Abstract
Light plays an essential role in regulating the growth, development, and metabolic activities of the edible mushroom Pleurotus ostreatus. In this research, the influence of white, blue, green, yellow, and red light, and darkness, on the global protein expression of P. ostreatus LGAM 1123 grown in submerged culture was explored. The growth of the fungus was not inhibited by light in any of the conditions tested compared with the dark. However, the mycelial protein content was reduced by 10% under blue and white light. Proteomic analysis revealed distinct proteomes for each light wavelength, with red and blue light presenting the most distinctive proteome profiles. (Data are available via ProteomeXchange with identifier PXD065402.) Blue light activates pathways such as the citrate cycle (TCA cycle), glycolysis/gluconeogenesis, and amino acid biosynthesis, while red light stimulates mRNA-related pathways. GC-MS analysis of the biomass revealed differences in the amino acids, sugars, and lipids produced. The distinct regulation of proteins and bioactive compounds under different light wavelengths suggests that specific wavelengths can direct the metabolism of P. ostreatus into biochemical pathways. These strategies could be beneficial for the food industry because particular nutrients can be increased during the fermentation of edible fungi without the need for genetic engineering of the strain.
1. Introduction
Autotrophic organisms use light for photosynthesis, while heterotrophic organisms use light as a source of information. Fungi can sense light using up to 11 photoreceptors and can alter the metabolism of carotenoids, polysaccharides, carbohydrates, fatty acids, and nucleotides [1,2]. They can sense red, far-red, blue, green, and near-ultraviolet light and regulate processes such as spore germination, phototropism, nutrient uptake, secondary metabolism, and the circadian clock [2]. Blue light receptors are separated into three categories: the cryptochrome and photolyase protein family; the blue-light sensor using FAD (BLUF) proteins; and the light, oxygen, and voltage (LOV) domain-containing proteins such as white collar 1 (WC-1) and white collar 2 (WC-2) or vivid (VVD) proteins. Transcription regulation in the nucleus is directly mediated by blue light photoreceptors. Opsins, retinal-binding proteins, can sense green light, while phytochromes, proteins with a linear tetrapyrrole, are the photoreceptors of red light [2]. Fungi not only sense light as a binary system to induce or suppress a function, but they can also sense the ratio of light wavelengths they receive and understand the time of day. It is well established that the habitat plays a crucial role in light perception. For instance, green light predominates in forest canopy environments, prompting fungi and plant pathogens to upregulate opsin proteins in response. Similarly, red light penetrates soil more effectively than blue light, influencing microbial and plant interactions belowground [2,3].
A recent review study comprehensively summarizes the influence of light on both preharvest and post-harvest stages in the production of edible mushrooms, highlighting its critical role in optimizing cultivation outcomes [4]. They concluded that for preharvest procedures, different wavelengths of ultraviolet, especially blue, improve the growth characteristics and nutritional value of mushrooms. Meanwhile, inhibition of microbial growth and regulation of physiological and energy metabolism can be achieved by using pulsed light and visible light (which enhances flavor), ultraviolet light (which improves nutrients and bioactive compounds), and γ-irradiation after the harvest of the mushrooms [4]. Additionally, blue light has been extensively studied for its ability to induce growth and morphological changes. Lentinula edodes cultivated under blue light and lightless conditions revealed a different proteome in each case, with 14 proteins downregulated and 8 proteins upregulated when blue light was used [5]. A similar study to ours, but with different species of Basidiomycetes, was conducted for Hypsizygus marmoreus in response to different light qualities. After a proteomic analysis, white light showed the most differentiated proteins, and they concluded that light stress was associated with primary metabolism [6]. For a Pleurotus genus strain, P. eryngii, it has also been confirmed that blue light induces mushroom primordium differentiation and fruiting body development via the regulation of glycolysis and gluconeogenesis, carbon metabolism, and biosynthesis of amino acid pathways [7]. Blue light upregulates both glycolysis and the pentose phosphate pathway in the pileus growth of Pleurotus ostreatus, with the three main organs of the mushroom growing better under blue light. Red light was found to slightly inhibit P. ostreatus pileus growth since it downregulates genes related to respiration metabolism [8]. Concerning P. ostreatus, two photoreceptor homologs induced by blue light have already been isolated: White Collar 1 (PoWC-1), and White Collar 2 (PoWC-2). Photoreceptors for other light wavelengths, such as opsins for green light and phytochromes for red light, have not yet been isolated.
Pleurotus ostreatus LGAM 1123 has already been studied by our team, and an optimization process revealed that the carbon and nitrogen sources mainly affect the biomass and protein production of the strain. The optimized glucose and yeast extract concentrations were found to be 54 g/L and 18 g/L, respectively [9]. Furthermore, glucose and xylose mixtures were used for the submerged cultivation of the strain and revealed an apparent clustering of specific amino acids with glucose, xylose, or mixtures of them [10]. In these two studies, agro-industrial residues were used in stirred tank bioreactors to produce high-protein biomass at low cost [9,10]. Additionally, a proteomic analysis of the produced cells under different glucose and xylose ratios in culture conditions revealed that the P. ostreatus proteome differs in each cultivation condition. Xylose was found to upregulate the tricarboxylic acid cycle (TCA), while glucose upregulates glycolysis [11]. In addition, high protein biomass and high laccase production have been accomplished by this strain via submerged fermentation using a wine industry sidestream, wine lees [12]. Finally, the same strain was used by our team to produce exopolysaccharides using red and green light in a 3.5 L stirred tank bioreactor [13]. Zerva et al. have shown that the same strain can grow and degrade olive mill wastewater, while the biomass was richer in total glucan when a semi-synthetic medium was used compared with the olive mill wastewater-based medium [14].
This work involves a comparative proteomic investigation of P. ostreatus LGAM 1123 mycelia grown in submerged culture under continuous illumination (white, blue, green, yellow, and red light) compared with dark. Mycelia were harvested during the mid-exponential phase of submerged cultures. A specific cell lysis and digestion procedure was adopted for the proteomic analysis of the strain using LC-MS/MS. A global protein network analysis using STRING was conducted on all differentially expressed proteins to obtain more protein information, and upregulated proteins were annotated through KEGG pathway mapping. Furthermore, the cells collected from the cultures were acid-hydrolyzed for nutrient analysis of the biomass, focusing on amino acids, sugars, and low-molecular-weight lipids, using GC-MS.
2. Materials and Methods
2.1. Chemicals and Reagents
Analytical-grade chemicals were utilized throughout this study. Yeast extract and potato dextrose agar (PDA) were sourced from Neogen Europe Ltd. (Ayr, UK). Various reagents, including N,O-bis(trimethylsilyl)trifluoroacetamide with 1% trimethylchlorosilane (BSTFA + 1% TMCS, analytical standard), zinc sulfate heptahydrate, glucose, manganese (II) sulfate heptahydrate, thiamine hydrochloride (Vitamin B1), EDTA disodium salt dihydrate, fructose, xylose, maltose, peptone, bovine serum albumin (BSA), and phenol were procured from Sigma-Aldrich (St. Louis, MO, USA). Sodium nitrate, di-potassium hydrogen phosphate (dibasic), potassium chloride, potassium nitrate, ammonium chloride, and ammonium sulfate were supplied by AppliChem (Darmstadt, Germany). Magnesium sulfate anhydrous, calcium chloride dihydrate, ferrous sulfate heptahydrate, ammonium molybdate tetrahydrate, sucrose, and urea were obtained from Fluka (Buchs, Switzerland). Sodium hydroxide was purchased from Panreac (Barcelona, Spain). Chloroform, acetonitrile, and methanol were obtained from Thermo Fisher Scientific (Waltham, MA, USA). Sulfuric acid and ethanol were supplied by Honeywell Riedel-de Haën, and hydrochloric acid (HCl) was procured from Merck (Darmstadt, Germany). HPLC grade n-hexane was obtained from CARLO ERBA Reagents (Milano, Italy). Iodoacetamide was provided by Acros Organics (Geel, Belgium). SeraMag carboxylate-modified beads were supplied by GE Life Sciences (Chicago, IL, USA), and MS-grade Trypsin/LysC was acquired from Promega (Madison, WI, USA).
2.2. Microorganism
Pleurotus ostreatus LGAM 1123, obtained from the Laboratory of General and Agricultural Microbiology at the Agricultural University of Athens (Athens, Greece), was used in this study. The handling and cultivation of the strain followed the previously described protocol [9].
2.3. Media and Growth Conditions
The strain was activated from the Petri cultures with submerged cultivations as described previously [9]. In all the experiments, the optimum cultivation conditions using a medium containing 54 g/L glucose and 18 g/L yeast extract were used [9]. The effects of five different light wavelengths (white: 6690 K, yellow: 2639 K, red: 1500 K, green: 8034 K, and blue: 25,000 K) and a control condition of darkness were investigated in 100 mL Erlenmeyer baffled flasks. The light intensity was maintained at a constant level using LED lighting at the bottom of the flask, with a rate of 50 μmol m−2 s−1, as shown in Figure S1. This measurement was taken using a US-SQS/L Submersible Spherical Quantum Sensor (Heinz Walz GmbH, Effeltrich, Germany). At the mid-exponential phase (day 4), all the contents of the flasks were harvested, and the biomass and protein content were estimated. Furthermore, the biomass was used for GC-MS and proteomic analysis.
2.4. Biomass Determination
The whole content of the flask that was harvested from the cultures was centrifuged at 4000 rpm for 10 min to separate the supernatant and cell pellets. Cell pellets were washed three times, and the samples were freeze-dried as previously described [9].
2.5. Protein Estimation
The total protein content in the powdered biomass was measured using the Dumas method, while soluble intracellular proteins were quantified via Pierce™ BCA protein assay kit (Thermo Fisher Scientific, Waltham, MA, USA) following the manufacturer’s protocol as previously described [15]. Extracellular protein levels were determined by the Bradford method using a BSA standard curve [16].
2.6. Proteomic Analysis
2.6.1. Cell Lysis and Protein Digestion
One hundred and fifty milligrams of P. ostreatus frozen cells derived from the harvested culture on the 4th day of cultivation were used. The cells were lysed, and the supernatant was processed through Sp3 (Single-Pot, Solid-Phase-Enhanced Sample Preparation) method as described previously [11]. The Sp3 (Single-Pot, Solid-Phase-Enhanced Sample Preparation) method is a paramagnetic bead-based technique used for rapid, robust, and efficient protein sample processing in proteomics. It involves nonselective protein binding onto hydrophilic paramagnetic beads achieved through ethanol-driven solvation capture. The supernatant was collected and processed according to the Sp3 protocol [17]. The extended proteomic analysis is described in the Supplementary Material.
2.6.2. LC-MS/MS Analysis
Samples were run on a liquid chromatography–tandem mass spectrometry (LC-MS/MS) setup consisting of a Dionex UltimateRSLC online with a Thermo Q Exactive HF-X Orbitrap mass spectrometer (Thermo Scientific, Waltham, MA, USA) as described previously and mentioned in the Supplementary Material [11].
2.6.3. Data Analysis
Orbitrap raw data were analyzed with DIA-NN 1.8.1 (Data-Independent Acquisition by Neural Networks) by searching against the Pleurotus ostreatus genome in the Uniprot Database [18]. The Pleurotus ostreatus strain PC15, (https://www.uniprot.org/taxonomy/1137138, accessed on 2 July 2025) was used, which contains 12,173 proteins, using the library-free mode of the software and allowing up to two tryptic missed cleavages. The analysis was performed as described previously and as can be seen in the Supplementary Material [11].
2.6.4. Deposition
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE [19] partner repository with the dataset identifier PXD065402.
2.7. GC-MS
2.7.1. Sample Preparation
Biomass hydrolysis was performed according to the method described by Bakratsas et al. [9]. Derivatization of the amino acids, sugars, and lipids to silylated analogs was conducted according to previous studies, with slight modifications [20,21]. A volume of 200 μL of BSTFA (with 1% TMCS) was added to the lyophilized samples, which were sealed tightly, vortexed vigorously, and then incubated at 75 °C for 4 h. Finally, the samples were placed at ambient temperature, diluted with an equal volume of n-hexane, and filtered through a 0.22 µm nylon membrane syringe filter.
2.7.2. GC-MS Analysis
GC-MS analysis was carried out using a Shimadzu GCMS-QP2010 SE system (Shimadzu, Tokyo, Japan) equipped with a MEGA-5 MS capillary column (30 m length × 0.25 mm internal diameter × 0.25 μm film thickness; MEGA, Legnano, Italy) as described in previous studies [22,23]. The system featured a single-quadrupole mass spectrometer operating in electron ionization mode at 70 eV. The ion source temperature was maintained at 200 °C and the interface temperature at 300 °C. The oven temperature was initially set at 100 °C, ramped to 190 °C at 2 °C/min, and then increased to 290 °C at 5 °C/min. High-purity helium (>99.999%) served as the carrier gas with a constant flow rate of 1.0 mL/min. Sample injection volume was 1 μL through an AOC-20i split/splitless injector operated in split mode (1:40) at 280 °C. Mass spectra were acquired in full-scan mode over m/z 35–500, and qualitative analysis utilized the NIST library with matches > 80%. Identification of derivatized amino acids, lipids (fatty acids and monoacylglycerols), and sugars relied on retention times of reference compounds and mass spectral library data. Representative GC-MS chromatograms are shown in Figure S2.
2.7.3. PCA
Principal Component Analysis (PCA) was applied to amino acid concentration data to identify potential groupings or clusters and to correlate these patterns with the experimental conditions for further analysis. The first principal component (PC1) accounts for the largest variance in the data, representing the most significant linear combination of variables. The second principal component (PC2) captures the maximum remaining variance after the effect of PC1 is removed, serving as the next-best linear combination of variables [24]. In this study, amino acid concentrations were treated as scores (variables), while light wavelengths were set as loadings, employing a PCA-X model with 6 observations and 16 variables.
2.8. Statistical Analysis
All experiments were performed in triplicate, and results are presented as mean ± standard deviation. Statistical analyses were conducted using one-way analysis of variance (ANOVA) followed by Tukey’s multiple range test to identify significant differences between groups. Differences with p-values less than 0.05 were considered statistically significant. These analyses were carried out using IBM SPSS Statistics version 28.0.1.0 (IBM Corporation, New York, NY, USA). Principal Component Analysis (PCA) was also performed using the same software version.
3. Results
3.1. Effect of Light Wavelength on the Growth of P. ostreatus LGAM 1123
P. ostreatus LGAM 1123 was submerged cultivated using continuous LED lighting at the bottom of the flasks, as shown in Figure S1. At the mid-exponential phase (day 4), cells were harvested from the cultures, and the biomass and protein content were estimated as described in Section 2.4 and Section 2.5 of the Section 2. No statistically significant differences were observed in the biomass produced by the fungus under different light and dark conditions (Table 1). The biomass of P. ostreatus reached values higher than 9 g/L for all studied conditions. Concerning the effect of light wavelength on the protein content of the mycelium, it was higher in darkness and in red and green light, as shown in Table 1. White, yellow, and blue light followed, with slightly lower protein contents. As concerns intracellular protein production and extracellular protein production, similar values were observed, with no inhibitory effect of lights compared with the control condition of darkness.

Table 1.
Effect of light wavelength on P. ostreatus LGAM 1123 growth, protein content, and intracellular and extracellular proteins. Same letter between two samples in each column indicates non-significant differences (p > 0.05) as determined by Tukey’s multiple range test.
3.2. Proteomic Analysis of Cells of P. ostreatus LGAM 1123 Submerged Cultivated Under Different Light Wavelengths
At the mid-exponential phase of P. ostreatus growth, biomass was withdrawn from the cultivation, and after cell lysis and digestion, a proteomic analysis was conducted using an LC-MS/MS. A total of 3844 expressed proteins were detected, with 1345 of them being significantly differentiated. As shown in Figure 1, each wavelength presented a distinct proteome, with specific proteins being upregulated and downregulated. Blue and white lights are clustered together, while green and yellow are observed in a different branch that connects to a branch connecting the red to the dark. Blue light upregulates the light blue cluster of 347 proteins and the orange cluster of 75 proteins. After a gene ontology annotation through the Perseus software (version 1.6.15.0), the orange cluster proteins are related to translation, and the light blue cluster proteins are related to small-molecule and cellular metabolic processes and oxidoreductase activity. White light upregulates some proteins of all the shown clusters, but it is grouped with the blue-light condition. The green light regulates 216 proteins from the olive-colored cluster, which consists of proteins related to intracellular transport and ribonucleotide binding, while the yellow light upregulates some proteins from all the clusters, with most of them being located in the pink and olive-colored clusters. The control condition of dark presents a distinct profile of upregulated proteins in the purple cluster, related to cytoplasmic proteins, and some others in the pink cluster, related to hydrolase activity proteins. In contrast, red light upregulates mostly the blue cluster of 307 proteins, as well as most of the pink cluster, which is composed of 314 proteins related to hydrolase activity.
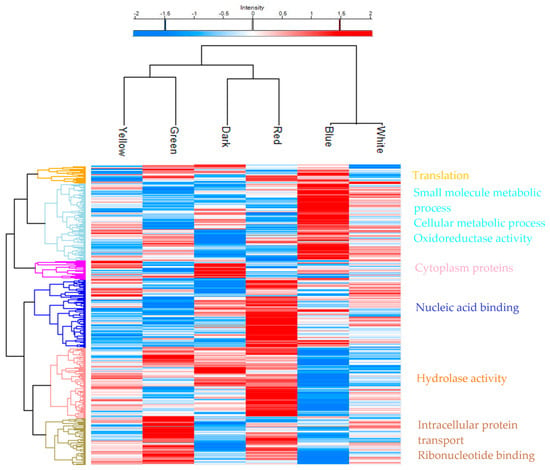
Figure 1.
Heatmap representing proteomic analysis of biomass from P. ostreatus LGAM 1123 exposed to different light wavelengths. Rows depict proteins clustered by average hierarchical clustering of ANOVA-significant proteins. LFQ intensity values were Log2 transformed, imputed, and z-scored. Upregulated proteins are shown in red, downregulated proteins in blue, and missing protein data in gray. Each column corresponds to a biological sample. Protein clusters were further characterized by gene ontology enrichment analysis using Perseus software.
As we mentioned above, the most distinctive differences between the tested conditions were those of red and dark with the condition of blue. For that reason, Volcano plots for the comparison between the red and blue and between dark and blue were created, as shown in Figure 2. From Figure 2a, we can observe that of 587 significantly altered proteins between red and blue, 337 are upregulated in the red condition. After a gene ontology analysis, it was found that proteins are related to spliceome and gene expression. In contrast, blue light favored the regulation of 181 that mostly correlated with carbon metabolism and the biosynthesis of secondary metabolites. From Figure 2b, we can see that the comparison between dark and blue shows 214 significantly altered proteins, of which 126 are favored by dark and are related to oxidoreductase activity, whereas 88 proteins are favored when blue light is used.
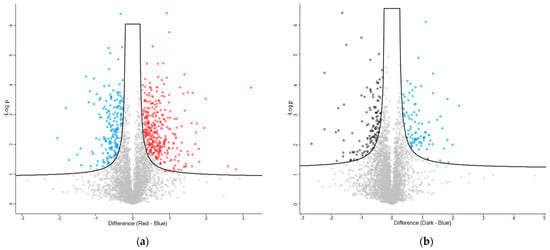
Figure 2.
Volcano plots display the magnitude of change (x-axis) against statistical significance (y-axis) for protein comparisons between red versus blue light and dark versus blue-light conditions. (a) Proteins significantly altered under red light are shown in red, those significantly altered under blue light in blue, and statistically insignificant proteins in gray. (b) Proteins significantly altered under blue light appear in blue, those significantly altered under dark conditions in black, and statistically insignificant proteins in gray.
After an enrichment analysis of gene ontology, proteins related to amino acid, carbohydrate, lipid, and nucleotide biosynthesis, as well as the tricarboxylic acid (TCA) and glycolysis cycles, are depicted in Table S1. As can be seen from the table, concerning amino acid biosynthesis, 21 proteins are observed, with 7 of them upregulated in blue light and 6 downregulated in red light. Concerning carbohydrate biosynthesis, seven proteins are detected, with three of them being upregulated under green light and most of them being downregulated under dark conditions. The proteins related to lipid biosynthesis were Squalene synthase, fatty acid synthase subunit alpha, and Biotin carboxylase, with green light used to upregulate all of them and blue light used to downregulate them. Twenty proteins were related to nucleotide biosynthesis, with five of them upregulated in blue and seven of them downregulated in green light conditions. Regarding the fundamental biological cycles of the TCA cycle and glycolysis, 13 significantly different proteins related to the TCA cycle were identified. Six of these proteins are upregulated under blue light, and five are downregulated under green light. Only three proteins related to glycolysis are observed, which are upregulated under blue light and downregulated when green light is used.
To connect the differentially expressed proteins with the related pathways from the KEGG pathway database, the genes related to the overexpressed proteins with a z-score > 1.5 for each tested condition were inserted into the STRING Database (https://string-db.org/, accessed on 3 July 2025). The proteins that were upregulated under red light were related to the mRNA surveillance pathway, which is crucial for maintaining cellular homeostasis and preventing the accumulation of damaged or misprocessed mRNAs. The cluster of proteins that were upregulated when blue light was used in the cultivation resulted in a total of 16 related pathways from KEGG, as shown in Table 2. These include pathways of specific molecules’ metabolism, such as methane and pyruvate metabolism, as well as the metabolism of specific amino acids, including cysteine and methionine, and glycine, serine, and threonine metabolism, among others. However, it also encompasses some of the most well-known biochemical pathways, including the TCA cycle, glycolysis and gluconeogenesis, and the biosynthesis of amino acids. Proteins that were upregulated when green light was used resulted in pathways related to mannose type O–glycan biosynthesis and RNA transport, while proteins upregulated by yellow light resulted in Arachidonic acid metabolism and oxidative phosphorylation. Dark and white upregulated proteins did not result in the stimulation of specific KEGG pathways.

Table 2.
KEGG pathways related to the proteins induced by the red, blue, green, and yellow light conditions used in the submerged cultivation of P. ostreatus LGAM 1123 were identified using the STRING database.
3.3. Biochemical Composition of P. ostreatus LGAM 1123 Determined Through GC-MS Analysis
3.3.1. Amino Acid Analysis by GC-MS
Sixteen amino acids were determined through GC-MS analysis of the biomass: seven of the nine essential amino acids, except for histidine and tryptophan; all the non-essential amino acids except for arginine, asparagine, and glutamine; and ornithine, a non-proteinogenic amino acid that plays a key role as an intermediate in metabolic pathways, particularly in the urea cycle (Table S2). As shown in Figure 3 and Table S2, the essential amino acids valine, isoleucine, and threonine appear to be increased in response to blue light compared with the other conditions. At the same time, lysine exists only in the presence of yellow light. The other essential amino acids do not seem to differentiate between the tested conditions. Regarding the umami amino acids responsible for the taste of meat, glutamic acid appears to be favored by yellow light, while aspartic acid does not show significant differences, with higher values detected in green, red, and dark conditions.
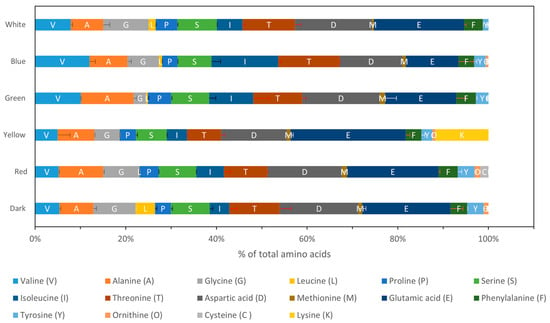
Figure 3.
Amino acid analysis of the biomass composition through GC-MS investigating the effect of different light wavelengths on P. ostreatus LGAM 1123 growth.
To investigate a correlation between light wavelength and specific amino acids and their clusters, a PCA was conducted (Figure 4). The first PC, which explains 86.0% of the variance, correlates positively with the amino acids aspartic acid (Asp, D), glutamic acid (Glu, E), threonine (Thr, T), alanine (Ala, A), serine (Ser, S), isoleucine (Ile, I), valine (Val, V), and glycine (Gly, G) and negatively with proline (Pro, P), phenylalanine (Phe, F), tyrosine (Tyr, Y), leucine (Leu, L), methionine (Met, M), ornithine (Orn, O), cysteine (Cys, C), and lysine (Lys, K). The second PC represents 9.0% of the variance, with isoleucine (Ile), valine (Val), and threonine (Thr) correlating positively, while Glu and Lys correlate negatively; the remaining amino acids do not contribute significantly to this component. From the loadings plot, we can see that all the light wavelengths (Figure S3a) correlate positively with the PC1. According to the scores plot (Figure S3b), we can summarize that Asp, Glu, Thr, Ala, Ser, Val, Ile, and Gly correlate positively with PC1, while Cys, Orn, Leu, Met, Tyr, Pro, and Phe correlate negatively with PC1. Concerning PC2, Ile, Val, and Thr correlate positively, while Glu and Lys correlate negatively with PC2. As can be seen from the biplot (Figure 4), three different clusters are observed across PC2: a cluster of biomass grown with green and blue light with Thr, Ile, and Val is detected; another with yellow light with Glu and Lys; and the last one of white, dark, and red with non-specific amino acids that are correlated across PC2.
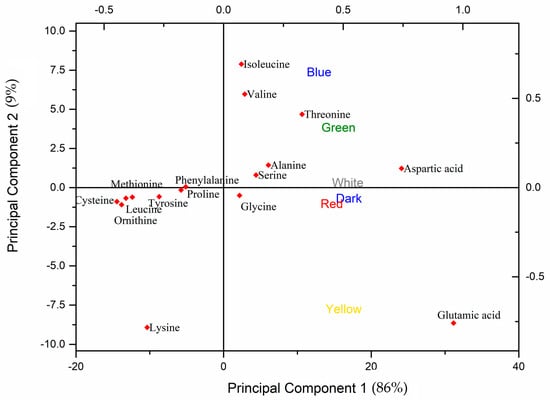
Figure 4.
PCA biplot for amino acid analysis from different light wavelengths’ effects on P. ostreatus LGAM 1123.
3.3.2. Sugar and Sugar Alcohols Analysis by GC-MS
As can be seen from Figure 5, three main sugars and sugar alcohols are detected in the biomass of P. ostreatus LGAM 1123: D-ribose, the component of RNA molecules; 1,5-anhydroglucitol, a sugar alcohol used as a sweetener; and D-glucosamine, a component of chitin. The dark condition produced biomass composed of seven different sugars: D-ribose; 1,5-anhydroglucitol; D-(+)-glucosamine; D-(+)-glucose, the primary energy source in cells; D-glucitol, a sugar alcohol used as a sweetener; erythritol, a low-calorie sweetener; and Myo-inositol, a component of cell signaling pathways; except for D-mannose, the component of glycoproteins. Yellow light produced biomass composed of only three main sugars: D-ribose, 1,5-anhydroglucitol, and D-glucosamine; blue light produced biomass composed of D-ribose, 1,5-anhydroglucitol, D-glucosamine, and Myo-inositol or inositol. Red light produced biomass composed mostly of D-ribose, 1,5-anhydroglucitol, D-glucitol, and Myo-inositol, whereas green light biomass was composed of D-ribose, 1,5-anhydroglucitol, D-glucosamine, glucose, mannose, and erythritol. Finally, the white wavelength produced a biomass composed mainly of D-ribose, 1,5-anhydroglucitol, D-glucosamine, D-glucitol, and Myo-inositol.
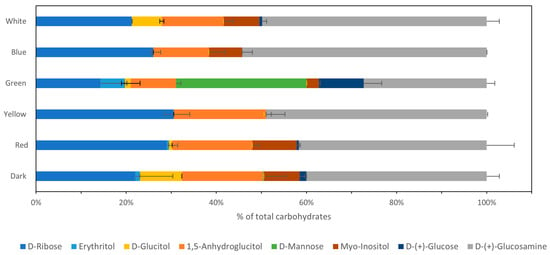
Figure 5.
Sugar analysis of the biomass composition through GC-MS investigating the effect of different light wavelengths on P. ostreatus LGAM 1123 growth.
3.3.3. Lipid Analysis Through GC-MS
As can be seen from Figure 6, five major lipids, three fatty acids, and two monoacylglycerols were observed in all tested conditions: palmitic acid (16:0); stearic acid (18:0); linoleic acid (18:2); 1-monopalmitin, a monoacylglycerol composed of one palmitic acid molecule esterified to a glycerol molecule at the sn-1 position; and glycerol monostearate, a monoester of stearic acid and glycerol. The dark condition, except for the main lipids, included also arachidic acid (20:0) and lignoceric acid (24:0). Red, yellow, and green light, in addition to the five main lipids, also included lignoceric acid, whereas white and blue light included only the five main lipids that were discussed above.
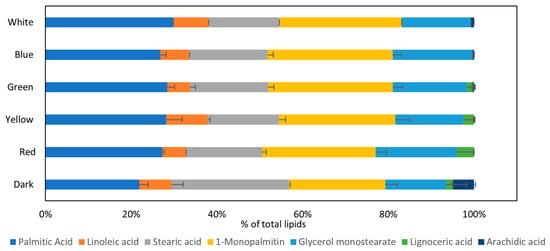
Figure 6.
Lipid analysis of the biomass composition by GC-MS investigating the effect of different light wavelengths on P. ostreatus LGAM 1123 growth.
4. Discussion
According to our results, P. ostreatus LGAM 1123 was able to grow under all tested light conditions, with biomass production reaching 10 g/L by the fourth day of submerged cultivation, comparable to growth in darkness. In contrast, previous studies on Pleurotus eryngii reported that light exposure generally imposed stress during submerged cultivation; however, only red light resulted in biomass yields similar to the dark control, with values close to 9.9 g/L, aligning with our findings [25]. The enhanced growth rate under red light was supported by a study on Cordyceps militaris cultivation with various light-emitting diodes, which reported a maximum specific growth rate of 1.47 day−1, exceeding that of the dark control, along with a biomass production of 17 g/L. Additionally, research on Pleurotus spp. in liquid cultivation demonstrated that among seven Pleurotus strains, growth under green light yielded lower biomass compared with the dark control condition [26]. Contrarily, Ha et al. reported that green light resulted in the highest biomass production, with a statistically significant increase compared with darkness, fluorescent light, UV-A, red, and blue light conditions [27]. A study investigating fruiting body production and mycelial growth of P. ostreatus in Petri dishes revealed that white and blue light promoted fruiting body formation, whereas red light and darkness significantly inhibited it. For mycelial cultivation, growth under red and blue light was reduced, whereas white light favored mycelial development. Notably, red light and dark conditions induced morphological differentiation in the mycelia. These findings underscore the differential effects of light wavelengths on distinct developmental stages of P. ostreatus [28]. A study of the strain Phanerochaete chrysosporium summarized that biomass production is favored by yellow and green light [29]. It is evident that for each fungal strain, even for strains from the same genus, different light wavelengths could favor biomass production, whereas sometimes light illumination could suppress biomass production. In our study, light illumination did not inhibit biomass production in P. ostreatus LGAM 1123—an observation of particular interest, as it suggests that specific light wavelengths could be strategically used to modulate the production of bioactive compounds without compromising growth.
Regarding the effect of light wavelength on protein content during the mid-exponential phase of cultivation, it appears to be higher for darkness and red and green light, with values of 56.2 ± 3.7%, 54.9 ± 2.2%, and 54.5 ± 2.8%, respectively. While intracellular and extracellular protein production does not seem to be affected by light wavelength, similar production to the control condition of darkness was achieved. In contrast, the scientific literature states that for the P. chrysosporium strain, a decrease in protein production was observed when red, blue, and UV lights were used in aerated cultivation, whereas in oxygenated cultivation, a protein decrease was observed in UV and white light. In addition, a study about Lentinus critinus mycelial biomass under different light wavelengths revealed that protein content decreased by 33%, 29%, and 15% for blue, green, and red light when compared with the control condition of dark [30]. P. ostreatus LGAM 1123 has been previously utilized for single-cell protein production, with process optimization conducted using response surface methodology. This optimization yielded a maximum protein production of 13.6 g/L at glucose and yeast extract concentrations of 54.1 g/L and 18.3 g/L, respectively, as determined through the software’s numerical optimization function. The optimization was confirmed in a 3.5 L stirred tank bioreactor with a protein content of 44.8 ± 0.8% [9]. These results confirm that biomass with similar or higher protein content can be achieved when light illumination is used for the submerged cultivation of P. ostreatus LGAM 1123 compared with the control condition of darkness. Consequently, light can be used for the submerged cultivation of P. ostreatus without the possibility of inhibiting biomass production or protein synthesis by the fungus.
Proteomic analysis of P. ostreatus LGAM 1123 cells was conducted on the fourth day of cultivation under different light wavelengths. A different cluster of proteins was upregulated when specific light wavelengths were used in submerged cultivation. Blue and white light were clustered together, while green was observed in a different branch with yellow light, which was connected to a branch that links red with dark, following exactly the correlation of the spectra that these colors have. Taking into account that white LED light contains a wide spectrum of 400–680 nm, with the highest peak at 478.7 nm, blue light at 465.4 nm, green at 522.6 nm, yellow at 581.9 nm, and red at 619.8 nm, the clustering of the colors is reasonable. Red and blue light wavelengths presented the most differentially expressed proteins. Blue light induced pathways such as the TCA cycle, glycolysis and gluconeogenesis, and the biosynthesis of amino acids, while red light induced mRNA-related pathways. Transcriptomic analysis of P. ostreatus exposed to blue and red light revealed that blue light enhanced the growth and development of all mushroom organs, particularly the pileus, while red light slightly inhibited pileus growth. When compared with dark conditions, blue light upregulated glycolysis, similar to our results, and the pentose phosphate pathway. In contrast, red light downregulated the regulation of many respiration metabolism genes in the pileus [8]. A new study about the effect of light on P. ostreatus mycelia cultivation in Petri dishes and fruiting bodies production through proteomic analysis revealed that, similar to our results, the difference between mycelia grown under darkness and blue light displayed the most differentiation. They also indicated that white light triggers oxidative phosphorylation (triggered by yellow light in our study), the tricarboxylic acid cycle (triggered by blue light in our study), the proteasome pathway and mRNA surveillance pathway (triggered by red light in our study), as well as pyruvate metabolism and endocytosis. Under all light conditions, endocytosis, purine metabolism, and oxidative phosphorylation were upregulated during the mycelial stage, whereas they were downregulated during fruiting body production, indicating that the blue light effect on mycelia was greater than in fruiting bodies [28]. The transcriptomic analysis of P. eryngii confirmed that pathways such as carbon metabolism, glycolysis/gluconeogenesis, and amino acid biosynthesis were regulated during blue light-induced primordia formation [7]. The proteomic analysis of another Basidiomycota, H. marmoreus, cultivated in Petri dishes revealed that the most differentiated proteins were observed under white light, in contrast to our result, which showed the blue light to present the most distinctive proteome; but similarly, the differentiated proteins under lights were related to mRNA-related processes, spliceome, and primary metabolism pathways [6]. To our knowledge, there is no other study of proteomic analysis of P. ostreatus submerged cultivation using different light wavelengths. A proteomic analysis of P. ostreatus LGAM 1123 has already been conducted by our group for submerged cultivation in different glucose/xylose mixtures. This study has confirmed that the strain exhibits different proteomes when supplemented with varying ratios of glucose and xylose in the medium. Specifically, xylose was found to upregulate the tricarboxylic acid cycle, whereas glucose upregulates glycolysis. Analysis using the STRING database revealed that xylose primarily upregulates proteins involved in amino acid biosynthesis in P. ostreatus, with the leucine, valine, and isoleucine biosynthetic pathway being the most significantly stimulated. This suggests a targeted metabolic response to xylose, favoring branched-chain amino acid production [11].
Afterward, a GC-MS analysis was conducted on the biomass of P. ostreatus LGAM 1123, which was cultivated under different light conditions. Concerning amino acid analysis, 16 amino acids were identified, including 7 of the 9 essential amino acids (except for histidine and tryptophan) and all the non-essential amino acids (except for arginine, asparagine, and glutamine). A PCA revealed three distinct clusters along PC2: one for green and blue light conditions, characterized by the amino acids Thr, Ile, and Val; another for yellow light, featuring Glu and Lys; and the last for white light, dark, and red light, with no specific amino acid correlation. Thr (threonine), Ile (isoleucine), and Val (valine) are essential amino acids important for muscle growth, immune support, and energy metabolism, with Thr also supporting connective tissue integrity. Glu (glutamic acid), a non-essential amino acid, acts as a neurotransmitter and a precursor to glutathione, aiding brain function and antioxidant defenses. Lys (lysine) is vital for protein synthesis, calcium absorption, and collagen formation, which supports bone and skin health [31,32]. The effect of light on protein and amino acid accumulation has already been reported, while the regulation of genes related to amino acid biosynthesis has been confirmed from a study of P. eryngii primordium differentiation into a fruiting body [7,30]. The metabolic profile of Cordyceps bassiana, as analyzed by GC-MS, has also been studied. This study showed different amino acid concentrations expressed in various cultivation media rather than a comparison of dark cultivation conditions and cultivation under light. Also, in the PCA of biomass extracts, the dark and light conditions were clustered together [33]. For the same strain, P. ostreatus LGAM 1123, our group has indicated that different amino acids are grouped according to specific cultivation conditions of glucose and xylose media, revealing a strong correlation between amino acid composition and glucose and xylose ratios [10].
Concerning carbohydrate analysis, three main sugars and sugar alcohols were detected in the biomass of P. ostreatus LGAM 1123 through GC-MS analysis: D-ribose, 1,5-anhydroglucitol, and D-glucosamine. Sugars like D-ribose, 1,5-anhydroglucitol, and D-glucosamine are important for energy production and cellular structure. Dark conditions produced biomass composed of seven different sugars, D-ribose, 1,5-anhydroglucitol, D-(+)-glucosamine, D-(+) glucose, D-glucitol, erythritol, and Myo-inositol, except D-mannose, whereas green light produced biomass composed of D-ribose, 1,5-anhydroglucitol, D-glucosamine, D-(+) glucose, D-mannose, and erythritol. D-ribose, the component of RNA molecules, was increased with red, blue, and yellow light; D-glucitol, a sugar alcohol used as a sweetener, was primarily observed in dark conditions and less in white light; while D-glucosamine was maximized under blue light. D-mannose, a component of glycoproteins, existed only under green illumination. These results were confirmed through proteomic analysis, which showed, according to the STRING database, that the mannose type O–glycan biosynthesis pathway is stimulated when green light is used in cultivation. In a study using GC-MS analysis of Cordyceps bassiana, erythritol, glycerol, mannitol, and glucose were increased when light was used in the cultivation, whereas glucitol and mannose were decreased when light was used [33]. To our knowledge, no other study has compared the biomass composition in sugars when Pleurotus ostreatus is grown under different light wavelengths. A study of Pleurotus pulmonarius has shown that when the strain was grown in glucose, the biomass was composed mainly of glucose, mannose, galactose, xylose, and fucose, whereas the composition ranged between different carbon sources that were used [34]. Also, our team has already shown that, as concerns polysaccharide production, red and green light can stimulate production, especially of exopolysaccharides that consist of beta-glucans [13].
Lipid analysis by GC-MS revealed that five major lipids were observed in all tested conditions: palmitic acid (16:0), stearic acid (18:0), linoleic acid (18:2), and the monoglycerides 1-monopalmitin and glycerol monostearate. Low-molecular-weight lipids such as palmitic acid (16:0), stearic acid (18:0), and linoleic acid (18:2) provide energy and maintain cell membrane integrity. The dark condition presents the most extensive distribution of lipids, with arachidic acid (20:0) and lignoceric acid (24:0) being detected in addition. Red, green, and yellow lights follow, while blue and white present only the five major lipids. A study of P. ostreatus analyzed through GC-MS confirmed that linoleic acid, palmitic acid, oleic acid, and stearic acid were the most abundant fatty acids in the Pleurotus ostreatus biomass [35]. In contrast, a study of P. pulmonarius cultivation in glucose has shown that the most abundant fatty acid was linoleic acid, followed by oleic acid, palmitic acid, and palmitoleic acid. At the same time, the distribution differed when different carbon sources were used [34]. To our knowledge, there is no other study about lipid distribution in the biomass of P. ostreatus grown under different light wavelengths. A study on the cultivation of L. critinus under different light wavelengths found that the major lipids produced were palmitic acid (16:0), oleic acid (18:1), and linoleic acid (18:2). Lipid amounts were generally lower under light illumination compared with darkness. Specifically, palmitic acid levels increased under green and red light, while linoleic acid increased under blue light [30]. GC-MS-based metabolic profiles of 100% n-hexane extracts of C. bassiana mycelium have shown that palmitic, stearic, linoleic, oleic, and palmitoleic acids were increased when the light of 3000 lux was used in cultivation compared with the dark condition [33]. In our study, palmitic acid and its esterified form, 1-monopalmitin, were increased in all light wavelengths compared with the dark, indicating that light favors the production of fatty acids with fewer carbon atoms. In contrast, the dark condition favored the production of larger carbon chain fatty acids like arachidic acid, lignoceric acid, and stearic acid equivalents, considering the sum of stearic acid and glycerol monostearate. Furthermore, it is worth mentioning that GC-MS analysis allows the detection of low-molecular-weight lipids such as fatty acids or glycerides of the broad lipid class.
5. Conclusions
In this study, we examined how different light wavelengths affect the global protein expression profile during submerged cultivation of P. ostreatus LGAM 1123 as well as the biochemical composition of the produced biomass. Our results showed that P. ostreatus could grow well under all tested light wavelengths and that the growth of the fungus, as well as its intracellular and extracellular protein production, was not inhibited by light in any of the conditions tested. Proteomic analysis revealed a distinct proteome profile for each light wavelength used, with red and blue light presenting the most distinctive proteome profiles. Blue light induces pathways such as the TCA cycle, glycolysis/gluconeogenesis, and biosynthesis of amino acids, while red light induces mRNA-related pathways. GC-MS analysis of biomass revealed differences in the produced amino acids, sugars, and lipids. The PCA identified three different clusters, one for green and blue light conditions with the amino acids Thr, Ile, and Val; another for yellow light with Glu and Lys; and the last one for white light, dark, and red light, which showed no specific correlation with individual amino acids. Regarding carbohydrate analysis, three major sugars were detected in P. ostreatus LGAM 1123 biomass: D-ribose, 1,5-anhydroglucitol, and D-glucosamine. The dark and green light conditions exhibited the greatest variety of sugars. From the fatty acids analysis, palmitic acid was found to increase in all the light wavelengths compared with darkness, whereas dark conditions favored the production of long-chain fatty acids like arachidic and lignoceric acid, while light induced the production of shorter-chain fatty acids. In summary, light wavelength affects Pleurotus ostreatus metabolism and the biochemical composition of the produced biomass. The ability to turn metabolism into specific pathways using light illumination could allow for improving the biochemical compound production, useful for the food industry.
The observed differences in amino acid and lipid composition in P. ostreatus have important nutritional and sensory implications. P. ostreatus contains a range of essential and non-essential amino acids. These amino acids play vital roles in metabolism, immune function, and protein synthesis, making the mushroom a valuable source of high-quality protein. The variation in lipid composition, including increased short-chain fatty acids under light and a preference for long-chain fatty acids in the dark, can influence both the flavor profile and nutritional value, as fatty acids contribute to taste, texture, and the bioavailability of fat-soluble nutrients. Therefore, controlling light conditions during cultivation could strategically enhance the nutritional quality and sensory characteristics of P. ostreatus, with potential benefits for food industry applications and human health. In parallel, a deeper analysis of proteomic data and the focus on certain metabolic pathways can help toward these industrial applications. Depending on the application, future studies could focus on the effect of light on the overall lipid, amino acid, and carbohydrate profiles of P. ostreatus LGAM 1123 by utilizing techniques such as LC-MS, while semi-pilot studies should be performed to ensure the application directly to the industrial scale.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/horticulturae11101234/s1, Extended Materials and Methods, Table S1: Protein analysis through gene ontology biological process related to amino acid, carbohydrate, lipid, and nucleotide biosynthesis, TCA, and the glycolysis cycle, with −Log p-value > 1.5. The highest values (z-scores) are marked in red, and the lowest are marked in blue; Figure S1: Cultivation of P. ostreatus LGAM 1123 using LED lights at the bottom of the flasks in a medium composed of 54 g/L glucose and 18 g/L yeast extract; Figure S2: GC-MS chromatograph from biomass produced from different light wavelengths effect on P. ostreatus LGAM 1123; Figure S3: Loading plot (a) and score plot (b) from PCA of the amino acid compositions in different light wavelengths. Table S2: Raw data of amino acid analysis of the biomass composition through GC-MS. Same letter between two samples in each column indicates non-significant differences (p > 0.05) as determined by Tukey’s multiple range test.
Author Contributions
Conceptualization, P.K.; methodology, G.B., M.S. and R.F.; software, M.S.; validation, G.B.; formal analysis, G.B., M.S. and R.F.; investigation, G.B. and R.F.; data curation, G.B.; writing—original draft preparation, G.B.; writing—review and editing, H.S. and P.K.; visualization, G.B.; supervision, H.S. and P.K.; funding acquisition, H.S. All authors have read and agreed to the published version of the manuscript.
Funding
Co-financed by the European Regional Development Fund of the European Union and Greek National Funds through the Operational Program Competitiveness, Entrepreneurship and Innovation, under the call RESEARCH–CREATE–INNOVATE (project code: T2EDK-02830).
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD065402.
Acknowledgments
We gratefully acknowledge Georgios I. Zervakis from the Laboratory of General and Agricultural Microbiology, Agricultural University of Athens, Iera Odos 75, 11855 Athens, Greece, for providing the fungal strain.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Tisch, D.; Schmoll, M. Light Regulation of Metabolic Pathways in Fungi. Appl. Microbiol. Biotechnol. 2010, 85, 1259–1277. [Google Scholar] [CrossRef]
- Smith, H. Light Quality, Photoperception, and Plant Strategy. Annu. Rev. Plant Physiol. 1982, 33, 481–518. [Google Scholar] [CrossRef]
- Wang, Z.; Wang, J.; Li, N.; Li, J.; Trail, F.; Dunlap, J.C.; Townsend, J.P. Light Sensing by Opsins and Fungal Ecology: NOP-1 Modulates Entry into Sexual Reproduction in Response to Environmental Cues. Mol. Ecol. 2018, 27, 216–232. [Google Scholar] [CrossRef]
- Feng, Y.; Xu, H.; Sun, Y.; Xia, R.; Hou, Z.; Li, Y.; Wang, Y.; Pan, S.; Li, L.; Zhao, C.; et al. Effect of Light on Quality of Preharvest and Postharvest Edible Mushrooms and Its Action Mechanism: A Review. Trends Food Sci. Technol. 2023, 139, 104119. [Google Scholar] [CrossRef]
- Park, Y.J.; Jang, M.J. Blue Light Induced Edible Mushroom (Lentinula edodes) Proteomic Analysis. J. Fungi 2020, 6, 127. [Google Scholar] [CrossRef]
- Zhu, L.; Su, Y.; Ma, Z.; Guo, L.; Yang, S.; Yu, H. Comparative Proteomic Analysis Reveals Differential Protein Expression of Hypsizygus Marmoreus in Response to Different Light Qualities. Int. J. Biol. Macromol. 2022, 223, 1320–1334. [Google Scholar] [CrossRef]
- Xie, C.; Gong, W.; Zhu, Z.; Yan, L.; Hu, Z.; Peng, Y. Comparative Transcriptomics of Pleurotus eryngii Reveals Blue-Light Regulation of Carbohydrate-Active Enzymes (CAZymes) Expression at Primordium Differentiated into Fruiting Body Stage. Genomics 2018, 110, 201–209. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Tong, X.; Tian, F.; Jia, C.; Li, C.; Li, Y. Transcriptomic Profiling Sheds Light on the Blue-Light and Red-Light Response of Oyster Mushroom (Pleurotus ostreatus). AMB Express 2020, 10, 10. [Google Scholar] [CrossRef] [PubMed]
- Bakratsas, G.; Polydera, A.; Nilson, O.; Κossatz, L.; Xiros, C.; Katapodis, P.; Stamatis, H. Single-Cell Protein Production by Pleurotus Ostreatus in Submerged Fermentation. Sustain. Food Technol. 2023, 1, 377–389. [Google Scholar] [CrossRef]
- Bakratsas, G.; Polydera, A.; Nilson, O.; Chatzikonstantinou, A.V.; Xiros, C.; Katapodis, P.; Stamatis, H. Mycoprotein Production by Submerged Fermentation of the Edible Mushroom Pleurotus ostreatus in a Batch Stirred Tank Bioreactor Using Agro-Industrial Hydrolysate. Foods 2023, 12, 2295. [Google Scholar] [CrossRef]
- Bakratsas, G.; Samiotaki, M.; Katapodis, P.; Stamatis, H. Proteomic Analysis of Pleurotus Ostreatus Grown on Glucose and Xylose Mixtures in Submerged Fermentation Provides Insights into Differentiated Mycelial Composition. Synth. Biol. Eng. 2024, 2, 10006. [Google Scholar] [CrossRef]
- Bakratsas, G.; Antoniadis, K.; Athanasiou, P.E.; Katapodis, P.; Stamatis, H. Laccase and Biomass Production via Submerged Cultivation of Pleurotus ostreatus Using Wine Lees. Biomass 2023, 4, 1–22. [Google Scholar] [CrossRef]
- Bakratsas, G.; Tsoumanis, C.; Stamatis, H.; Katapodis, P. Exopolysaccharide Production in Submerged Fermentation of Pleurotus ostreatus under Red and Green Light. Fermentation 2024, 10, 313. [Google Scholar] [CrossRef]
- Zerva, A.; Papaspyridi, L.M.; Christakopoulos, P.; Topakas, E. Valorization of Olive Mill Wastewater for the Production of β-Glucans from Selected Basidiomycetes. Waste Biomass Valorization 2017, 8, 1721–1731. [Google Scholar] [CrossRef]
- Smith, P.K.; Krohn, R.I.; Hermanson, G.T.; Mallia, A.K.; Gartner, F.H.; Provenzano, M.D.; Fujimoto, E.K.; Goeke, N.M.; Olson, B.J.; Klenk, D.C. Measurement of Protein Using Bicinchoninic Acid. Anal. Biochem. 1985, 150, 76–85. [Google Scholar] [CrossRef] [PubMed]
- Kruger, N.J. The Bradford Method for Protein Quantitation. In The Protein Protocols Handbook; Springer Protocols Handbooks: Totowa, NJ, USA, 1996; Volume 32, pp. 15–20. [Google Scholar]
- Hughes, C.S.; Moggridge, S.; Müller, T.; Sorensen, P.H.; Morin, G.B.; Krijgsveld, J. Single-Pot, Solid-Phase-Enhanced Sample Preparation for Proteomics Experiments. Nat. Protoc. 2019, 14, 68–85. [Google Scholar] [CrossRef]
- Demichev, V.; Messner, C.B.; Vernardis, S.I.; Lilley, K.S.; Ralser, M. DIA-NN: Neural Networks and Interference Correction Enable Deep Proteome Coverage in High Throughput. Nat. Methods 2020, 17, 41–44. [Google Scholar] [CrossRef]
- Perez-Riverol, Y.; Bandla, C.; Kundu, D.J.; Kamatchinathan, S.; Bai, J.; Hewapathirana, S.; John, N.S.; Prakash, A.; Walzer, M.; Wang, S.; et al. The PRIDE Database at 20 Years: 2025 Update. Nucleic Acids Res. 2025, 53, D543–D553. [Google Scholar] [CrossRef]
- Du, F.; Zou, Y.; Hu, Q.; Jing, Y.; Yang, X. Metabolic Profiling of Pleurotus tuoliensis during Mycelium Physiological Maturation and Exploration on a Potential Indicator of Mycelial Maturation. Front. Microbiol. 2019, 10, 3274. [Google Scholar] [CrossRef]
- Gao, S.; Xu, B.; Zheng, X.; Wan, X.; Zhang, X.; Wu, G.; Cong, Z. Developing an Analytical Method for Free Amino Acids in Atmospheric Precipitation Using Gas Chromatography Coupled with Mass Spectrometry. Atmos. Res. 2021, 256, 105579. [Google Scholar] [CrossRef]
- Wezgowiec, J.; Wieczynska, A.; Wieckiewicz, W.; Kulbacka, J.; Saczko, J.; Pachura, N.; Wieckiewicz, M.; Gancarz, R.; Wilk, K.A. Polish Propolis-Chemical Composition and Biological Effects in Tongue Cancer Cells and Macrophages. Molecules 2020, 25, 2426. [Google Scholar] [CrossRef] [PubMed]
- Spyrou, S.; Chatzikonstantinou, A.; Giannakopoulou, A.; Fotiadou, R.; Priska, S.; Simos, Y.; Tsakni, A.; Peschos, D.; Houhoula, D.; Voutsas, E.; et al. Fungal Laccase-Mediated Enhancement of the Bioactivity of Green Algae Extracts. Catal. Res. 2023, 3, 1–29. [Google Scholar] [CrossRef]
- Lauria, A.; Ippolito, M.; Almerico, A.M. Principal Component Analysis on Molecular Descriptors as an Alternative Point of View in the Search of New Hsp90 Inhibitors. Comput. Biol. Chem. 2009, 33, 386–390. [Google Scholar] [CrossRef]
- Wu, J.; Chen, H.; Chen, M.; Kan, S.; Shieh, C.; Liu, Y. Quantitative Analysis of LED Effects on Edible Mushroom Pleurotus Eryngii in Solid and Submerged Cultures. J. Chem. Technol. Biotechnol. 2013, 88, 1841–1846. [Google Scholar] [CrossRef]
- Araújo, N.L.; Avelino, K.V.; Halabura, M.I.W.; Marim, R.A.; Kassem, A.S.S.; Linde, G.A.; Colauto, N.B.; do Valle, J.S. Use of Green Light to Improve the Production of Lignocellulose-Decay Enzymes by Pleurotus Spp. in Liquid Cultivation. Enzyme Microb. Technol. 2021, 149, 109860. [Google Scholar] [CrossRef]
- Ha, S.Y.; Jung, J.Y.; Yang, J.K. Effect of Light-Emitting Diodes on Cordycepin Production in Submerged Culture of Paecilomyces japonica. J. Korean Wood Sci. Technol. 2020, 48, 548–561. [Google Scholar] [CrossRef]
- Zhu, L.; Su, Y.; Ma, S.; Guo, L.; Yang, S.; Yu, H. Comparative Proteomic Analysis Reveals Candidate Pathways Related to the Effect of Different Light Qualities on the Development of Mycelium and Fruiting Body of Pleurotus ostreatus. J. Agric. Food Chem. 2023, 72, 1361–1375. [Google Scholar] [CrossRef]
- Ramírez, D.A.; Muñoz, S.V.; Atehortua, L.; Michel, F.C. Effects of Different Wavelengths of Light on Lignin Peroxidase Production by the White-Rot Fungi Phanerochaete chrysosporium Grown in Submerged Cultures. Bioresour. Technol. 2010, 101, 9213–9220. [Google Scholar] [CrossRef]
- Halabura, M.I.W.; Avelino, K.V.; Araújo, N.L.; Kassem, A.S.S.; Seixas, F.A.V.; Barros, L.; Fernandes, Â.; Liberal, Â.; Ivanov, M.; Soković, M.; et al. Light Conditions Affect the Growth, Chemical Composition, Antioxidant and Antimicrobial Activities of the White-Rot Fungus Lentinus Crinitus Mycelial Biomass. Photochem. Photobiol. Sci. 2023, 22, 669–686. [Google Scholar] [CrossRef]
- Neinast, M.; Murashige, D.; Arany, Z. Branched Chain Amino Acids. Annu. Rev. Physiol. 2019, 81, 139–164. [Google Scholar] [CrossRef] [PubMed]
- Bach, F.; Helm, C.V.; Bellettini, M.B.; Maciel, G.M.; Haminiuk, C.W.I. Edible Mushrooms: A Potential Source of Essential Amino Acids, Glucans and Minerals. Int. J. Food Sci. Technol. 2017, 52, 2382–2392. [Google Scholar] [CrossRef]
- Hyun, S.H.; Lee, S.Y.; Park, S.J.; Kim, D.Y.; Chun, Y.J.; Sung, G.H.; Kim, S.H.; Choi, H.K. Alteration of Media Composition and Light Conditions Change Morphology, Metabolic Profile, and Beauvericin Biosynthesis in Cordyceps bassiana Mycelium. J. Microbiol. Biotechnol. 2013, 23, 47–55. [Google Scholar] [CrossRef]
- Smiderle, F.R.; Olsen, L.M.; Ruthes, A.C.; Czelusniak, P.A.; Santana-Filho, A.P.; Sassaki, G.L.; Gorin, P.A.J.; Iacomini, M. Exopolysaccharides, Proteins and Lipids in Pleurotus pulmonarius Submerged Culture Using Different Carbon Sources. Carbohydr. Polym. 2012, 87, 368–376. [Google Scholar] [CrossRef] [PubMed]
- Hadar, Y.; Cohen-Arazi, E. Chemical Composition of the Edible Mushroom Pleurotus ostreatus Produced by Fermentation. Appl. Environ. Microbiol. 1986, 51, 1352–1354. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).