De Novo Assembly and Comparative Analysis of the Mitochondrial Genomes for Six Rubus Species
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Acquisition, Genome Assembly, and Annotation
2.2. Repeat Sequence Identification
2.3. RNA Editing Predictions
2.4. Ka/Ks Value Analysis
2.5. Codon Usage Bias Analysis
2.6. Mitochondrial and Plastid Sequence Migration Analysis
2.7. Phylogenetic Analysis
2.8. Collinearity Analysis of Mitochondrial Genome
3. Results
3.1. Characteristics of Rubus Mitogenomes
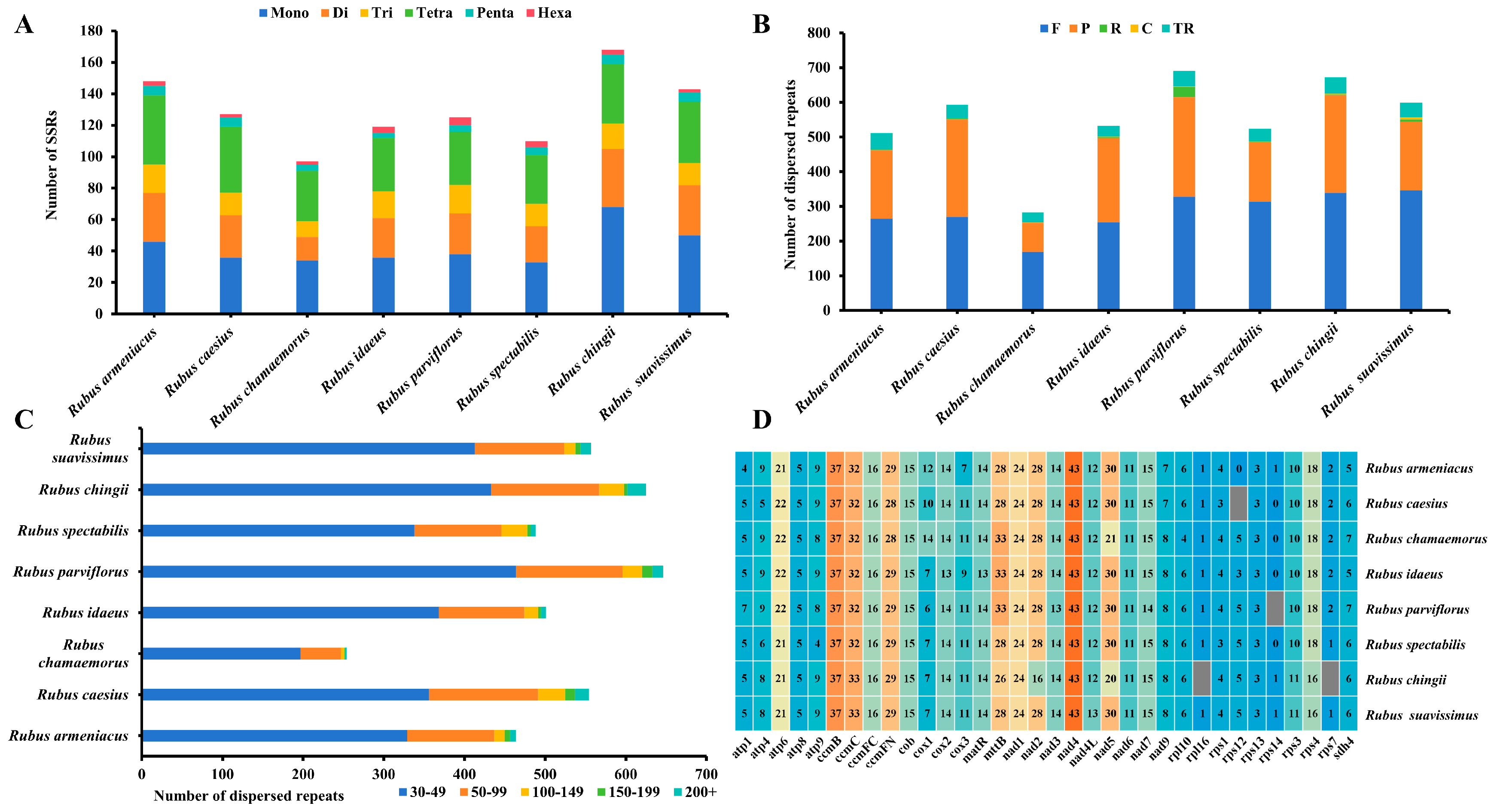
3.2. Repeat Sequence Analysis and RNA Editing Prediction
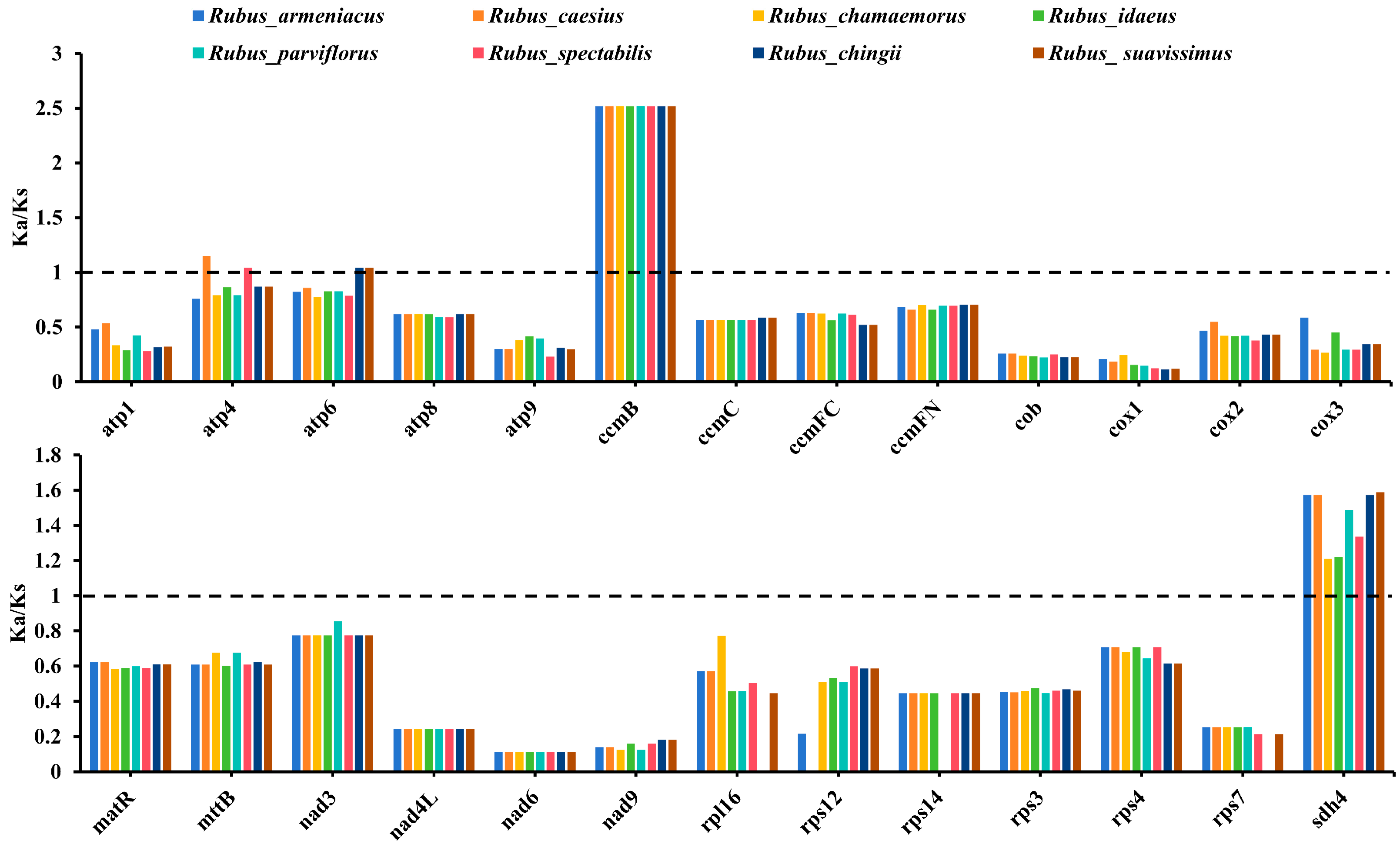
3.3. Analysis of Selection Pressure on Shared PCGs Among Rubus Species
3.4. Codon Usage Bias Analysis of Rubus mitogenomes
3.5. Sequence Migration Analysis
3.6. Multidimensional Systemic Generation Analysis
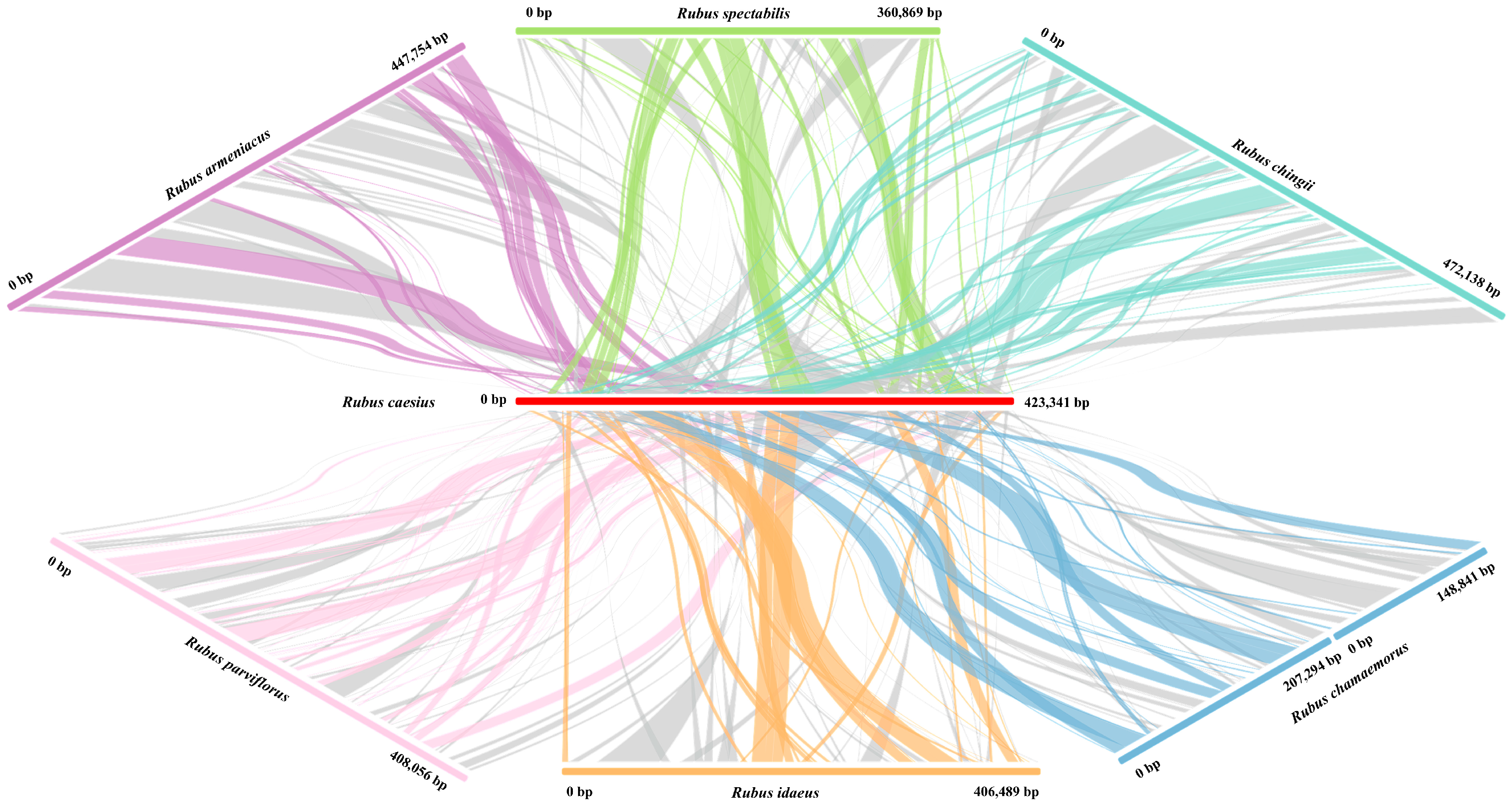
3.7. Collinearity Analysis of Mitochondrial Genome
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Carter, K.A.; Liston, A.; Bassil, N.V.; Alice, L.A.; Bushakra, J.M.; Sutherland, B.L.; Mockler, T.C.; Bryant, D.W.; Hummer, K.E. Target Capture Sequencing Unravels Rubus Evolution. Front. Plant Sci. 2019, 10, 1615. [Google Scholar] [CrossRef] [PubMed]
- Huang, T.; Chen, J.; Hummer, K.E.; Alice, L.A.; Wang, W.; He, Y.; Yu, S.; Yang, M.; Chai, T.; Zhu, X.; et al. Phylogeny of Rubus (Rosaceae): Integrating Molecular and Morphological Evidence into an Infrageneric Revision. Taxon 2023, 72, 278–306. [Google Scholar] [CrossRef]
- Zhang, H.; Yan, M.; Li, L.; Jiang, Z.; Xiong, Y.; Wang, Y.; Akogwu, C.O.; Tolulope, O.M.; Zhou, H.; Sun, Y.; et al. Assembly and Comparative Analysis of the Complete Mitochondrial Genome of Red Raspberry (Rubus idaeus L.) Revealing Repeat-Mediated Recombination and Gene Transfer. BMC Plant Biol. 2025, 25, 85. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Wu, Y.; Zhang, S.; Yang, H.; Wu, W.; Lyu, L.; Li, W. Overexpression of RuFLS2 Enhances Flavonol-Related Substance Contents and Gene Expression Levels. Int. J. Mol. Sci. 2022, 23, 14230. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Zhang, K.; Xiao, Y.; Zhang, L.; Huang, Y.; Li, X.; Chen, S.; Peng, Y.; Yang, S.; Liu, Y.; et al. Genome Assembly and Population Resequencing Reveal the Geographical Divergence of Shanmei (Rubus corchorifolius). Genom. Proteom. Bioinform. 2022, 20, 1106–1118. [Google Scholar] [CrossRef]
- Hua, Y.; Dai, B.; Luo, Y.; Ding, Y. Integrated Analysis of Multiple Metabolome and Transcriptome Revealed the Accumulation of Flavonoids and Associated Molecular Regulation Mechanisms in Rubus chingii Hu at Different Developmental Stages. Plant Physiol. Biochem. 2023, 204, 108085. [Google Scholar] [CrossRef]
- Kumar, V.; Sharma, A.; Sharma, N.; Saini, R.; Dev, K.; El-Shazly, M.; Bari, A.B.A. A Review of Botany, Traditional Applications, Phytochemistry, Pharmacological Applications, and Toxicology of Rubus ellipticus Smith Fruits. Naunyn-Schmiedeberg’s Arch. Pharmacol. 2024, 397, 4483–4497. [Google Scholar] [CrossRef]
- Tao, Y.; Bao, J.; Zhu, F.; Pan, M.; Liu, Q.; Wang, P. Ethnopharmacology of Rubus idaeus Linnaeus: A Critical Review on Ethnobotany, Processing Methods, Phytochemicals, Pharmacology and Quality Control. J. Ethnopharmacol. 2023, 302, 115870. [Google Scholar] [CrossRef]
- Alice, L.; Campbell, C. Hybridization and Gene Flow Between Distantly Related Species of Rubus (Rosaceae): Evidence from Nuclear Ribosomal DNA Internal Transcribed Spacer Region Sequences. Syst. Bot. 2001, 26, 769–778. [Google Scholar]
- Sochor, M.; Vašut, R.J.; Sharbel, T.F.; Trávníček, B. How Just a Few Makes a Lot: Speciation via Reticulation and Apomixis on Example of European Brambles (Rubus subgen. Rubus, Rosaceae). Mol. Phylogenetics Evol. 2015, 89, 13–27. [Google Scholar] [CrossRef]
- Thompson, M.M.; Zhao, C.M. Chromosome Numbers of Rubus species in southwest China. Acta Hortic. 1993, 352, 493–502. [Google Scholar] [CrossRef]
- Focke, W.O. Species Ruborum: Monographiae Generis Rubi Prodromus; Botanica, B., Ed.; Schweizerbart: Stuttgart, Germany, 1914. [Google Scholar]
- Lu, L.; David, E.B. Flora of China; Rubus Linnaeus; 1: 492. 1753; Science Press: Beijing, China, 2003; Volume 9, pp. 195–285. [Google Scholar]
- Xiong, X.-H.; Zhou, X.-M.; Li, M.; Xu, B.; Deng, H.-N.; Yu, Q.; Gao, X.-F. Pollen Morphology in Rubus (Rosaceae) and Its Taxonomic Implications. Plant Syst. Evol. 2019, 305, 705–716. [Google Scholar] [CrossRef]
- Wang, Y.; Chen, Q.; Chen, T.; Tang, H.; Liu, L.; Wang, X. Phylogenetic Insights into Chinese Rubus (Rosaceae) from Multiple Chloroplast and Nuclear DNAs. Front. Plant Sci. 2016, 7, 968. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wang, X.; Chen, Q.; Zhang, L.; Tang, H.; Luo, Y.; Liu, Z. Phylogenetic Insight into Subgenera Idaeobatus and Malachobatus (Rubus, Rosaceae) Inferring from ISH Analysis. Mol. Cytogenet. 2015, 8, 11. [Google Scholar] [CrossRef]
- Birky, C.W. The Inheritance of Genes in Mitochondria and Chloroplasts: Laws, Mechanisms, and Models. Annu. Rev. Genet. 2001, 35, 125–148. [Google Scholar] [CrossRef]
- Shen, B.; Shen, A.; Liu, L.; Tan, Y.; Li, S.; Tan, Z. Assembly and Comparative Analysis of the Complete Multichromosomal Mitochondrial Genome of Cymbidium ensifolium, an Orchid of High Economic and Ornamental Value. BMC Plant Biol. 2024, 24, 255. [Google Scholar] [CrossRef]
- Rydin, C.; Wikström, N.; Bremer, B. Conflicting Results from Mitochondrial Genomic Data Challenge Current Views of Rubiaceae Phylogeny. Am. J. Bot. 2017, 104, 1522–1532. [Google Scholar] [CrossRef]
- Gray, M.W.; Cedergren, R.; Abel, Y.; Sankoff, D. On the Evolutionary Origin of the Plant Mitochondrion and Its Genome. Proc. Natl. Acad. Sci. USA 1989, 86, 2267–2271. [Google Scholar] [CrossRef]
- Wu, Z.; Liao, X.; Zhang, X.; Tembrock, L.R.; Broz, A. Genomic Architectural Variation of Plant Mitochondria—A Review of Multichromosomal Structuring. J. Syst. Evol. 2022, 60, 160–168. [Google Scholar] [CrossRef]
- Kozik, A.; Rowan, B.A.; Lavelle, D.; Berke, L.; Schranz, M.E.; Michelmore, R.W.; Christensen, A.C. The Alternative Reality of Plant Mitochondrial DNA: One Ring Does Not Rule Them All. PLoS Genet. 2019, 15, e1008373. [Google Scholar] [CrossRef]
- Jin, J.-J.; Yu, W.-B.; Yang, J.-B.; Song, Y.; dePamphilis, C.W.; Yi, T.-S.; Li, D.-Z. GetOrganelle: A Fast and Versatile Toolkit for Accurate de Novo Assembly of Organelle Genomes. Genome Biol. 2020, 21, 241. [Google Scholar] [CrossRef]
- He, W.; Xiang, K.; Chen, C.; Wang, J.; Wu, Z. Master Graph: An Essential Integrated Assembly Model for the Plant Mitogenome Based on a Graph-Based Framework. Brief. Bioinform. 2023, 24, bbac522. [Google Scholar] [CrossRef] [PubMed]
- Bi, C.; Shen, F.; Han, F.; Qu, Y.; Hou, J.; Xu, K.; Xu, L.; He, W.; Wu, Z.; Yin, T. PMAT: An Efficient Plant Mitogenome Assembly Toolkit Using Low-Coverage HiFi Sequencing Data. Hortic. Res. 2024, 11, uhae023. [Google Scholar] [CrossRef]
- Sun, M.; Zhang, M.; Chen, X.; Liu, Y.; Liu, B.; Li, J.; Wang, R.; Zhao, K.; Wu, J. Rearrangement and Domestication as Drivers of Rosaceae Mitogenome Plasticity. BMC Biol. 2022, 20, 181. [Google Scholar] [CrossRef] [PubMed]
- VanBuren, R.; Bryant, D.; Bushakra, J.M.; Vining, K.J.; Edger, P.P.; Rowley, E.R.; Priest, H.D.; Michael, T.P.; Lyons, E.; Filichkin, S.A.; et al. The Genome of Black Raspberry (Rubus occidentalis). Plant J. 2016, 87, 535–547. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Lei, T.; Han, G.; Yue, J.; Zhang, X.; Yang, Q.; Ruan, H.; Gu, C.; Zhang, Q.; Qian, T.; et al. The Chromosome-scale Reference Genome of Rubus chingii Hu Provides Insight into the Biosynthetic Pathway of Hydrolyzable Tannins. Plant J. 2021, 107, 1466–1477. [Google Scholar] [CrossRef]
- Wang, Y.; Guan, J.; Zhang, Q. Chromosome-Scale Genome, Together with Transcriptome and Metabolome, Provides Insights into the Evolution and Anthocyanin Biosynthesis of Rubus rosaefolius Sm. (Rosaceae). Hortic. Res. 2024, 11, uhae064. [Google Scholar] [CrossRef]
- Price, R.J.; Davik, J.; Fernandéz Fernandéz, F.; Bates, H.J.; Lynn, S.; Nellist, C.F.; Buti, M.; Røen, D.; Šurbanovski, N.; Alsheikh, M.; et al. Chromosome-Scale Genome Sequence Assemblies of the ‘Autumn Bliss’ and ‘Malling Jewel’ Cultivars of the Highly Heterozygous Red Raspberry (Rubus idaeus L.) Derived from Long-Read Oxford Nanopore Sequence Data. PLoS ONE 2023, 18, e0285756. [Google Scholar] [CrossRef]
- Wu, W.; Wang, L.; Huang, W.; Zhang, X.; Li, Y.; Guo, W. A High-Quality Genome Assembly Reveals Adaptations Underlying Glossy, Wax-Coated Leaves in the Heat-Tolerant Wild Raspberry Rubus leucanthus. DNA Res. 2024, 31, dsae024. [Google Scholar] [CrossRef]
- Brůna, T.; Aryal, R.; Dudchenko, O.; Sargent, D.J.; Mead, D.; Buti, M.; Cavallini, A.; Hytönen, T.; Andrés, J.; Pham, M.; et al. A Chromosome-Length Genome Assembly and Annotation of Blackberry (Rubus argutus, cv. “Hillquist”). G3 2023, 13, jkac289. [Google Scholar] [CrossRef]
- Shi, Y.; Chen, Z.; Jiang, J.; Wu, W.; Xin, Y.; Zeng, W. Assembly and Comparative Analysis of the Complete Mitogenome of Rubus chingii var. suavissimus, an Exceptional Berry Plant Possessing Sweet Leaves. Front. Plant Sci. 2024, 15, 1504687. [Google Scholar] [CrossRef]
- Bi, C.; Qu, Y.; Hou, J.; Wu, K.; Ye, N.; Yin, T. Deciphering the Multi-Chromosomal Mitochondrial Genome of Populus simonii. Front. Plant Sci. 2022, 13, 914635. [Google Scholar] [CrossRef]
- Wick, R.R.; Schultz, M.B.; Zobel, J.; Holt, K.E. Bandage: Interactive Visualization of de Novo Genome Assemblies. Bioinformatics 2015, 31, 3350–3352. [Google Scholar] [CrossRef] [PubMed]
- Li, H. New Strategies to Improve Minimap2 Alignment Accuracy. Bioinformatics 2021, 37, 4572–4574. [Google Scholar] [CrossRef]
- Li, J.; Ni, Y.; Lu, Q.; Chen, H.; Liu, C. PMGA: A Plant Mitochondrial Genome Annotator. Plant Commun. 2025, 6, 101191. [Google Scholar] [CrossRef] [PubMed]
- Tillich, M.; Lehwark, P.; Pellizzer, T.; Ulbricht-Jones, E.S.; Fischer, A.; Bock, R.; Greiner, S. GeSeq—Versatile and Accurate Annotation of Organelle Genomes. Nucleic Acids Res. 2017, 45, W6–W11. [Google Scholar] [CrossRef] [PubMed]
- Shi, L.; Chen, H.; Jiang, M.; Wang, L.; Wu, X.; Huang, L.; Liu, C. CPGAVAS2, an Integrated Plastome Sequence Annotator and Analyzer. Nucleic Acids Res. 2019, 47, W65–W73. [Google Scholar] [CrossRef]
- Chan, P.P.; Lin, B.Y.; Mak, A.J.; Lowe, T.M. tRNAscan-SE 2.0: Improved Detection and Functional Classification of Transfer RNA Genes. Nucleic Acids Res. 2021, 49, 9077–9096. [Google Scholar] [CrossRef]
- Chen, Y.; Ye, W.; Zhang, Y.; Xu, Y. High Speed BLASTN: An Accelerated MegaBLAST Search Tool. Nucleic Acids Res. 2015, 43, 7762–7768. [Google Scholar] [CrossRef]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An Integrated and Extendable Desktop Software Platform for the Organization and Analysis of Sequence Data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef]
- Zhang, X.; Chen, H.; Ni, Y.; Wu, B.; Li, J.; Burzyński, A.; Liu, C. Plant Mitochondrial Genome Map (PMGmap): A Software Tool for the Comprehensive Visualization of Coding, Noncoding and Genome Features of Plant Mitochondrial Genomes. Mol. Ecol. Resour. 2024, 24, e13952. [Google Scholar] [CrossRef] [PubMed]
- Beier, S.; Thiel, T.; Münch, T.; Scholz, U.; Mascher, M. MISA-Web: A Web Server for Microsatellite Prediction. Bioinformatics 2017, 33, 2583–2585. [Google Scholar] [CrossRef]
- Benson, G. Tandem Repeats Finder: A Program to Analyze DNA Sequences. Nucleic Acids Res. 1999, 27, 573–580. [Google Scholar] [CrossRef]
- Kurtz, S. REPuter: The Manifold Applications of Repeat Analysis on a Genomic Scale. Nucleic Acids Res. 2001, 29, 4633–4642. [Google Scholar] [CrossRef]
- Zhang, D.; Gao, F.; Jakovlić, I.; Zou, H.; Zhang, J.; Li, W.X.; Wang, G.T. PhyloSuite: An Integrated and Scalable Desktop Platform for Streamlined Molecular Sequence Data Management and Evolutionary Phylogenetics Studies. Mol. Ecol. Resour. 2020, 20, 348–355. [Google Scholar] [CrossRef] [PubMed]
- Edera, A.A.; Small, I.; Milone, D.H.; Sanchez-Puerta, M.V. Deepred-Mt: Deep Representation Learning for Predicting C-to-U RNA Editing in Plant Mitochondria. Comput. Biol. Med. 2021, 136, 104682. [Google Scholar] [CrossRef]
- Katoh, K. MAFFT: A Novel Method for Rapid Multiple Sequence Alignment Based on Fast Fourier Transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Zhang, Y.; Zhang, Z.; Zhu, J.; Yu, J. KaKs_Calculator 2.0: A Toolkit Incorporating Gamma-Series Methods and Sliding Window Strategies. Genom. Proteom. Bioinform. 2010, 8, 77–80. [Google Scholar] [CrossRef]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y.; et al. TBtools-II: A “One for All, All for One” Bioinformatics Platform for Biological Big-Data Mining. Mol. Plant 2023, 16, 1733–1742. [Google Scholar] [CrossRef]
- Xie, J.; Chen, Y.; Cai, G.; Cai, R.; Hu, Z.; Wang, H. Tree Visualization By One Table (tvBOT): A Web Application for Visualizing, Modifying and Annotating Phylogenetic Trees. Nucleic Acids Res. 2023, 51, W587–W592. [Google Scholar] [CrossRef]
- Marçais, G.; Delcher, A.L.; Phillippy, A.M.; Coston, R.; Salzberg, S.L.; Zimin, A. MUMmer4: A Fast and Versatile Genome Alignment System. PLoS Comput. Biol. 2018, 14, e1005944. [Google Scholar] [CrossRef] [PubMed]
- He, W.; Yang, J.; Jing, Y.; Xu, L.; Yu, K.; Fang, X. NGenomeSyn: An Easy-to-Use and Flexible Tool for Publication-Ready Visualization of Syntenic Relationships across Multiple Genomes. Bioinformatics 2023, 39, btad121. [Google Scholar] [CrossRef]
- Liu, M.; Fan, R.; Wang, C.; Dai, L.; Chu, S. Complete Analysis and Phylogenetic Analysis of Polygonatum sibiricum Mitochondria. BMC Plant Biol. 2025, 25, 471. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Wen, J.; Crabbe, M.J.C.; Chen, C.; Ren, Z. Complete Chloroplast Genome Characterization of Three Plagiomnium Species and the Phylogeny of Family Mniaceae. Genetica 2025, 153, 6. [Google Scholar] [CrossRef]
- Angiosperm Phylogeny Group; Chase, M.W.; Christenhusz, M.J.; Fay, M.F.; Byng, J.; Judd, W.; Soltis, D.; Mabberley, D.; Sennikov, A.; Soltis, P. An Update of the Angiosperm Phylogeny Group Classification for the Orders and Families of Flowering Plants: APG IV. Bot. J. Linn. Soc. 2016, 181, 1–20. [Google Scholar] [CrossRef]
- Wang, H.; Wu, Z.; Li, T.; Zhao, J. Highly Active Repeat-Mediated Recombination in the Mitogenome of the Aquatic Grass Hygroryza aristata. BMC Plant Biol. 2024, 24, 644. [Google Scholar] [CrossRef]
- Wang, Z.; Wang, R.; Sang, Y.; Wang, T.; Su, Y.; Liao, W. Comparative Analysis of Mitochondrial Genomes of Invasive Weed Mikania micrantha and Its Indigenous Congener Mikania cordata. Int. J. Biol. Macromol. 2024, 281, 136357. [Google Scholar] [CrossRef]
- Xu, C.; Bi, W.; Ma, R.; Li, P.; Liu, F.; Liu, Z. Assembly and Comparative Analysis of the Complete Mitochondrial of Spodiopogon sagittifolius, an Endemic and Protective Species from Yunnan, China. BMC Plant Biol. 2025, 25, 373. [Google Scholar] [CrossRef]
- Gao, C.; Wang, S.; Huang, Y.; Deng, Y. Assembly and Comparative Analysis of the Complete Mitochondrial Genome of Echinacanthus longipes (Acanthaceae), Endemic to the Sino-Vietnamese Karst Flora. BMC Genom. 2025, 26, 251. [Google Scholar] [CrossRef]
- Mower, J.P. Variation in Protein Gene and Intron Content among Land Plant Mitogenomes. Mitochondrion 2020, 53, 203–213. [Google Scholar] [CrossRef]
- Alverson, A.J.; Rice, D.W.; Dickinson, S.; Barry, K.; Palmer, J.D. Origins and Recombination of the Bacterial-Sized Multichromosomal Mitochondrial Genome of Cucumber. Plant Cell 2011, 23, 2499–2513. [Google Scholar] [CrossRef] [PubMed]
- Skippington, E.; Barkman, T.J.; Rice, D.W.; Palmer, J.D. Miniaturized Mitogenome of the Parasitic Plant Viscum scurruloideum Is Extremely Divergent and Dynamic and Has Lost All Nad Genes. Proc. Natl. Acad. Sci. USA 2015, 112, E3515–E3524. [Google Scholar] [CrossRef] [PubMed]
- Putintseva, Y.A.; Bondar, E.I.; Simonov, E.P.; Sharov, V.V.; Oreshkova, N.V.; Kuzmin, D.A.; Konstantinov, Y.M.; Shmakov, V.N.; Belkov, V.I.; Sadovsky, M.G.; et al. Siberian larch (Larix sibirica Ledeb.) Mitochondrial Genome Assembled Using Both Short and Long Nucleotide Sequence Reads Is Currently the Largest Known Mitogenome. BMC Genom. 2020, 21, 654. [Google Scholar] [CrossRef]
- Han, F.; Bi, C.; Zhao, Y.; Gao, M.; Wang, Y.; Chen, Y. Unraveling the Complex Evolutionary Features of the Cinnamomum camphora Mitochondrial Genome. Plant Cell Rep. 2024, 43, 183. [Google Scholar] [CrossRef] [PubMed]
- Dong, S.; Chen, L.; Liu, Y.; Wang, Y.; Zhang, S.; Yang, L.; Lang, X.; Zhang, S. The Draft Mitochondrial Genome of Magnolia biondii and Mitochondrial Phylogenomics of Angiosperms. PLoS ONE 2020, 15, e0231020. [Google Scholar] [CrossRef]
- Du, M.-Z.; Liu, S.; Zeng, Z.; Alemayehu, L.A.; Wei, W.; Guo, F.-B. Amino Acid Compositions Contribute to the Proteins’ Evolution under the Influence of Their Abundances and Genomic GC Content. Sci. Rep. 2018, 8, 7382. [Google Scholar] [CrossRef]
- Kubo, T.; Newton, K.J. Angiosperm Mitochondrial Genomes and Mutations. Mitochondrion 2008, 8, 5–14. [Google Scholar] [CrossRef]
- Jiang, M.; Ni, Y.; Zhang, J.; Li, J.; Liu, C. Complete Mitochondrial Genome of Mentha spicata L. Reveals Multiple Chromosomal Configurations and RNA Editing Events. Int. J. Biol. Macromol. 2023, 251, 126257. [Google Scholar] [CrossRef]
- Fajardo, D.; Schlautman, B.; Steffan, S.; Polashock, J.; Vorsa, N.; Zalapa, J. The American Cranberry Mitochondrial Genome Reveals the Presence of Selenocysteine (tRNA-Sec and SECIS) Insertion Machinery in Land Plants. Gene 2014, 536, 336–343. [Google Scholar] [CrossRef]
- Li, Y.; Hu, X.; Xiao, M.; Huang, J.; Lou, Y.; Hu, F.; Fu, X.; Li, Y.; He, H.; Cheng, J. An Analysis of Codon Utilization Patterns in the Chloroplast Genomes of Three Species of Coffea. BMC Genom. Data 2023, 24, 42. [Google Scholar] [CrossRef]
- Liu, D.; Guo, H.; Zhu, J.; Qu, K.; Chen, Y.; Guo, Y.; Ding, P.; Yang, H.; Xu, T.; Jing, Q.; et al. Complex Physical Structure of Complete Mitochondrial Genome of Quercus acutissima (Fagaceae): A Significant Energy Plant. Genes 2022, 13, 1321. [Google Scholar] [CrossRef] [PubMed]
- Parvathy, S.T.; Udayasuriyan, V.; Bhadana, V. Codon Usage Bias. Mol. Biol. Rep. 2022, 49, 539–565. [Google Scholar] [CrossRef] [PubMed]
- Bock, H.; Brennicke, A.; Schuster, W. Rps3 and Rpll6 Genes Do Not Overlap in Oenothera Mitochondria: GTG as a Potential Translation Initiation Codon in Plant Mitochondria? Plant Mol. Biol. 1994, 24, 811–818. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Cui, G.; He, K.; Xu, K.; Liu, W.; Wang, Y.; Wang, Z.; Liu, S.; Bi, C. Assembly and Comparative Analysis of the Complete Mitochondrial Genome of Ilex rotunda Thunb. Forests 2024, 15, 1117. [Google Scholar] [CrossRef]
- Sun, N.; Han, F.; Wang, S.; Shen, F.; Liu, W.; Fan, W.; Bi, C. Comprehensive Analysis of the Lycopodium japonicum Mitogenome Reveals Abundant tRNA Genes and Cis-Spliced Introns in Lycopodiaceae Species. Front. Plant Sci. 2024, 15, 1446015. [Google Scholar] [CrossRef]
- Liu, Q.; Wu, Z.; Tian, C.; Yang, Y.; Liu, L.; Feng, Y.; Li, Z. Complete Mitochondrial Genome of the Endangered Prunus pedunculata (Prunoideae, Rosaceae) in China: Characterization and Phylogenetic Analysis. Front. Plant Sci. 2023, 14, 1266797. [Google Scholar] [CrossRef]
- Zhang, Y.; Guo, X.; Ma, R.; Yu, M.; Xu, J.; Shen, Z. Assembly and Analysis of the Complete Mitochondrial Genome of Prunus davidiana (Rosaceae). Front. Plant Sci. 2025, 16, 1558619. [Google Scholar] [CrossRef]
- Sloan, D.B.; Wu, Z.; Sharbrough, J. Correction of Persistent Errors in Arabidopsis Reference Mitochondrial Genomes. Plant Cell 2018, 30, 525–527. [Google Scholar] [CrossRef]
- Bi, C.; Lu, N.; Xu, Y.; He, C.; Lu, Z. Characterization and Analysis of the Mitochondrial Genome of Common Bean (Phaseolus vulgaris) by Comparative Genomic Approaches. Int. J. Mol. Sci. 2020, 21, 3778. [Google Scholar] [CrossRef]
- Sun, S.; Xiao, N.; Sha, Z. Complete mitochondrial genomes of four deep-sea echinoids: Conserved mitogenome organization and new insights into the phylogeny and evolution of Echinoidea. PeerJ. 2022, 10, e13730. [Google Scholar] [CrossRef]
- Wang, D.; Liu, F.; Wang, L.; Huang, S.; Yu, J. Nonsynonymous Substitution Rate (Ka) Is a Relatively Consistent Parameter for Defining Fast-Evolving and Slow-Evolving Protein-Coding Genes. Biol. Direct 2011, 6, 13. [Google Scholar] [CrossRef]
- Zhou, P.; Zhang, Q.; Li, F.; Huang, J.; Zhang, M. Assembly and Comparative Analysis of the Complete Mitochondrial Genome of Ilex metabaptista (Aquifoliaceae), a Chinese Endemic Species with a Narrow Distribution. BMC Plant Biol. 2023, 23, 393. [Google Scholar] [CrossRef]
- Li, Z.-Z.; Wang, Y.; He, X.-Y.; Li, W.-G. The Taihangia Mitogenome Provides New Insights into Its Adaptation and Organelle Genome Evolution in Rosaceae. Planta 2025, 261, 59. [Google Scholar] [CrossRef] [PubMed]
- Ma, Q.; Wang, Y.; Li, S.; Wen, J.; Zhu, L.; Yan, K.; Du, Y.; Ren, J.; Li, S.; Chen, Z.; et al. Assembly and Comparative Analysis of the First Complete Mitochondrial Genome of Acer truncatum Bunge: A Woody Oil-Tree Species Producing Nervonic Acid. BMC Plant Biol. 2022, 22, 29. [Google Scholar] [CrossRef]
- Christensen, A.C. Plant Mitochondrial Genome Evolution Can Be Explained by DNA Repair Mechanisms. Genome Biol. Evol. 2013, 5, 1079–1086. [Google Scholar] [CrossRef] [PubMed]
- Yisilam, G.; Liu, Z.; Turdi, R.; Chu, Z.; Luo, W.; Tian, X. Assembly and Comparative Analysis of the Complete Mitochondrial Genome of Isopyrum anemonoides (Ranunculaceae). PLoS ONE 2023, 18, e0286628. [Google Scholar] [CrossRef]
- Yong, Y.; Wang, Y.; Wang, D.; Yuan, X.; Zhang, Q. The Organelle Genomes of the Endangered Seagrass Zostera caespitosa Reveal Sequence Divergences, Massive Gene Transfer, and Uncommon RNA Editing Types. Front. Plant Sci. 2025, 16, 1550467. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.-C.; Chen, H.; Yang, D.; Liu, C. Diversity of Mitochondrial Plastid DNAs (MTPTs) in Seed Plants. Mitochondrial DNA Part A 2018, 29, 635–642. [Google Scholar] [CrossRef]
- Bi, C.; Paterson, A.H.; Wang, X.; Xu, Y.; Wu, D.; Qu, Y.; Jiang, A.; Ye, Q.; Ye, N. Analysis of the Complete Mitochondrial Genome Sequence of the Diploid Cotton Gossypium raimondii by Comparative Genomics Approaches. BioMed Res. Int. 2016, 2016, 5040598. [Google Scholar] [CrossRef]
- Gao, C.; Ren, X.; Mason, A.S.; Liu, H.; Xiao, M.; Li, J.; Fu, D. Horizontal Gene Transfer in Plants. Funct. Integr. Genom. 2014, 14, 23–29. [Google Scholar] [CrossRef]
- Clifton, S.W.; Minx, P.; Fauron, C.M.-R.; Gibson, M.; Allen, J.O.; Sun, H.; Thompson, M.; Barbazuk, W.B.; Kanuganti, S.; Tayloe, C.; et al. Sequence and Comparative Analysis of the Maize NB Mitochondrial Genome. Plant Physiol. 2004, 136, 3486–3503. [Google Scholar] [CrossRef] [PubMed]
- Unseld, M.; Marienfeld, J.R.; Brandt, P.; Brennicke, A. The Mitochondrial Genome of Arabidopsis thaliana Contains 57 Genes in 366,924 Nucleotides. Nat. Genet. 1997, 15, 57–61. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.-X.; Li, Z.-H.; Schuiteman, A.; Chase, M.W.; Li, J.-W.; Huang, W.-C.; Hidayat, A.; Wu, S.-S.; Jin, X.-H. Phylogenomics of Orchidaceae Based on Plastid and Mitochondrial Genomes. Mol. Phylogenetics Evol. 2019, 139, 106540. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Ferguson, D.K.; Yang, Y. New Insights into the Plastome Evolution of Lauraceae Using Herbariomics. BMC Plant Biol. 2023, 23, 387. [Google Scholar] [CrossRef]
- Liu, C.; Chen, H.-H.; Tang, L.-Z.; Khine, P.K.; Han, L.-H.; Song, Y.; Tan, Y.-H. Plastid Genome Evolution of a Monophyletic Group in the Subtribe lauriineae (Laureae, Lauraceae). Plant Divers. 2022, 44, 377–388. [Google Scholar] [CrossRef]
- Dong, W.; Xu, C.; Cheng, T.; Lin, K.; Zhou, S. Sequencing Angiosperm Plastid Genomes Made Easy: A Complete Set of Universal Primers and a Case Study on the Phylogeny of Saxifragales. Genome Biol. Evol. 2013, 5, 989–997. [Google Scholar] [CrossRef] [PubMed]
- Qiu, Y.-L.; Li, L.; Wang, B.; Xue, J.-Y.; Hendry, T.A.; Li, R.-Q.; Brown, J.W.; Liu, Y.; Hudson, G.T.; Chen, Z.-D. Angiosperm Phylogeny Inferred from Sequences of Four Mitochondrial Genes. J. Syst. Evol. 2010, 48, 391–425. [Google Scholar] [CrossRef]
- Pearl, S.A.; Welch, M.E.; McCauley, D.E. Mitochondrial Heteroplasmy and Paternal Leakage in Natural Populations of Silene vulgaris, a Gynodioecious Plant. Mol. Biol. Evol. 2009, 26, 537–545. [Google Scholar] [CrossRef][Green Version]
- Howarth, D.G.; Gardner, D.E.; Morden, C.W. Phylogeny of Rubus subgenus Idaeobatus (Rosaceae) and Its Implications Toward Colonization of the Hawaiian Islands. Syst. Bot. 1997, 22, 433–441. [Google Scholar] [CrossRef]
- Wang, X.; Wang, J.; Jin, D.; Guo, H.; Lee, T.-H.; Liu, T.; Paterson, A.H. Genome Alignment Spanning Major Poaceae Lineages Reveals Heterogeneous Evolutionary Rates and Alters Inferred Dates for Key Evolutionary Events. Mol. Plant 2015, 8, 885–898. [Google Scholar] [CrossRef]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shi, Y.; Chen, Z.; Jiang, J.; Li, Q.; Zeng, W. De Novo Assembly and Comparative Analysis of the Mitochondrial Genomes for Six Rubus Species. Horticulturae 2025, 11, 559. https://doi.org/10.3390/horticulturae11050559
Shi Y, Chen Z, Jiang J, Li Q, Zeng W. De Novo Assembly and Comparative Analysis of the Mitochondrial Genomes for Six Rubus Species. Horticulturae. 2025; 11(5):559. https://doi.org/10.3390/horticulturae11050559
Chicago/Turabian StyleShi, Yujie, Zhen Chen, Jingyong Jiang, Qianfan Li, and Wei Zeng. 2025. "De Novo Assembly and Comparative Analysis of the Mitochondrial Genomes for Six Rubus Species" Horticulturae 11, no. 5: 559. https://doi.org/10.3390/horticulturae11050559
APA StyleShi, Y., Chen, Z., Jiang, J., Li, Q., & Zeng, W. (2025). De Novo Assembly and Comparative Analysis of the Mitochondrial Genomes for Six Rubus Species. Horticulturae, 11(5), 559. https://doi.org/10.3390/horticulturae11050559





