Origin and Possible Members of the ‘Malvasia’ Family: The New Fuencaliente de La Palma Hypothesis on the True ‘Malvasia’
Abstract
1. Introduction
2. Materials and Methods
2.1. The Term ‘Malvasia’ in the Cultivar and Prime Names
2.2. SSR or Microsatellites Used
2.3. Data Analysis
3. Results
3.1. The Names of the Term ‘Malvasia’
3.2. Genetic Structure of the Population of the “Malvasia” Group Varieties (Genetic Strategy)
3.3. Distribution of the ‘Malvasia’ Variety Group Population According to Geographical Criteria
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Crespan, M.; Cabello, F.; Giannetto, S.; Ibáñez, J.; Kontic, J.K.; Maletic, E.; Peji’c, I.; Rodriguez-Torres, I.; Antonacci, D. Malvasia delle Lipari, Malvasia di Sardegna, Greco di Gerace, Malvasia de Sitges and Malvasia dubrovacka-synonyms of an old and famous grape cultivar. Vitis 2006, 45, 69–73. [Google Scholar] [CrossRef]
- Maul, E.; Röckel, R. Vitis International Variety Catalogue (VIVC): A cultivar database referenced by genetic profiles and morphology. In BIO Web of Conferences, Proceedings of the 38th World Congress of Vine and Wine, Mainz, Germany, 5–10 July 2015; EDP Sciences: Hulis, France, 2015; Volume 5, p. 01009. [Google Scholar] [CrossRef]
- Vitis International Variety Catalogue. Available online: http://www.vivc.de (accessed on 1 March 2025).
- Lacômbe, T.; Boursiquot, J.M.; Laucou, V.; Dechesne, F.; Varès, D.; This, P. Relationships and genetic diversity within the accessions related to Malvasia held in the Domaine de Vassal grape germplasm repository. Am. J. Enol. Vitic. 2007, 58, 124–131. [Google Scholar] [CrossRef]
- Galet, P. Dictionnaire Encylcopédique Des Cépages, 1st ed.; Hachette: Paris, France, 2000. [Google Scholar]
- Calò, A.; Costacurta, A.; Scienza, A. Vitigni d’Italia; Calderini Edagricole: Milano, Italy, 2001. [Google Scholar]
- Crespan, M. Exploration and Evaluation of Grapevine Biodiversity Using Molecular Markers. Mittellungen Klosterneuberg 2010, 60, 310–315. Available online: https://www.researchgate.net/publication/298851588_Exploration_and_evaluation_of_grapevine_biodiversity_using_molecular_markers (accessed on 3 March 2025).
- Dong, Y.; Duan, S.; Xia, Q.; Liang, Z.; Dong, X.; Margaryan, K.; Musayev, M.; Goryslavets, S.; Zdunić, G.; Bert, P.-F.; et al. Dual domestications and origin of traits in grapevine evolution. Science 2023, 379, 892–901. [Google Scholar] [CrossRef]
- Borrego, J.; de Andres, M.T.; Gomez, J.L.; Ibáñez, J. Genetic study of Malvasia and Torrontes groups through molecular markers. Am. J. Enol. Vitic. 2002, 53, 125–130. [Google Scholar] [CrossRef]
- Rodriguez-Torres, I.; Ibáñez, J.; De Andrés, M.T.; Rubio, C.; Borrego, J.; Cabello, F.; Zerolo, J.; Muñoz-Organero, G. Synonyms and homonyms of ‘Malvasía’ cultivars (Vitis vinifera L.) existing in Spain. Span. J. Agric. Res. 2009, 7, 563–571. [Google Scholar] [CrossRef]
- Thomas, M.R.; Scott, N.S. Microsatellite repeats in grapevine reveal DNA polymorphisms when analyzed as sequence-tagged sites (STSs). Theor. Appl. Genet. 1993, 86, 985–990. [Google Scholar] [CrossRef]
- Bowers, J.E.; Dangl, G.S.; Vignani, R.; Meredith, C.P. Isolation and characterization of new polymorphic simple sequence repeat loci in grape (Vitis vinifera L.). Genome 1996, 39, 628–633. [Google Scholar] [CrossRef]
- Bowers, J.E.; Dangl, G.S.; Meredith, C.P. Development and characterization of additional microsatellite DNA markers for grape. Am. J. Enol. Vitic. 1999, 50, 243–246. [Google Scholar] [CrossRef]
- Sefc, K.M.; Regner, F.; Turetschek, E.; Glössl, J.; Steinkellner, H. Identification of microsatellite sequences in Vitis riparia and their applicability for genotyping of different Vitis species. Genome 1999, 42, 367–373. [Google Scholar] [CrossRef]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef] [PubMed]
- Falush, D.; Stephens, M.; Pritchard, J.K. Inference of population structure using multilocus genotype data: Linked loci and correlated allele frequencies. Genetics 2003, 164, 1567–1587. [Google Scholar] [CrossRef] [PubMed]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software structure: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef] [PubMed]
- Peakall, R.; Smouse, P.E. GenAlEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research-an update. Bioinformatics 2012, 28, 2537–2539. [Google Scholar] [CrossRef] [PubMed]
- Paetkau, D.; Calvert, W.; Stirling, I.; Strobeck, C. Microsatellite analysis of population structure in Canadian polar bears. Mol. Ecol. 1995, 4, 347–354. [Google Scholar] [CrossRef]
- Paetkau, D.; Slade, R.; Burden, M.; Estoup, A. Genetic assignment methods for the direct, real-time estimation of migration rate: A simulation-based exploration of accuracy and power. Mol. Ecol. 2004, 13, 55–65. [Google Scholar] [CrossRef]
- VanderPlas, J. Three-Dimensional Plotting in Matplotlib. In Python Data Science Handbook; O’Reilly Media, Inc.: Sevastopol, CA, USA, 2016; Available online: https://jakevdp.github.io/PythonDataScienceHandbook/04.12-three-dimensional-plotting.html (accessed on 21 August 2023).
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic threes. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [CrossRef]
- Žulj Mihaljević, M.; Maletić, E.; Preiner, D.; Zdunić, G.; Bubola, M.; Zyprian, E.; Pejić, I. Genetic Diversity, Population Structure, and Parentage Analysis of Croatian Grapevine Germplasm. Genes 2020, 11, 737. [Google Scholar] [CrossRef]
- Italian Vitis Database. Available online: https://vitisdb.it/varieties/list?page=8 (accessed on 8 March 2025).
- Vine to Wine Cercle (Portugal and Spain). Available online: http://www.vinetowinecircle.com/en/ (accessed on 8 March 2025).
- Rodriguez-Torres, I. Albillo Forastero. In Variedades de vid Cultivadas en Canarias. Descriptores Morfológicos. Caracterización Morfológica, Molecular, Agronómica y Enológica, 2nd ed.; Instituto Canario de Investigaciones Agrarias, Ed.; Gobierno de Canarias: Santa Cruz de Tenerife, Spain, 2018; Available online: https://www.icia.es/icia/download/Publicaciones/Variedades_Vid_Canarias.pdf (accessed on 4 March 2025).
- Meneghetti, S.; Bavaresco, L.; Calò, A.; Costacurta, A. Inter- and Intra-Varietal Genetic Variability in Vitis vinifera L. In The Mediterrean Genetic Code-Grapevine and Olive; Sladonja, B., Ed.; IntechOpen: London, UK, 2013; ISBN 978-953-51-4251-5. [Google Scholar] [CrossRef][Green Version]
- Mendez, J.J. Acerca del Canary Wine. In Compendio de la Vitivinicultura del Archipiélago Canario, 2nd ed.; Asociación de Viticultores y Bodegueros de Canarias AVIBO, Ed.; Centro de la Cultura Popular Canaria (CCPC): Canary Island, Spain, 2024. [Google Scholar]
- Marsal, G.; Mateo, J.M.; Canals, J.M.; Zamora, F.; Fort, F. SSR analysis of 338 accessions planted in Penedes (Spain) reveals 28 unreported molecular profiles of Vitis vinifera L. Am. J. Enol. Vitic. 2016, 67, 466–470. [Google Scholar] [CrossRef]
- Marsal, G.; Bota, J.; Martorell, A.; Canals, J.M.; Zamora, F.; Fort, F. Local cultivars of Vitis vinifera L. in Spanish Islands: Balearic Archipelago. Sci. Hortic. 2017, 226, 122–132. [Google Scholar] [CrossRef]
- Marsal, G.; Mendez, J.J.; Mateo-Sanz, J.M.; Ferrer, S.; Canals, J.M.; Zamora, F.; Fort, F. Molecular characterization of Vitis vinifera L. local cultivars from volcanic areas (the Canary Islands and Madeira) using SSR markers. OENO One 2019, 53, 667–680. [Google Scholar] [CrossRef]
- Fort, F.; Marsal, G.; Mateo-Sanz, J.M.; Pena, V.; Canals, J.M.; Zamora, F. Molecular characterisation of the current cultivars of Vitis vinifera L. in Lanzarote (Canary Islands, Spain) reveals nine individuals which correspond to eight new varieties and two new sports. OENO One 2022, 56, 281–295. [Google Scholar] [CrossRef]
- Fort, F.; Lin-Yang, Q.; Suárez-Abreu, L.R.; Sancho-Galán, P.; Canals, J.M.; Zamora, F. Study of Molecular Biodiversity and Population Structure of Vitis vinifera L. ssp. vinifera on the Volcanic Island of El Hierro (Canary Islands, Spain) by Using Microsatellite Markers. Horticulturae 2023, 9, 1297. [Google Scholar] [CrossRef]
- Fort, F.; Lin-Yang, Q.; Valls, C.; Sancho-Galán, P.; Canals, J.M.; Zamora, F. Characterisation and Identification of Vines from Fuerteventura (Canary Volcanic Archipelago (Spain)) Using Simple Sequence Repeat Markers. Horticulturae 2023, 9, 1301. [Google Scholar] [CrossRef]
- Fort, F.; Lin-Yang, Q.; Valls, C.; Sancho-Galán, P.; Canals, J.M.; Zamora, F. Analysis of the Diversity Presented by Vitis vinifera L. in the Volcanic Island of La Gomera (Canary Archipelago, Spain) Using Simple Sequence Repeats (SSRs) as Molecular Markers. Horticulturae 2024, 10, 14. [Google Scholar] [CrossRef]
- García-Muñoz, S.; Lacômbe, T.; De Andrés, M.T.; Gaforio, L.; Muñoz-Organero, G.; Laucou, V.; This, P.; Cabello, F. Grape varieties (Vitis vinifera L.) from the Balearic Islands: Genetic characterization and relationship with Iberian Peninsula and Mediterranean Basin. Genet. Resour. Crop Evol. 2012, 59, 589–605. [Google Scholar] [CrossRef]
- Arroyo-Garcia, R.; Ruiz-García, L.; Bolling, L.; Ocete, R.; López, M.A.; Arnold, C.; Ergul, A.; Söylemezoğlu, G.; Uzun, H.I.; Cabello, F.; et al. Multiple origins of cultivated grape-vine (Vitis vinifera L. ssp. sativa) based on chloroplast DNA polymorphisms. Mol. Ecol. 2006, 15, 3707–3714. [Google Scholar] [CrossRef]
- Méndez, J.J.; Marsal, G.; Canals, N.; Canals, J.M.; Zamora, F.; Fort, F. Del Origen de la familia Vitaceae hasta la aparición de la especie Vitis vinifera L. In Jornadas Históricas “Canary Wine”; Cátedra de Agroturismo y Enoturismo de Canarias, Ed.; Instituto Canario de Calidad Agroalimentaria y Universidad de La Laguna: Santa Cruz de Tenerife, Spain, 2021; pp. 88–89. [Google Scholar]
- Massonnet, M.; Cochetel, N.; Minio, A.; Vondras, A.M.; Lin, J.; Muyle, A.; Garcia, J.F.; Zhou, Y.; Delledonne, M.; Riaz, S.; et al. The genetic basis of sex determination in grapes. Nat. Commun. 2020, 11, 2902. [Google Scholar] [CrossRef]
- D’Onofrio, C.; Tumino, G.; Gardiman, M.; Crespan, M.; Bignami, C.; de Palma, L.; Barbagallo, M.G.; Muganu, M.; Morcia, C.; Novello, V.; et al. Parentage atlas of Italian grapevine varieties as inferred from SNP genotyping. Front. Plant Sci. 2021, 11, 605934. [Google Scholar] [CrossRef]
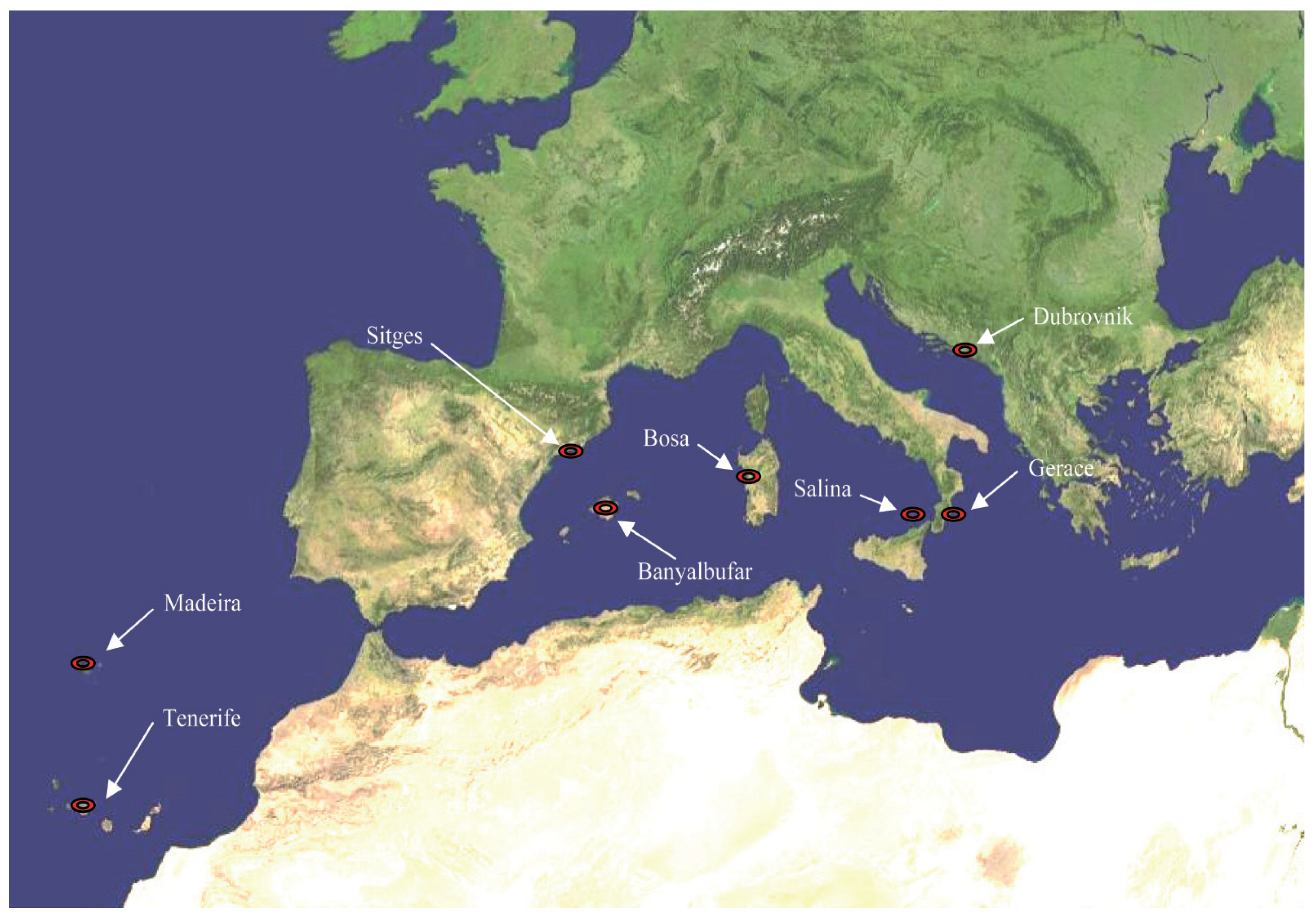
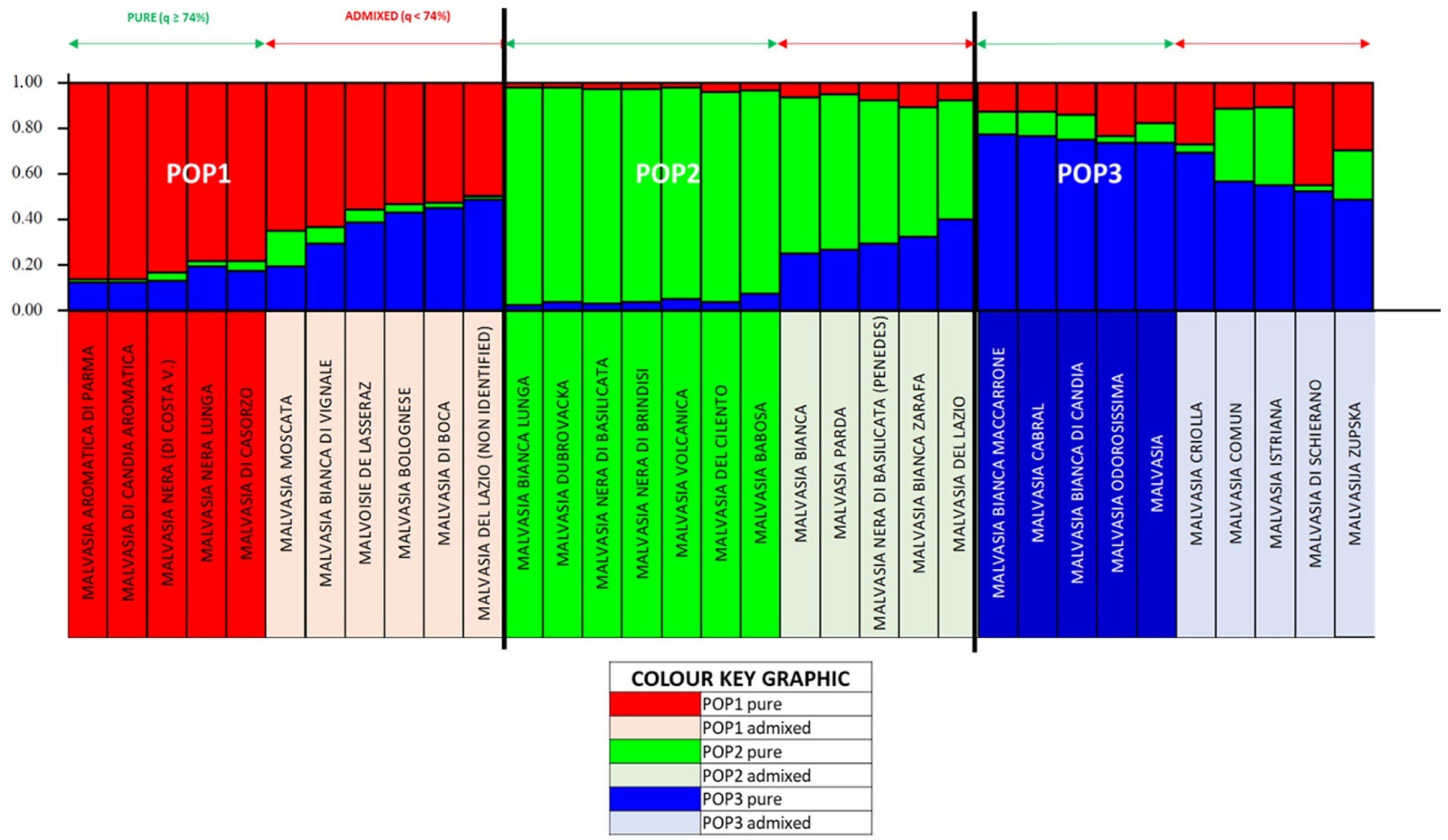
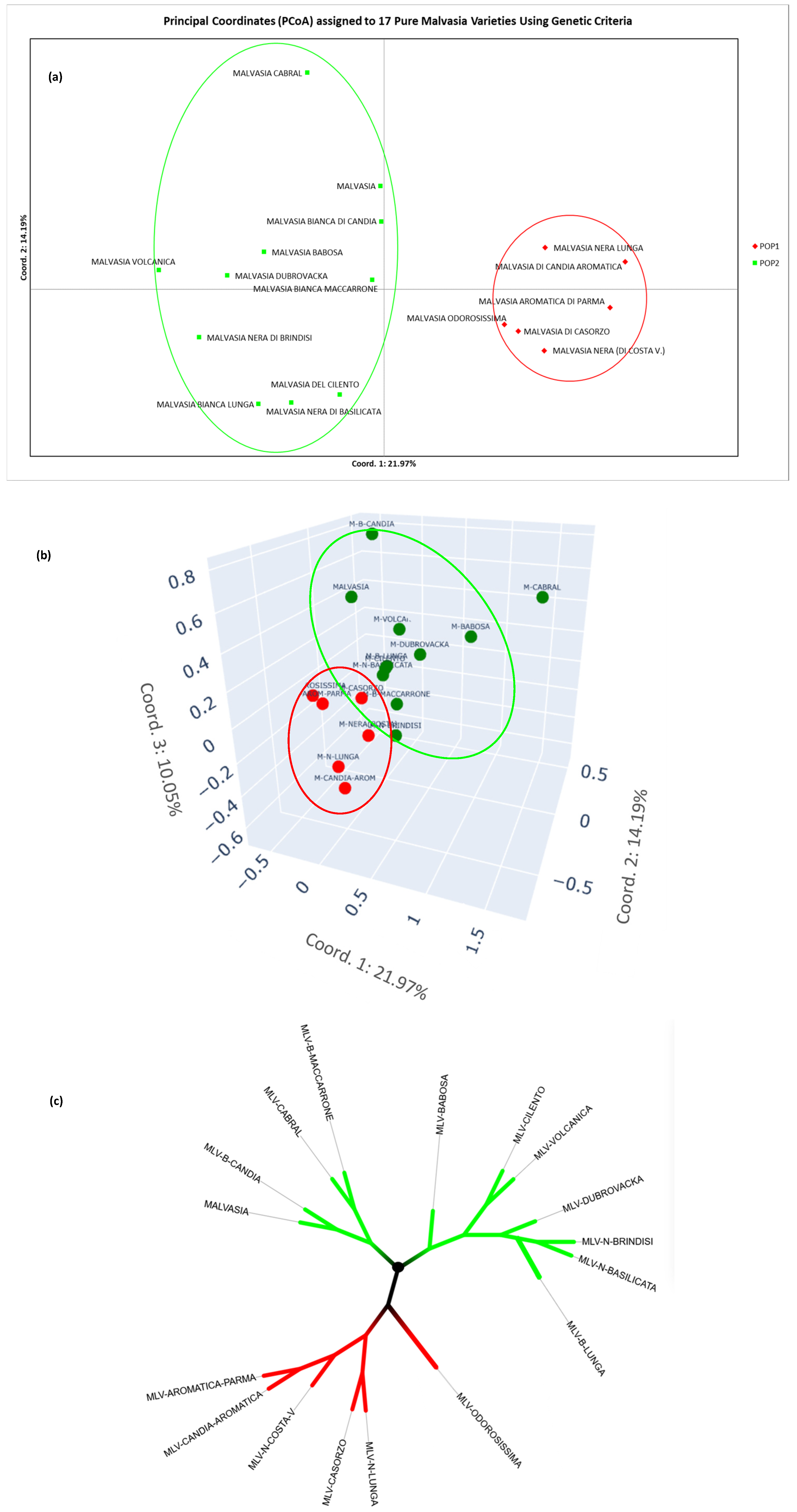


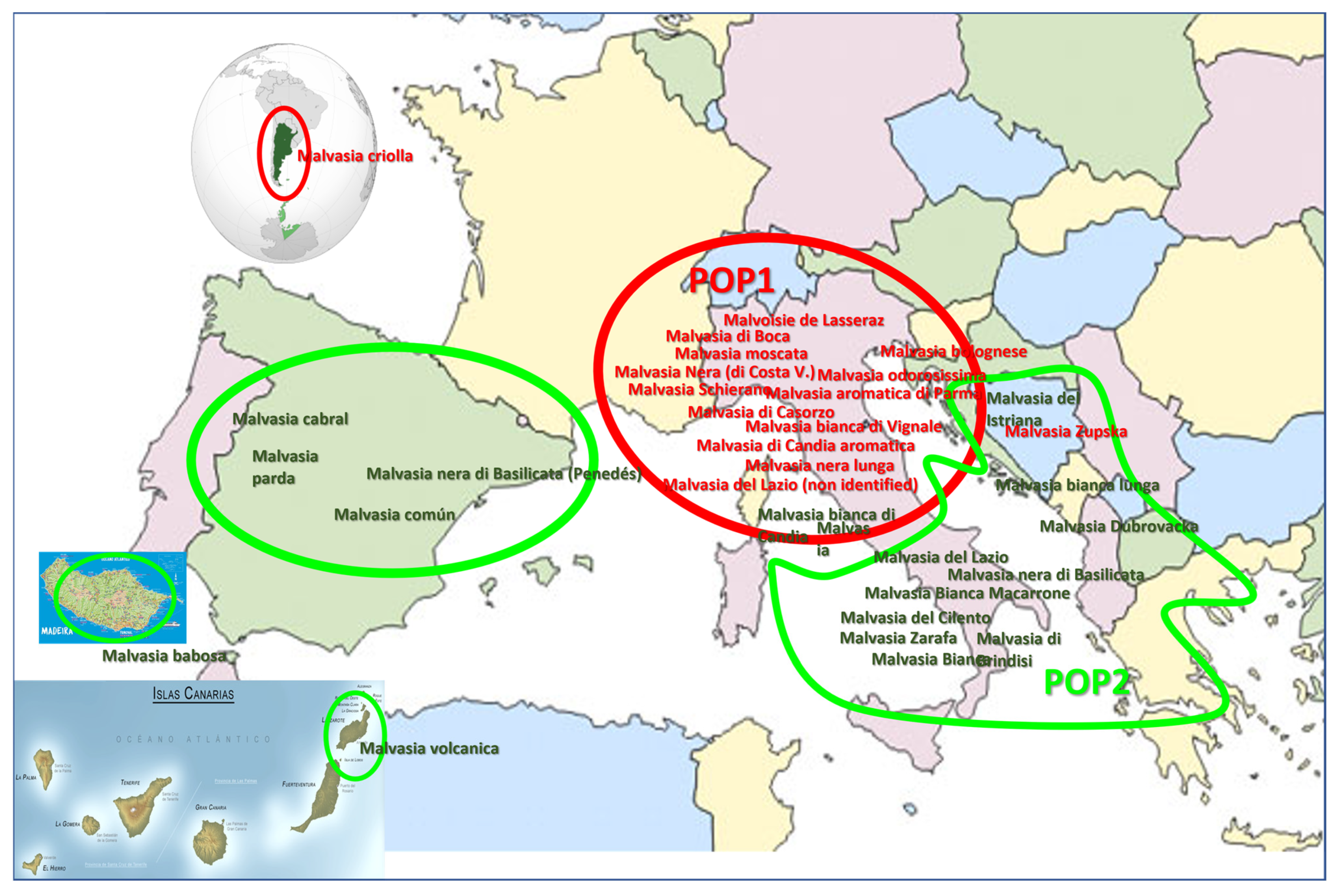


| NAME | CULTIVARS NAME (No. of Accesions) | PRINCIPAL NAME (No. of Accessions) |
|---|---|---|
| MALVASIA | 290 | 203 |
| MALVOISIE | 52 | 13 |
| MALVASIER | 15 | 3 |
| MALVAZIYA | 14 | 8 |
| MALVAZIJA | 13 | 9 |
| MALVAZIA | 11 | 2 |
| MALMSEY | 6 | 3 |
| MALVAGIA | 4 | 4 |
| MALVASIJA | 4 | 4 |
| MALVASIE | 2 | 0 |
| MALVASIKA | 1 | 1 |
| MALVASIYA | 1 | 0 |
| TOTAL No. OF NAMES | 413 | 250 |
| GENERAL INFORMATION | PEDIGREE | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| PRIME NAME | COUNTRY ORIGIN | CHLOROTYPE | SEED FORMATION | SEX | TASTE | COLOR | USE | PARENT 1 | PARENT 2 |
| MALVASIA AROMATICA DI PARMA | FRA139/MEX062/USA 06/USA028 | D | COMPLETE | FEMALE | MUSCAT | W | W | MUSCAT A PETITS GRAINS BLANCS | NEBBIOLO x BOTTAGERA (FALSE) |
| MALVASIA DI CANDIA AROMATICA | ITALY | D | COMPLETE | HERMAFRODITE | MUSCAT | W | T/W | MALVASIA AROMATICA DI PARMA (MUSCAT A PETITS GRAINS BLANCS x NEBBIOLO X BOTTAGERA (FALSE)) | ? |
| MALVASIA NERA (DI COSTA V.) | ITALY | D | COMPLETE | HERMAFRODITE | MUSCAT | B | ? | MALVASIA AROMATICA DI PARMA (MUSCAT A PETITS GRAINS BLANCS x NEBBIOLO X BOTTAGERA (FALSE)) | LAMBRUSCA DI ALESSANDRIA (CROVIN x NERETTO DI MARENGO (LAMBRUSCA DI ALESSANDRIA x ?)) |
| MALVASIA NERA LUNGA | ITALY | D | COMPLETE | HERMAFRODITE | ARMATIC | B | W | MALVASIA AROMATICA DI PARMA (MUSCAT A PETITS GRAINS BLANCS x NEBBIOLO X BOTTAGERA (FALSE)) | FREISA (NEBBIOLO x ?) |
| MALVASIA DI CASORZO | ITALY | D | COMPLETE | HERMAFRODITE | AROMATIC | B | W | MALVASIA AROMATICA DI PARMA (MUSCAT A PETITS GRAINS BLANCS x NEBBIOLO X BOTTAGERA (FALSE)) | LAMBRUSCA DI ALESSANDRIA (CROVIN x NERETTO DI MARENGO (LAMBRUSCA DI ALESSANDRIA x ?)) |
| MALVASIA MOSCATA | ITALY | D | COMPLETE | HERMAFRODITE | MUSCAT | W | W | MALVASIA AROMATICA DI PARMA (MUSCAT A PETITS GRAINS BLANCS x NEBBIOLO X BOTTAGERA (FALSE)) | ? |
| MALVASIA BIANCA DI VIGNALE | ITALY | D | COMPLETE | HERMAFRODITE | AROMATIC | W | W | MALVASIA AROMATICA DI PARMA (MUSCAT A PETITS GRAINS BLANCS x NEBBIOLO X BOTTAGERA (FALSE)) | COCCALONA BIANCA |
| MALVOISIE DE LASSERAZ | FRANCE | ? | COMPLETE | FEMALE | NONE | W | W | ? | ? |
| MALVASIA BOLOGNESE | ITALY | ? | COMPLETE | ? | ? | W | W | ? | ? |
| MALVASIA DI BOCA | ITALY | ? | COMPLETE | HERMAFRODITE | AROMATIC | W | W | ? | ? |
| MALVASIA DEL LAZIO (NON-IDENTIFIED) | ITALY | ? | COMPLETE | HERMAFRODITE | MUSCAT | W | W | MUSCAT OF ALEXANDRIA (MUSCAT A PETITS GRAINS BLANCS x HEPTALIKO) | SCHIAVA GROSSA |
| MALVASIA BIANCA LUNGA | ITALY | D | COMPLETE | HERMAFRODITE | NONE | W | W | ? | ? |
| MALVASIA DUBROVACKA | SPAIN | A | COMPLETE | HERMAFRODITE | AROMATIC | W | W | ? | ? |
| MALVASIA NERA DI BASILICATA | ITALY | ? | COMPLETE | HERMAFRODITE | ARMATIC | B | W | MALVASIA BIANCA LUNGA | SOMARELLO NERO (UVA SACRA (ACHLADI x ?) x GARGANEGA) |
| MALVASIA NERA DI BRINDISI | ITALY | ? | COMPLETE | HERMAFRODITE | NONE | B | T/W | MALVASIA BIANCA LUNGA | NEGRO AMARO (? x MAIOLICA (? x VISPAROLA)) |
| MALVASIA VOLCANICA | SPAIN | ? | COMPLETE | HERMAFRODITE | OTHER FLAVOR THAN MUSCAT, FOXY OR HERBACEOUS | W | W | MALVASIA DUBROVACKA | BERMEJUELA |
| MALVASIA DEL CILENTO | ITALY | ? | ? | ? | ? | B | W | ? | ? |
| MALVASIA BABOSA | PORTUGAL | A | COMPLETE | ? | ? | W | W | HEBEN | MALVASIA DUBROVACKA |
| MALVASIA BIANCA | ITALY | ? | COMPLETE | HERMAFRODITE | NONE | W | W | SCIACCARELLO | ? |
| MALVASIA PARDA | PORTUGAL | ? | COMPLETE | HERMAFRODITE | NONE | W | W | ? | ? |
| MALVASIA NERA DI BASILICATA (PENEDES) | SPAIN | ? | COMPLETE | ? | NONE | B | W | ? | ? |
| MALVASIA BIANCA ZARAFA | ITALY | ? | COMPLETE | ? | MUSCAT | W | W | ? | ? |
| MALVASIA DEL LAZIO | ITALY | ? | COMPLETE | HERMAFRODITE | NONE | W | W | ? | ? |
| MALVASIA BIANCA MACCARRONE | ITALY | ? | COMPLETE | ? | MUSCAT | W | W | ? | ? |
| MALVASIA CABRAL | PRT051 | ? | COMPLETE | HERMAFRODITE | NONE | Rs | W | ? | ? |
| MALVASIA BIANCA DI CANDIA | ITALY | D | COMPLETE | HERMAFRODITE | NONE | W | W | GARGANEGA | ? |
| MALVASIA ODOROSISSIMA | ITALY | ? | COMPLETE | FEMALE | MUSCAT | W | W | ? | ? |
| MALVASIA | ITALY | ? | COMPLETE | ? | ? | B | W | ? | ? |
| MALVASIA CRIOLLA | ARGENTINA | ? | COMPLETE | HERMAFRODITE | MUSCAT | W | W | LISTAN PRIETO | MUSCAT OF ALEXANDRIA (MUSCAT A PETITS GRAINS BLANCS x HEPTALIKO) |
| MALVASIA COMUN | SPAIN | A | COMPLETE | HERMAFRODITE | NONE | W | W | HEBEN | ? |
| MALVASIA ISTRIANA | CROATIA | D | COMPLETE | HERMAFRODITE | NONE | W | W | ? | ? |
| MALVASIA DI SCHIERANO | ITALY | D | COMPLETE | HERMAFRODITE | MUSCAT | B | W | MUSCAT A PETITS GRAINS BLANCS | ? |
| MALVASIJA ZUPSKA | CROATIA | D | COMPLETE | ? | ? | W | W | HEUNISCH WEISS | ? |
| GENERAL INFORMATION | PEDIGREE | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| PRIME NAME | COUNTRY ORIGIN | CHLOROTYPE | SEED FORMATION | SEX | TASTE | COLOR | USE | PARENT 1 | PARENT 2 |
| MALVASIA AROMATICA DI PARMA | ITA | D | COMPLETE | FEMALE | MUSCAT | W | W | MUSCAT A PETITS GRAINS BLANCS | NEBBIOLO x BOTTAGERA (FALSE) |
| MALVASIA DI CANDIA AROMATICA | ITA | D | COMPLETE | HERMAFRODITE | MUSCAT | W | T/W | MALVASIA AROMATICA DI PARMA (MUSCAT A PETITS GRAINS BLANCS x NEBBIOLO X BOTTAGERA (FALSE)) | ? |
| MALVASIA NERA LUNGA | ITA | D | COMPLETE | HERMAFRODITE | ARMATIC | B | W | MALVASIA AROMATICA DI PARMA (MUSCAT A PETITS GRAINS BLANCS x NEBBIOLO X BOTTAGERA (FALSE)) | FREISA (NEBBIOLO x ?) |
| MALVASIA BIANCA DI VIGNALE | ITA | D | COMPLETE | HERMAFRODITE | AROMATIC | W | W | MALVASIA AROMATICA DI PARMA (MUSCAT A PETITS GRAINS BLANCS x NEBBIOLO X BOTTAGERA (FALSE)) | COCCALONA BIANCA |
| MALVASIA DI CASORZO | ITA | D | COMPLETE | HERMAFRODITE | AROMATIC | B | W | MALVASIA AROMATICA DI PARMA (MUSCAT A PETITS GRAINS BLANCS x NEBBIOLO X BOTTAGERA (FALSE)) | LAMBRUSCA DI ALESSANDRIA (CROVIN x NERETTO DI MARENGO (LAMBRUSCA DI ALESSANDRIA x ?)) |
| MALVASIA NERA (DI COSTA V.) | ITA | D | COMPLETE | HERMAFRODITE | MUSCAT | W | ? | MALVASIA AROMATICA DI PARMA (MUSCAT A PETITS GRAINS BLANCS x NEBBIOLO X BOTTAGERA (FALSE)) | LAMBRUSCA DI ALESSANDRIA (CROVIN x NERETTO DI MARENGO (LAMBRUSCA DI ALESSANDRIA x ?)) |
| MALVASIA MOSCATA | ITA | D | COMPLETE | HERMAFRODITE | MUSCAT | W | W | MALVASIA AROMATICA DI PARMA (MUSCAT A PETITS GRAINS BLANCS x NEBBIOLO X BOTTAGERA (FALSE)) | ? |
| MALVASIA DI SCHIERANO | ITA | D | COMPLETE | HERMAFRODITE | MUSCAT | B | W | MUSCAT A PETITS GRAINS BLANCS | ? |
| MALVASIA DEL LAZIO (NON-IDENTIFIED) | ITA | ? | COMPLETE | HERMAFRODITE | MUSCAT | W | W | MUSCAT OF ALEXANDRIA (MUSCAT A PETITS GRAINS BLANCS x HEPTALIKO) | SCHIAVA GROSSA |
| MALVASIA CRIOLLA | ARG | ? | COMPLETE | HERMAFRODITE | MUSCAT | W | W | LISTAN PRIETO | MUSCAT OF ALEXANDRIA (MUSCAT A PETITS GRAINS BLANCS x HEPTALIKO) |
| MALVASIA BOLOGNESE | ITA | ? | COMPLETE | ? | ? | W | W | ? | ? |
| MALVASIA DI BOCA | ITA | ? | COMPLETE | HERMAFRODITE | AROMATIC | W | W | ? | ? |
| MALVOISIE DE LASSERAZ | FRA | ? | COMPLETE | FEMALE | NONE | W | W | ? | ? |
| MALVASIA ODOROSISSIMA | ITA | ? | COMPLETE | FEMALE | MUSCAT | W | W | ? | ? |
| MALVASIJA ZUPSKA | HRV | D | COMPLETE | ? | ? | W | W | HEUNISCH WEISS | ? |
| MALVASIA | ITA | ? | COMPLETE | ? | ? | B | W | ? | ? |
| MALVASIA BIANCA DI CANDIA | ITA | D | COMPLETE | HERMAFRODITE | NONE | W | W | GARGANEGA | ? |
| MALVASIA DUBROVACKA | ESP | A | COMPLETE | HERMAFRODITE | AROMATIC | W | W | ? | ? |
| MALVASIA BABOSA | PRT | A | COMPLETE | ? | ? | W | W | HEBEN | MALVASIA DUBROVACKA |
| MALVASIA VOLCANICA | ESP | ? | COMPLETE | HERMAFRODITE | OTHER FLAVOR THAN MUSCAT, FOXY OR HERBACEOUS | W | W | MALVASIA DUBROVACKA | BERMEJUELA |
| MALVASIA BIANCA LUNGA | ITA | D | COMPLETE | HERMAFRODITE | NONE | W | W | ? | ? |
| MALVASIA NERA DI BRINDISI | ITA | ? | COMPLETE | HERMAFRODITE | NONE | B | T/W | MALVASIA BIANCA LUNGA | NEGRO AMARO (? x MAIOLICA (? x VISPAROLA)) |
| MALVASIA NERA DI BASILICATA | ITA | ? | COMPLETE | HERMAFRODITE | ARMATIC | B | W | MALVASIA BIANCA LUNGA | SOMARELLO NERO (UVA SACRA (ACHLADI x ?) x GARGANEGA) |
| MALVASIA DEL CILENTO | ITA | ? | ? | ? | ? | B | W | ? | ? |
| MALVASIA ISTRIANA | HRV | D | COMPLETE | HERMAFRODITE | NONE | W | W | ? | ? |
| MALVASIA COMUN | ESP | A | COMPLETE | HERMAFRODITE | NONE | W | W | HEBEN | ? |
| MALVASIA BIANCA | ITA | ? | COMPLETE | NONE | W | W | SCIACCARELLO | ? | |
| MALVASIA BIANCA ZARAFA | ITA | ? | COMPLETE | ? | MUSCAT | W | W | ? | ? |
| MALVASIA PARDA | PRT | ? | COMPLETE | HERMAFRODITE | NONE | W | W | ? | ? |
| MALVASIA BIANCA MACCARRONE | ITA | ? | COMPLETE | ? | MUSCAT | W | W | ? | ? |
| MALVASIA CABRAL | PRT051 | ? | COMPLETE | HERMAFRODITE | NONE | Rs | W | ? | ? |
| MALVASIA NERA DI BASILICATA (PENEDES) | ESP | ? | COMPLETE | ? | NONE | B | W | ? | ? |
| MALVASIA DEL LAZIO | ITA | ? | COMPLETE | HERMAFRODITE | NONE | W | W | ? | ? |
| GENERAL INFORMATION | PEDIGREE | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| PRIME NAME | COUNTRY ORIGIN | CHLOROTYPE | SEED FORMATION | SEX | TASTE | COLOR | USE | PARENT 1 | PARENT 2 |
| MALVASIA DUBROVACKA | ESP | A | COMPLETE | HERMAFRODITE | AROMATIC | W | W | ? | ? |
| MALVASIA BABOSA | PRT | A | COMPLETE | ? | ? | W | W | HEBEN | MALVASIA DUBROVACKA |
| MALVASIA VOLCANICA | ESP | ? | COMPLETE | HERMAFRODITE | OTHER FLAVOR THAN MUSCAT, FOXY OR HERBACEOUS | W | W | MALVASIA DUBROVACKA | BERMEJUELA |
| MALVASIA BIANCA LUNGA | ITA | D | COMPLETE | HERMAFRODITE | NONE | W | W | ? | ? |
| MALVASIA NERA DI BRINDISI | ITA | ? | COMPLETE | HERMAFRODITE | NONE | B | T/W | MALVASIA BIANCA LUNGA | NEGRO AMARO (? x MAIOLICA (? x VISPAROLA)) |
| MALVASIA NERA DI BASILICATA | ITA | ? | COMPLETE | HERMAFRODITE | ARMATIC | B | W | MALVASIA BIANCA LUNGA | SOMARELLO NERO (UVA SACRA (ACHLADI x ?) x GARGANEGA) |
| MALVASIA DEL CILENTO | ITA | ? | ? | ? | ? | B | W | ? | ? |
| MALVASIA ISTRIANA | HRV | D | COMPLETE | HERMAFRODITE | NONE | W | W | ? | ? |
| MALVASIJA ZUPSKA | HRV | D | COMPLETE | ? | ? | W | W | HEUNISCH WEISS | ? |
| MALVASIA AROMATICA DI PARMA | ITA | D | COMPLETE | FEMALE | MUSCAT | W | W | MUSCAT A PETITS GRAINS BLANCS | NEBBIOLO x BOTTAGERA (FALSE) |
| MALVASIA DI CANDIA AROMATICA | ITA | D | COMPLETE | HERMAFRODITE | MUSCAT | W | T/W | MALVASIA AROMATICA DI PARMA (MUSCAT A PETITS GRAINS BLANCS x NEBBIOLO X BOTTAGERA (FALSE)) | ? |
| MALVASIA NERA LUNGA | ITA | D | COMPLETE | HERMAFRODITE | ARMATIC | B | W | MALVASIA AROMATICA DI PARMA (MUSCAT A PETITS GRAINS BLANCS x NEBBIOLO X BOTTAGERA (FALSE)) | FREISA (NEBBIOLO x ?) |
| MALVASIA BIANCA DI VIGNALE | ITA | D | COMPLETE | HERMAFRODITE | AROMATIC | W | W | MALVASIA AROMATICA DI PARMA (MUSCAT A PETITS GRAINS BLANCS x NEBBIOLO X BOTTAGERA (FALSE)) | COCCALONA BIANCA |
| MALVASIA DI CASORZO | ITA | D | COMPLETE | HERMAFRODITE | AROMATIC | N | W | MALVASIA AROMATICA DI PARMA (MUSCAT A PETITS GRAINS BLANCS x NEBBIOLO X BOTTAGERA (FALSE)) | LAMBRUSCA DI ALESSANDRIA (CROVIN x NERETTO DI MARENGO (LAMBRUSCA DI ALESSANDRIA x ?)) |
| MALVASIA NERA (DI COSTA V.) | ITA | D | COMPLETE | HERMAFRODITE | MUSCAT | W | ? | MALVASIA AROMATICA DI PARMA (MUSCAT A PETITS GRAINS BLANCS x NEBBIOLO X BOTTAGERA (FALSE)) | LAMBRUSCA DI ALESSANDRIA (CROVIN x NERETTO DI MARENGO (LAMBRUSCA DI ALESSANDRIA x ?)) |
| MALVASIA MOSCATA | ITA | D | COMPLETE | HERMAFRODITE | MUSCAT | W | W | MALVASIA AROMATICA DI PARMA (MUSCAT A PETITS GRAINS BLANCS x NEBBIOLO X BOTTAGERA (FALSE)) | ? |
| MALVASIA DI SCHIERANO | ITA | D | COMPLETE | HERMAFRODITE | MUSCAT | B | W | MUSCAT A PETITS GRAINS BLANCS | ? |
| MALVASIA DEL LAZIO (NON-IDENTIFIED) | ITA | ? | COMPLETE | HERMAFRODITE | MUSCAT | W | W | MUSCAT OF ALEXANDRIA (MUSCAT A PETITS GRAINS BLANCS x HEPTALIKO) | SCHIAVA GROSSA |
| MALVASIA CRIOLLA | ARG | ? | COMPLETE | HERMAFRODITE | MUSCAT | W | W | LISTAN PRIETO | MUSCAT OF ALEXANDRIA (MUSCAT A PETITS GRAINS BLANCS x HEPTALIKO) |
| MALVASIA BOLOGNESE | ITA | ? | COMPLETE | ? | ? | W | W | ? | ? |
| MALVASIA DI BOCA | ITA | ? | COMPLETE | HERMAFRODITE | AROMATIC | W | W | ? | ? |
| MALVOISIE DE LASSERAZ | FRA | ? | COMPLETE | FEMALE | NONE | W | W | ? | ? |
| MALVASIA ODOROSISSIMA | ITA | ? | COMPLETE | FEMALE | MUSCAT | W | W | ? | ? |
| MALVASIA COMUN | ESP | A | COMPLETE | HERMAFRODITE | NONE | W | W | HEBEN | ? |
| MALVASIA BIANCA | ITA | ? | COMPLETE | HERMAFRODITE | NONE | W | W | SCIACCARELLO | ? |
| MALVASIA BIANCA ZARAFA | ITA | ? | COMPLETE | ? | MUSCAT | W | W | ? | ? |
| MALVASIA | ITA | ? | COMPLETE | ? | ? | B | W | ? | ? |
| MALVASIA BIANCA DI CANDIA | ITA | D | COMPLETE | HERMAFRODITE | NONE | W | W | GARGANEGA | ? |
| MALVASIA PARDA | PRT | ? | COMPLETE | HERMAFRODITE | NONE | W | W | ? | ? |
| MALVASIA BIANCA MACCARRONE | ITA | ? | COMPLETE | ? | MUSCAT | W | W | ? | ? |
| MALVASIA CABRAL | PRT051 | ? | COMPLETE | HERMAFRODITE | NONE | Rs | W | ? | ? |
| MALVASIA NERA DI BASILICATA (PENEDES) | ESP | ? | COMPLETE | ? | NONE | B | W | ? | ? |
| MALVASIA DEL LAZIO | ITA | ? | COMPLETE | HERMAFRODITE | NONE | W | W | ? | ? |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fort, F.; Suárez-Abreu, L.R.; Lin-Yang, Q.; Asenjo, J.; Deis, L.; Canals, J.M.; Zamora, F. Origin and Possible Members of the ‘Malvasia’ Family: The New Fuencaliente de La Palma Hypothesis on the True ‘Malvasia’. Horticulturae 2025, 11, 561. https://doi.org/10.3390/horticulturae11060561
Fort F, Suárez-Abreu LR, Lin-Yang Q, Asenjo J, Deis L, Canals JM, Zamora F. Origin and Possible Members of the ‘Malvasia’ Family: The New Fuencaliente de La Palma Hypothesis on the True ‘Malvasia’. Horticulturae. 2025; 11(6):561. https://doi.org/10.3390/horticulturae11060561
Chicago/Turabian StyleFort, Francesca, Luis Ricardo Suárez-Abreu, Qiying Lin-Yang, Juancho Asenjo, Leonor Deis, Joan Miquel Canals, and Fernando Zamora. 2025. "Origin and Possible Members of the ‘Malvasia’ Family: The New Fuencaliente de La Palma Hypothesis on the True ‘Malvasia’" Horticulturae 11, no. 6: 561. https://doi.org/10.3390/horticulturae11060561
APA StyleFort, F., Suárez-Abreu, L. R., Lin-Yang, Q., Asenjo, J., Deis, L., Canals, J. M., & Zamora, F. (2025). Origin and Possible Members of the ‘Malvasia’ Family: The New Fuencaliente de La Palma Hypothesis on the True ‘Malvasia’. Horticulturae, 11(6), 561. https://doi.org/10.3390/horticulturae11060561








