Lightweight CNN–Transformer Hybrid Network with Contrastive Learning for Few-Shot Noxious Weed Recognition
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Collection
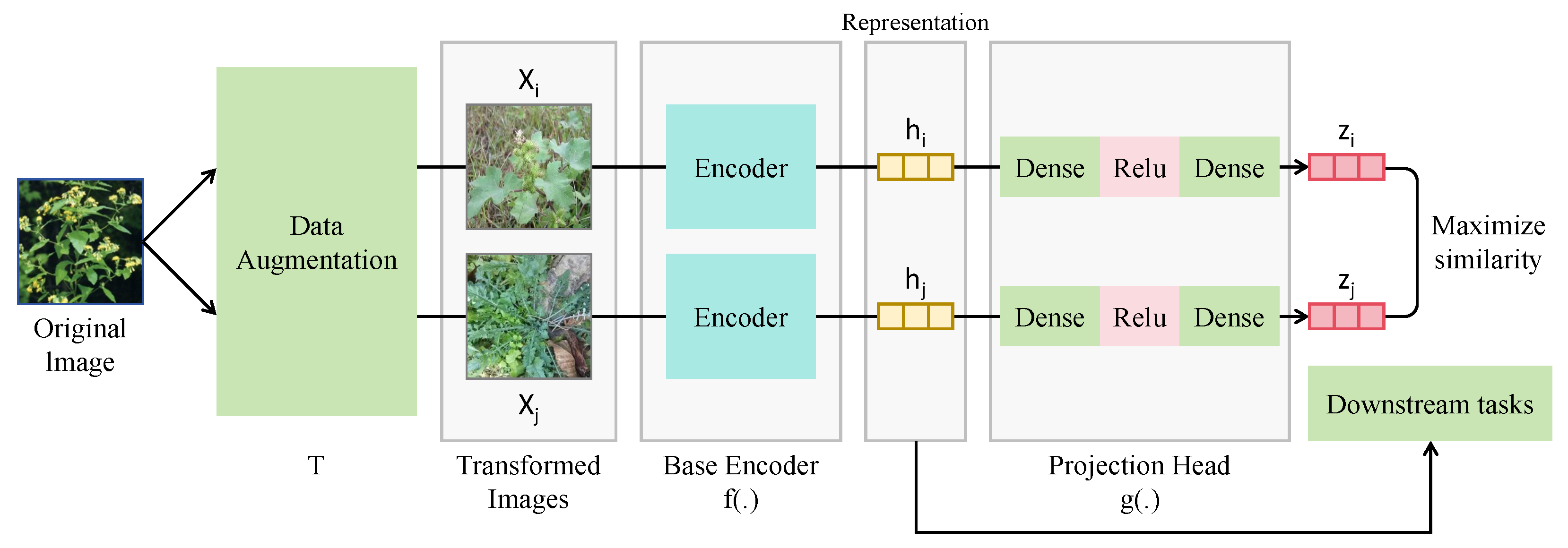
2.2. Data Augmentation
2.3. Proposed Method
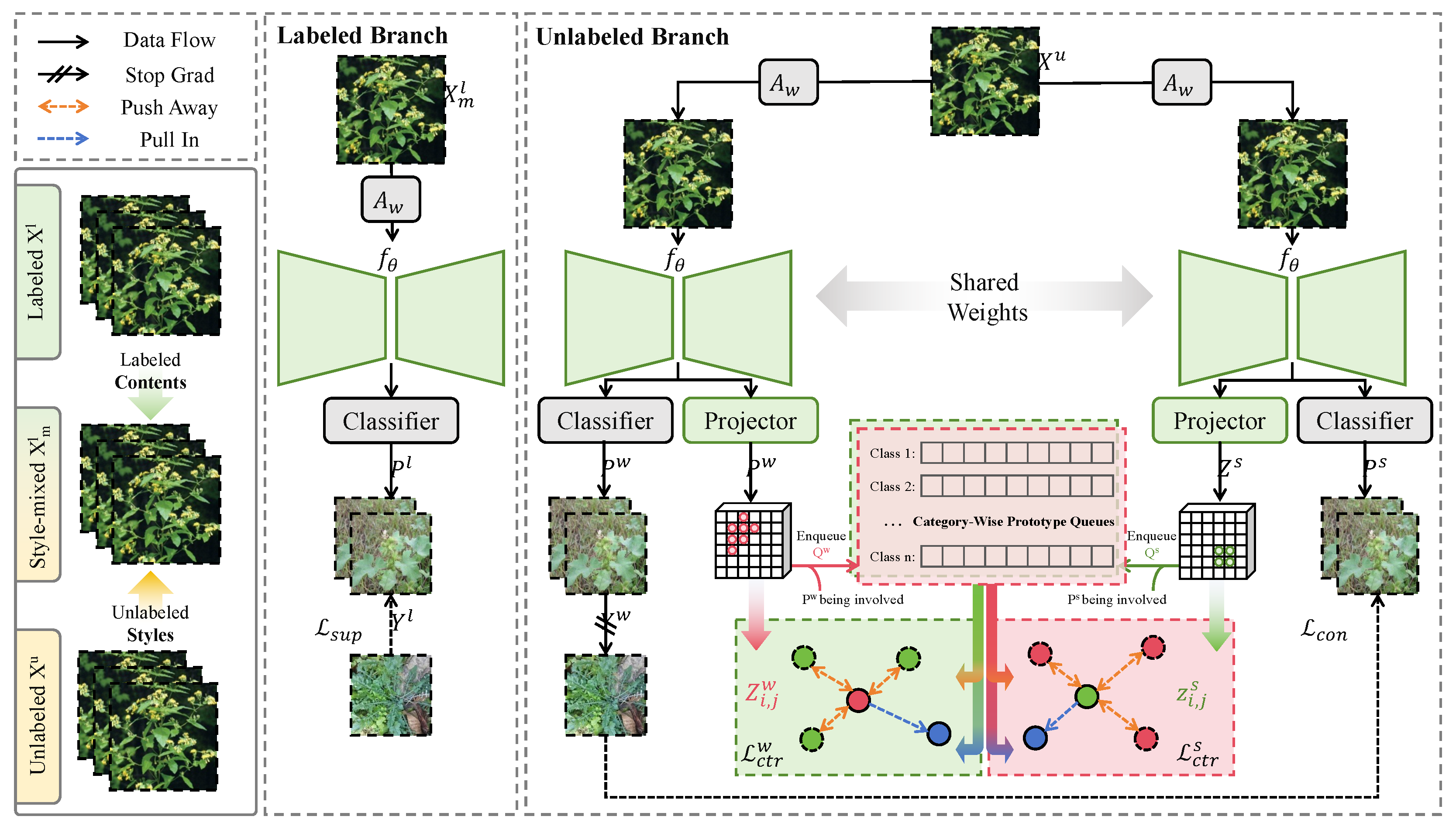
2.3.1. Overall
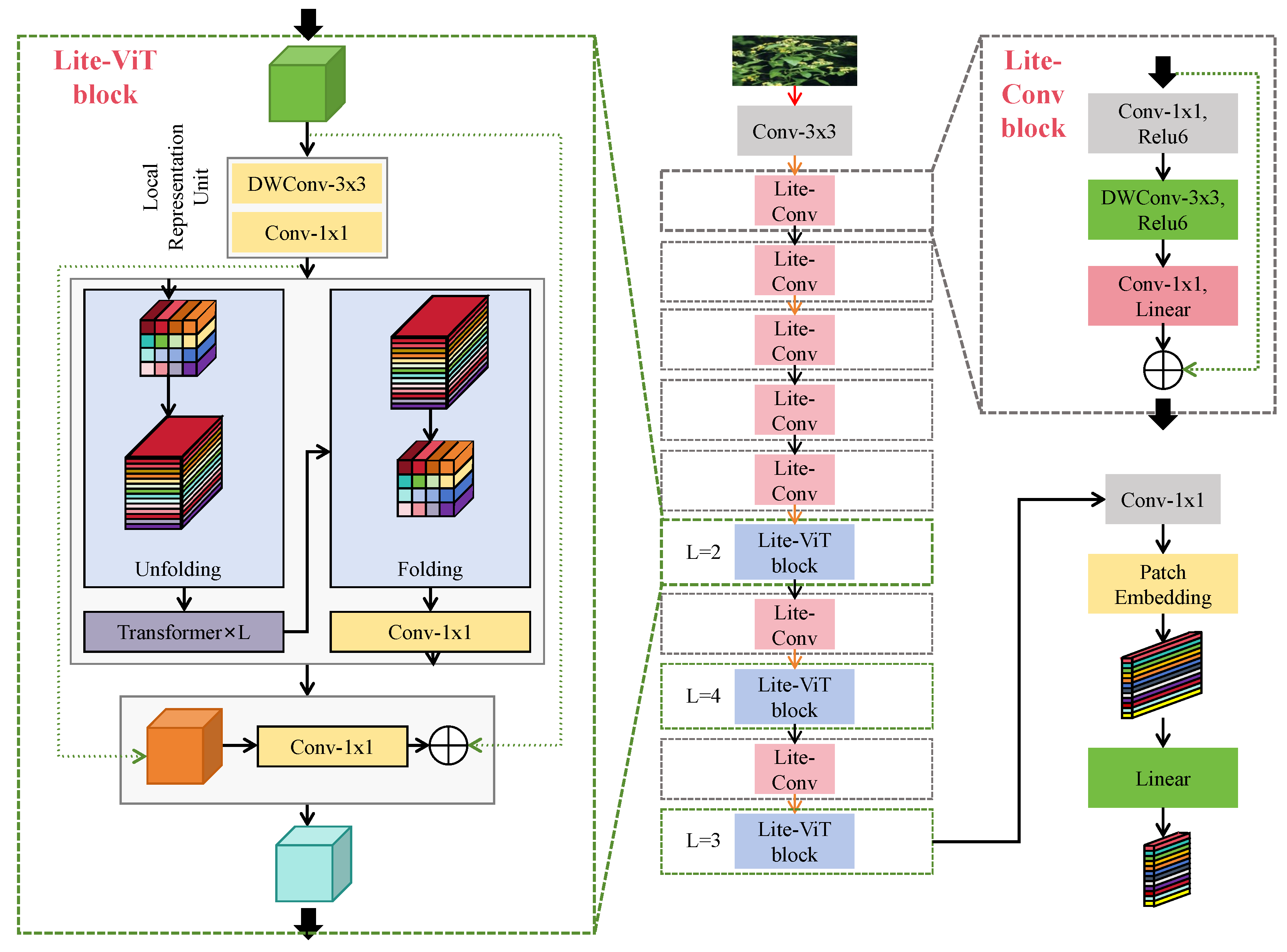
2.3.2. Lightweight Hybrid Backbone
2.3.3. Pseudo-Label Generator
2.3.4. Contrastive and Boundary Enhancement
3. Results
3.1. Experimental Setup
3.1.1. Experimental Platform and Hyperparameter Settings
3.1.2. Baseline
3.1.3. Evaluation Metrics
3.2. Classification Performance Comparison of Different Models
3.3. Model Efficiency Comparison on Jetson Nano
3.4. Performance on Rare Weed Categories Under Few-Shot Setting
4. Discussion
4.1. Practical Deployment Analysis
- Stellera chamaejasme (wolf poison): One of the most destructive alpine toxic weeds with massive root systems and high aboveground biomass. It contains potent diterpenoid toxins and exhibits aggressive expansion, competing fiercely with high-quality forage.
- Aconitum gymnandrum (aconite): Contains highly toxic aconitine alkaloids that can cause livestock death even upon minimal ingestion.
- Pedicularis kansuensis: A semi-parasitic species that thrives in degraded meadows; it is non-lethal but has extremely poor palatability and strong competitive ability.
- Astragalus adsurgens: A leguminous poisonous weed that contains cyanogenic glycosides, inducing chronic poisoning if overgrazed.
- Cicuta virosa (water hemlock): Distributed in meadow wetland margins; its tuber contains cicutoxin, a powerful neurotoxin that causes respiratory paralysis.
4.2. Limitations and Future Work
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Jin, X.; Che, J.; Chen, Y. Weed identification using deep learning and image processing in vegetable plantation. IEEE Access 2021, 9, 10940–10950. [Google Scholar] [CrossRef]
- Wang, S.; Cheng, W.; Tan, H.; Guo, B.; Han, X.; Wu, C.; Yang, D. Study on Diversity of Poisonous Weeds in Grassland of the Ili Region in Xinjiang. Agronomy 2024, 14, 330. [Google Scholar] [CrossRef]
- Mishra, A.M.; Gautam, V. Weed Species Identification in Different Crops Using Precision Weed Management: A Review. In Proceedings of the ISIC, New Delhi, India, 25–27 February 2021; pp. 180–194. [Google Scholar]
- Coleman, G.R.; Bender, A.; Hu, K.; Sharpe, S.M.; Schumann, A.W.; Wang, Z.; Bagavathiannan, M.V.; Boyd, N.S.; Walsh, M.J. Weed detection to weed recognition: Reviewing 50 years of research to identify constraints and opportunities for large-scale cropping systems. Weed Technol. 2022, 36, 741–757. [Google Scholar] [CrossRef]
- Sinlae, A.A.J.; Alamsyah, D.; Suhery, L.; Fatmayati, F. Classification of Broadleaf Weeds Using a Combination of K-Nearest Neighbor (KNN) and Principal Component Analysis (PCA). Sink. J. Dan Penelit. Tek. Inform. 2021, 6, 93–100. [Google Scholar]
- Chen, S.; Memon, M.S.; Shen, B.; Guo, J.; Du, Z.; Tang, Z.; Guo, X.; Memon, H. Identification of weeds in cotton fields at various growth stages using color feature techniques. Ital. J. Agron. 2024, 19, 100021. [Google Scholar] [CrossRef]
- Janneh, L.L.; Zhang, Y.; Cui, Z.; Yang, Y. Multi-level feature re-weighted fusion for the semantic segmentation of crops and weeds. J. King Saud Univ.-Comput. Inf. Sci. 2023, 35, 101545. [Google Scholar] [CrossRef]
- Zhang, Y.; Wa, S.; Liu, Y.; Zhou, X.; Sun, P.; Ma, Q. High-accuracy detection of maize leaf diseases CNN based on multi-pathway activation function module. Remote Sens. 2021, 13, 4218. [Google Scholar]
- Zhou, X.; Chen, S.; Ren, Y.; Zhang, Y.; Fu, J.; Fan, D.; Lin, J.; Wang, Q. Atrous Pyramid GAN Segmentation Network for Fish Images with High Performance. Electronics 2022, 11, 911. [Google Scholar] [CrossRef]
- Espejo-Garcia, B.; Mylonas, N.; Athanasakos, L.; Vali, E.; Fountas, S. Combining generative adversarial networks and agricultural transfer learning for weeds identification. Biosyst. Eng. 2021, 204, 79–89. [Google Scholar] [CrossRef]
- Shao, Y.; Guan, X.; Xuan, G.; Gao, F.; Feng, W.; Gao, G.; Wang, Q.; Huang, X.; Li, J. GTCBS-YOLOv5s: A lightweight model for weed species identification in paddy fields. Comput. Electron. Agric. 2023, 215, 108461. [Google Scholar] [CrossRef]
- Islam, M.D.; Liu, W.; Izere, P.; Singh, P.; Yu, C.; Riggan, B.; Zhang, K.; Jhala, A.J.; Knezevic, S.; Ge, Y.; et al. Towards real-time weed detection and segmentation with lightweight CNN models on edge devices. Comput. Electron. Agric. 2025, 237, 110600. [Google Scholar] [CrossRef]
- Hu, R.; Su, W.H.; Li, J.L.; Peng, Y. Real-time lettuce-weed localization and weed severity classification based on lightweight YOLO convolutional neural networks for intelligent intra-row weed control. Comput. Electron. Agric. 2024, 226, 109404. [Google Scholar] [CrossRef]
- Muthulakshmi, M.; Sharvani, S.; Varshini, S.; MS, S. Weed Crop Identification and Classification Using Transfer Learning with Variants of MobileNet and DenseNet. In Proceedings of the 2025 International Conference in Advances in Power, Signal, and Information Technology (APSIT), Bhubaneswar, India, 23–25 May 2025; pp. 1–6. [Google Scholar]
- Liu, Z.; Jung, C. Deep sparse depth completion using multi-scale residuals and channel shuffle. IEEE Access 2024, 12, 18189–18197. [Google Scholar] [CrossRef]
- Liu, X.; Sui, Q.; Chen, Z. Real time weed identification with enhanced mobilevit model for mobile devices. Sci. Rep. 2025, 15, 27323. [Google Scholar] [CrossRef]
- Chen, D.; Lu, Y.; Li, Z.; Young, S. Performance evaluation of deep transfer learning on multi-class identification of common weed species in cotton production systems. Comput. Electron. Agric. 2022, 198, 107091. [Google Scholar] [CrossRef]
- Fang, B.; Li, X.; Han, G.; He, J. Rethinking pseudo-labeling for semi-supervised facial expression recognition with contrastive self-supervised learning. IEEE Access 2023, 11, 45547–45558. [Google Scholar] [CrossRef]
- Mishra, A.M.; Kaur, P.; Singh, M.P.; Singh, S.P. A self-supervised overlapped multiple weed and crop leaf segmentation approach under complex light condition. Multimed. Tools Appl. 2024, 83, 68993–69018. [Google Scholar] [CrossRef]
- Cascante-Bonilla, P.; Tan, F.; Qi, Y.; Ordonez, V. Curriculum labeling: Revisiting pseudo-labeling for semi-supervised learning. In Proceedings of the AAAI Conference on Artificial Intelligence, Virtually, 2–9 February 2021; Volume 35, pp. 6912–6920. [Google Scholar]
- Güldenring, R.; Nalpantidis, L. Self-supervised contrastive learning on agricultural images. Comput. Electron. Agric. 2021, 191, 106510. [Google Scholar] [CrossRef]
- Ran, L.; Li, Y.; Liang, G.; Zhang, Y. Pseudo labeling methods for semi-supervised semantic segmentation: A review and future perspectives. IEEE Trans. Circuits Syst. Video Technol. 2024, 35, 3054–3080. [Google Scholar] [CrossRef]
- Lin, X.; Wa, S.; Zhang, Y.; Ma, Q. A dilated segmentation network with the morphological correction method in farming area image Series. Remote Sens. 2022, 14, 1771. [Google Scholar] [CrossRef]
- Kage, P.; Rothenberger, J.C.; Andreadis, P.; Diochnos, D.I. A review of pseudo-labeling for computer vision. arXiv 2024, arXiv:2408.07221. [Google Scholar] [CrossRef]
- Guo, Z.; Xue, Y.; Wang, C.; Geng, Y.; Lu, R.; Li, H.; Sun, D.; Lou, Z.; Chen, T.; Shi, J.; et al. Efficient weed segmentation in maize fields: A semi-supervised approach for precision weed management with reduced annotation overhead. Comput. Electron. Agric. 2025, 229, 109707. [Google Scholar] [CrossRef]
- Li, R.; Wang, X.; Cui, Y.; Xu, Y.; Zhou, Y.; Tang, X.; Jiang, C.; Song, Y.; Dong, H.; Yan, S. A Semi-Supervised Diffusion-Based Framework for Weed Detection in Precision Agricultural Scenarios Using a Generative Attention Mechanism. Agriculture 2025, 15, 434. [Google Scholar] [CrossRef]
- Li, J.; Chen, D.; Yin, X.; Li, Z. Performance evaluation of semi-supervised learning frameworks for multi-class weed detection. Front. Plant Sci. 2024, 15, 1396568. [Google Scholar] [CrossRef]
- Huang, Y.; Bais, A. Unsupervised domain adaptation for weed segmentation using greedy pseudo-labelling. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Seattle, WA, USA, 16–22 June 2024; pp. 2484–2494. [Google Scholar]
- Zhang, L.; Zhang, Y.; Ma, X. A new strategy for tuning ReLUs: Self-adaptive linear units (SALUs). In Proceedings of the ICMLCA 2021; 2nd International Conference on Machine Learning and Computer Application, Shenyang, China, 17–19 December 2021; 2021; pp. 1–8. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Sandler, M.; Howard, A.; Zhu, M.; Zhmoginov, A.; Chen, L.C. Mobilenetv2: Inverted residuals and linear bottlenecks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–22 June 2018; pp. 4510–4520. [Google Scholar]
- Szegedy, C.; Liu, W.; Jia, Y.; Sermanet, P.; Reed, S.E.; Anguelov, D.; Erhan, D.; Vanhoucke, V.; Rabinovich, A. Going Deeper with Convolutions. arXiv 2014, arXiv:1409.4842. [Google Scholar] [CrossRef]
- Mehta, S.; Rastegari, M. Mobilevit: Light-weight, general-purpose, and mobile-friendly vision transformer. arXiv 2021, arXiv:2110.02178. [Google Scholar]
- Zhang, X.; Zhou, X.; Lin, M.; Sun, J. Shufflenet: An extremely efficient convolutional neural network for mobile devices. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–22 June 2018; pp. 6848–6856. [Google Scholar]
- Pinasthika, K.; Laksono, B.S.P.; Irsal, R.B.P.; Shabiyya, S.; Yudistira, N. SparseSwin: Swin transformer with sparse transformer block. Neurocomputing 2024, 580, 127433. [Google Scholar] [CrossRef]
- Cui, J.; Zhang, Y.; Chen, H.; Zhang, Y.; Cai, H.; Jiang, Y.; Ma, R.; Qi, L. CSWin-MBConv: A dual-network fusing CNN and Transformer for weed recognition. Eur. J. Agron. 2025, 164, 127528. [Google Scholar] [CrossRef]




| Weed Species | Close-Up Images | Wide-Angle Images | UAV Images | Public Dataset Supplements |
|---|---|---|---|---|
| Datura stramonium | 230 | 180 | 140 | 250 |
| Xanthium sibiricum | 220 | 175 | 135 | 260 |
| Solanum nigrum | 225 | 180 | 130 | 255 |
| Alopecurus aequalis | 215 | 190 | 145 | 250 |
| Stellera chamaejasme | 210 | 185 | 150 | 255 |
| Echinochloa crus-galli | 235 | 175 | 135 | 255 |
| Portulaca oleracea | 228 | 182 | 140 | 250 |
| Setaria viridis | 230 | 178 | 145 | 247 |
| Sonchus oleraceus | 232 | 180 | 138 | 250 |
| Eleusine indica | 226 | 176 | 142 | 246 |
| Total | 2271 | 1791 | 1400 | 2568 |
| Grand Total | 7030 | |||
| Model | Precision (%) | Recall (%) | F1-Score (%) | Accuracy (%) |
|---|---|---|---|---|
| ResNet18 | 85.42 ± 0.35 | 83.67 ± 0.41 | 84.53 ± 0.38 | 84.91 ± 0.33 |
| MobileNetV2 | 86.15 ± 0.28 | 82.04 ± 0.36 | 84.04 ± 0.32 | 84.38 ± 0.30 |
| GoogLeNet | 83.19 ± 0.42 | 80.37 ± 0.39 | 81.76 ± 0.41 | 82.11 ± 0.35 |
| MobileViT | 87.28 ± 0.31 | 85.51 ± 0.34 | 86.38 ± 0.29 | 86.02 ± 0.27 |
| ShuffleNet | 84.92 ± 0.37 | 81.63 ± 0.40 | 83.24 ± 0.36 | 83.51 ± 0.33 |
| SparseSwin | 88.02 ± 0.29 | 86.73 ± 0.33 | 87.37 ± 0.31 | 87.12 ± 0.28 |
| CSWin-MBConvand | 88.56 ± 0.27 | 87.04 ± 0.30 | 87.79 ± 0.29 | 87.51 ± 0.27 |
| Ours | 89.64 ± 0.25 | 87.91 ± 0.28 | 88.76 ± 0.27 | 88.43 ± 0.26 |
| Model | Size (MB) | FLOPs (G) | FPS | Latency (ms) | Power (W) |
|---|---|---|---|---|---|
| ResNet18 | 44.6 | 1.82 | 11.3 | 88.5 | 6.2 |
| MobileNetV2 | 14.0 | 0.31 | 14.9 | 67.1 | 5.4 |
| GoogLeNet | 22.5 | 0.72 | 16.7 | 59.9 | 5.9 |
| MobileViT | 26.3 | 0.98 | 13.2 | 75.7 | 6.0 |
| ShuffleNet | 10.3 | 0.26 | 17.4 | 57.5 | 4.8 |
| SparseSwin | 24.7 | 0.65 | 15.8 | 63.3 | 5.6 |
| CSWin-MBConv | 21.9 | 0.58 | 16.2 | 61.7 | 5.3 |
| Ours | 18.6 | 0.41 | 18.9 | 52.9 | 5.1 |
| Method | Precision (%) | Recall (%) | F1-Score (%) | Accuracy (%) |
|---|---|---|---|---|
| Ours (w/o PL) | 67.66 ± 0.72 | 64.52 ± 0.81 | 66.06 ± 0.76 | 65.87 ± 0.69 |
| Ours (w/o CL) | 68.30 ± 0.68 | 65.41 ± 0.79 | 66.83 ± 0.73 | 66.63 ± 0.65 |
| Ours (CNN only) | 69.57 ± 0.74 | 66.26 ± 0.83 | 67.88 ± 0.77 | 67.83 ± 0.71 |
| Ours (full) | 82.40 ± 0.52 | 78.52 ± 0.61 | 80.41 ± 0.58 | 80.32 ± 0.54 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, R.; Yu, B.; Zhang, B.; Ma, H.; Qin, Y.; Lv, X.; Yan, S. Lightweight CNN–Transformer Hybrid Network with Contrastive Learning for Few-Shot Noxious Weed Recognition. Horticulturae 2025, 11, 1236. https://doi.org/10.3390/horticulturae11101236
Li R, Yu B, Zhang B, Ma H, Qin Y, Lv X, Yan S. Lightweight CNN–Transformer Hybrid Network with Contrastive Learning for Few-Shot Noxious Weed Recognition. Horticulturae. 2025; 11(10):1236. https://doi.org/10.3390/horticulturae11101236
Chicago/Turabian StyleLi, Ruiheng, Boda Yu, Boming Zhang, Hongtao Ma, Yihan Qin, Xinyang Lv, and Shuo Yan. 2025. "Lightweight CNN–Transformer Hybrid Network with Contrastive Learning for Few-Shot Noxious Weed Recognition" Horticulturae 11, no. 10: 1236. https://doi.org/10.3390/horticulturae11101236
APA StyleLi, R., Yu, B., Zhang, B., Ma, H., Qin, Y., Lv, X., & Yan, S. (2025). Lightweight CNN–Transformer Hybrid Network with Contrastive Learning for Few-Shot Noxious Weed Recognition. Horticulturae, 11(10), 1236. https://doi.org/10.3390/horticulturae11101236









