates fruit ripening via the MKK1-MPK6-3/11-4-bZIP21 module. Cell Rep. 2023, 42, 112832. [Google Scholar] [CrossRef]
- Fan, Z.; Ba, L.; Shan, W.; Xiao, Y.; Lu, W.; Kuang, J.; Chen, J. A banana R2R3-MYB transcription factor MaMYB3 is involved in fruit ripening through modulation of starch degradation by repressing starch degradation-related genes and MabHLH6. Plant J. 2018, 96, 1191–1205. [Google Scholar] [CrossRef]
- Liu, F.; Dou, T.; Hu, C.; Zhong, Q.; Sheng, O.; Yang, Q.; Deng, G.; He, W.; Gao, H.; Li, C.; et al. WRKY transcription factor MaWRKY49 positively regulates pectate lyase genes during fruit ripening of Musa acuminata. Plant Physiol. Biochem. 2023, 194, 643–650. [Google Scholar] [CrossRef]
- Wu, C.J.; Cai, D.L.; Li, J.; Lin, Z.X.; Wei, W.; Shan, W.; Chen, J.Y.; Lu, W.J.; Su, X.G.; Kuang, J.F. Banana MabHLH28 positively regulates the expression of softening-related genes to mediate fruit ripening independently or via cooperating with MaWRKY49/111. Hortic. Res. 2024, 11, uhae053. [Google Scholar] [CrossRef]
- Wei, W.; Yang, Y.-Y.; Wu, C.-J.; Kuang, J.-F.; Lu, W.-J.; Chen, J.-Y.; Shan, W. MaNAC19–MaXB3 regulatory module mediates sucrose synthesis in banana fruit during ripening. Int. J. Biol. Macromol. 2023, 253, 127144. [Google Scholar] [CrossRef] [PubMed]
- Wei, W.; Yang, Y.-Y.; Chen, J.-Y.; Lakshmanan, P.; Kuang, J.-F.; Lu, W.-J.; Shan, W. MaNAC029 modulates ethylene biosynthesis and fruit quality and undergoes MaXB3-mediated proteasomal degradation during banana ripening. J. Adv. Res. 2022, 53, 33–47. [Google Scholar] [CrossRef] [PubMed]
- Wei, W.; Yang, Y.-Y.; Wu, C.-J.; Kuang, J.-F.; Chen, J.-Y.; Lu, W.-J.; Shan, W. MaMADS1–MaNAC083 transcriptional regulatory cascade regulates ethylene biosynthesis during banana fruit ripening. Hortic. Res. 2023, 10, uhad177. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Sun, X.L.; He, J.; Shan, J.J.; Sheng, O.; Dou, T.X.; Li, Y.Y.; He, W.D.; Yang, Q.S.; Hu, C.H.; et al. Membrane-associated NAC transcription factor MaNAC169 is a positive regulator during banana fruit ripening. Postharvest Biol. Tec. 2025, 223, 113451. [Google Scholar] [CrossRef]
- Fu, M.; Zheng, Y.; Zhang, J.; Deng, C.; Zhang, J.; Jia, C.; Miao, H.; Wang, J.; Zheng, S.; Jin, Z.; et al. MaEIL4-MaMADS36-MaACS7 module transcriptionally regulates ethylene biosynthesis during banana fruit ripening. Hortic. Res. 2024, 12, uhae345. [Google Scholar] [CrossRef]
- Li, H.; Chen, Z.; Zhu, W.; Ni, X.; Wang, J.; Fu, L.; Chen, J.; Li, T.; Tang, L.; Yang, Y.; et al. The MaNAP1-MaMADS1 transcription factor module mediates ethylene-regulated peel softening and ripening in banana. Plant Cell 2024, 37. [Google Scholar] [CrossRef]
- Quevillon, E.; Silventoinen, V.; Pillai, S.; Harte, N.; Mulder, N.; Apweiler, R.; Lopez, R. InterProScan: Protein domains identi-fier. Nucleic Acids Res. 2005, 33, W116–W120. [Google Scholar] [CrossRef]
- Wang, J.Y.; Chitsaz, F.; Derbyshire, M.K.; Gonzales, N.R.; Gwadz, M.; Lu, S.N.; Marchler, G.H.; Song, J.S.; Thanki, N.; Yam-ashita, R.A.; et al. The conserved domain database in 2023. Nucleic Acids Res. 2023, 51, D384–D388. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. 20 years of the SMART protein domain annotation resource. Nucleic Acids Res. 2018, 46, D493–D496. [Google Scholar] [CrossRef]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, w202–w208. [Google Scholar] [CrossRef]
- Chen, C.J.; Wu, Y.; Li, J.W.; Wang, X.; Zeng, Z.H.; Xu, J.; Liu, Y.L.; Feng, J.T.; Chen, H.; He, Y.H.; et al. TBtools-II: A “one for all, all for one” bioinformatics platform for biological big-data mining. Mol. Plant. 2023, 16, 1733–1742. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; DeBarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.-H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef] [PubMed]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Wang, Z.; Miao, H.; Liu, J.; Xu, B.; Yao, X.; Xu, C.; Zhao, S.; Fang, X.; Jia, C.; Wang, J.; et al. Musa balbisiana genome reveals subgenome evolution and functional divergence. Nat. Plants 2019, 5, 810–821. [Google Scholar] [CrossRef]
- Chen, L.; Zhong, H.-Y.; Kuang, J.-F.; Li, J.-G.; Lu, W.-J.; Chen, J.-Y. Validation of reference genes for RT-qPCR studies of gene expression in banana fruit under different experimental conditions. Planta 2011, 234, 377–390. [Google Scholar] [CrossRef]
- Wang, H.; Wang, H.Y. The miR156/SPL module, a regulatory hub and versatile toolbox, gears up crops for enhanced agronomic traits. Mol. Plant. 2015, 8, 677–688. [Google Scholar] [CrossRef]
- Tian, J.; Wang, C.; Xia, J.; Wu, L.; Xu, G.; Wu, W.; Li, D.; Qin, W.; Han, X.; Chen, Q.; et al. Teosinte ligule allele narrows plant architecture and enhances high-density maize yields. Science 2019, 365, 658–664. [Google Scholar] [CrossRef]
- Hu, C.; Liu, F.; Sheng, O.; Yang, Q.; Dou, T.; Dong, T.; Li, C.; Gao, H.; He, W.; Liu, S.; et al. Efficient and transgene-free genome editing in banana using a REG-2 promoter–driven gene-deletion system. Mol. Hortic. 2023, 3, 1–4. [Google Scholar] [CrossRef]
- Cheng, C.; Wu, S.; Deng, G.; Sheng, O.; Yi, G.; Yang, Q. Recent advances and future directions in banana molecular biology and breeding. Mol. Hortic. 2024, 4, 1–25. [Google Scholar] [CrossRef]
- Li, Y.; Huang, W.; Gao, H.; Yi, G.; Yan, S. Regulation of starch metabolism in banana fruit: Mechanisms shaping the nutritional quality. Curr. Opin. Plant Biol. 2025, 84, 102698. [Google Scholar] [CrossRef]
- Miao, H.; Zhang, J.; Zheng, Y.; Jia, C.; Hu, Y.; Wang, J.; Zhang, J.; Sun, P.; Jin, Z.; Zhou, Y.; et al. Shaping the future of bananas: Advancing genetic trait regulation and breeding in the postgenomics era. Hortic. Res. 2025, 12, uhaf044. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, B.; Dou, T.; Sheng, O.; He, W.; Deng, G.; Bi, F.; Li, C.; Dong, T.; Yang, Q.; Hu, C.; et al. Characterization of SPL Family Genes in Banana Reveals the Critical Role of MaSPL1 in Regulating Fruit Ripening. Horticulturae 2025, 11, 576. https://doi.org/10.3390/horticulturae11060576
Wang B, Dou T, Sheng O, He W, Deng G, Bi F, Li C, Dong T, Yang Q, Hu C, et al. Characterization of SPL Family Genes in Banana Reveals the Critical Role of MaSPL1 in Regulating Fruit Ripening. Horticulturae. 2025; 11(6):576. https://doi.org/10.3390/horticulturae11060576
Chicago/Turabian StyleWang, Bo, Tongxin Dou, Ou Sheng, Weidi He, Guiming Deng, Fangcheng Bi, Chunyu Li, Tao Dong, Qiaosong Yang, Chunhua Hu, and et al. 2025. "Characterization of SPL Family Genes in Banana Reveals the Critical Role of MaSPL1 in Regulating Fruit Ripening" Horticulturae 11, no. 6: 576. https://doi.org/10.3390/horticulturae11060576
APA StyleWang, B., Dou, T., Sheng, O., He, W., Deng, G., Bi, F., Li, C., Dong, T., Yang, Q., Hu, C., Gao, H., Liu, S., Liu, C., Liu, J., Yi, G., & Li, Y. (2025). Characterization of SPL Family Genes in Banana Reveals the Critical Role of MaSPL1 in Regulating Fruit Ripening. Horticulturae, 11(6), 576. https://doi.org/10.3390/horticulturae11060576






