Comprehensive Identification and Expression Profiling of the NAC Family During Female Cone Development in Torreya grandis
Abstract
1. Introduction
2. Materials and Methods
2.1. Identification and Structural Analysis of NAC Family
2.2. Phylogenetic Analysis
2.3. Expression Analysis of NAC Genes in T. grandis
2.4. Promoter Bioinformatics Analysis
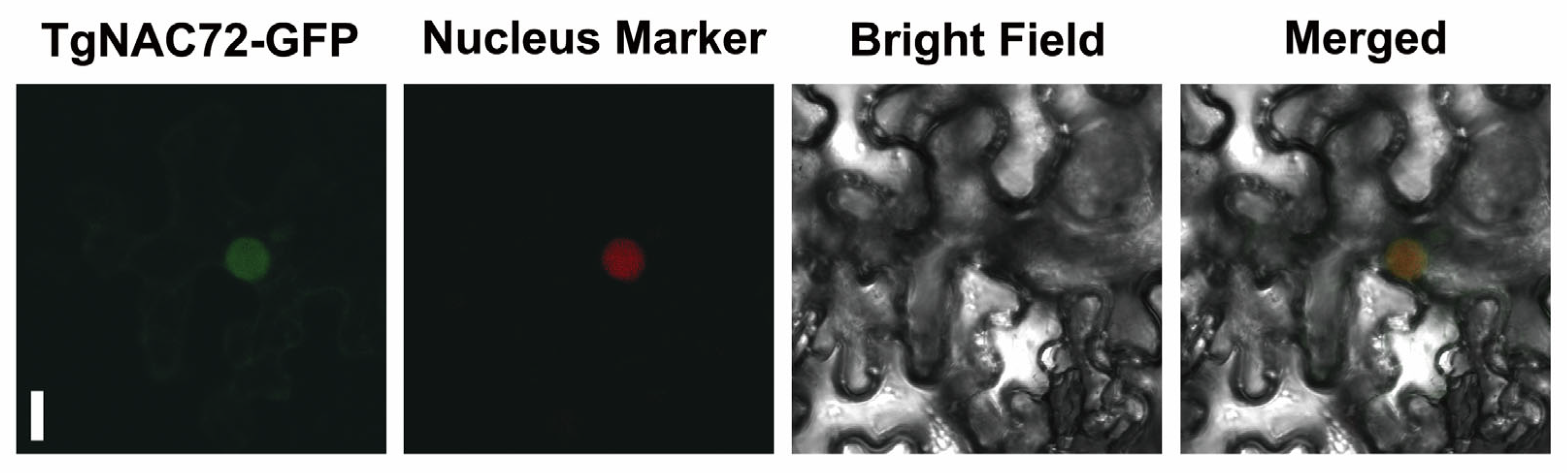
2.5. Subcellular Localization
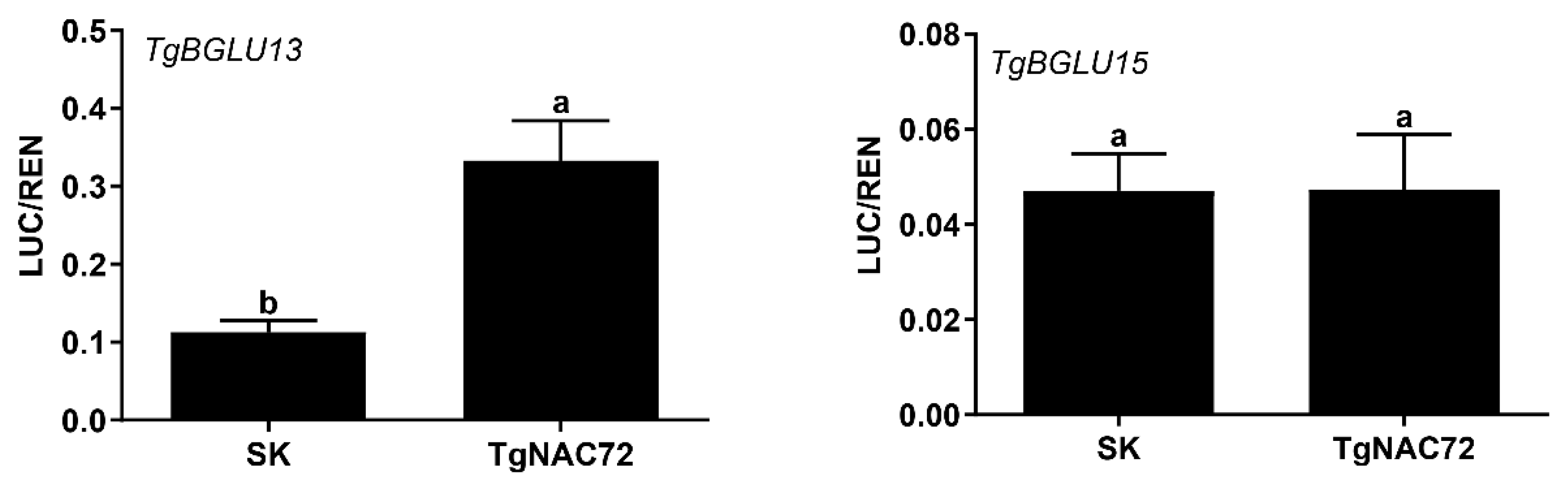
2.6. Luciferase (LUC) Assays in Tobacco
2.7. Statistical Analysis
3. Results
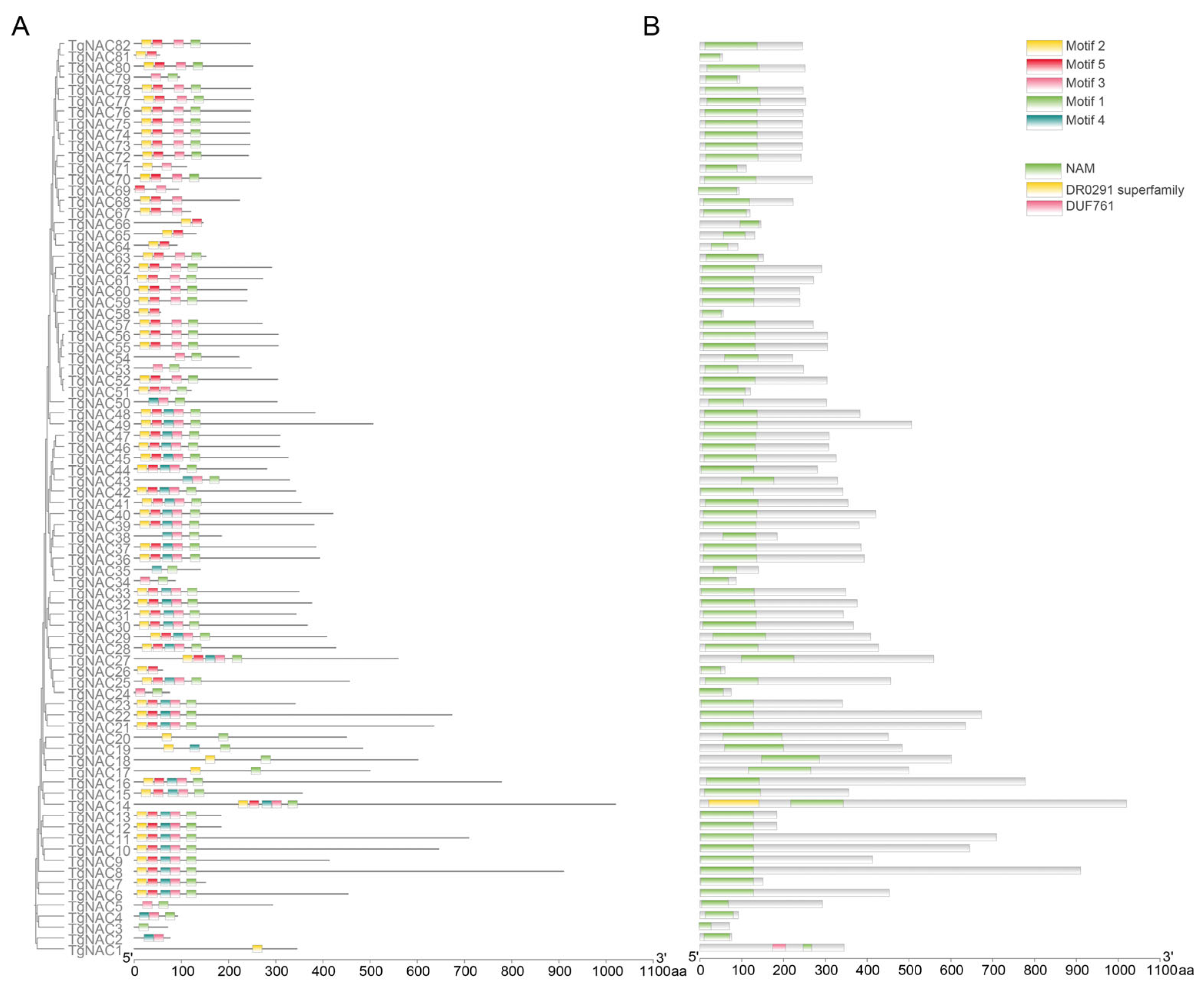
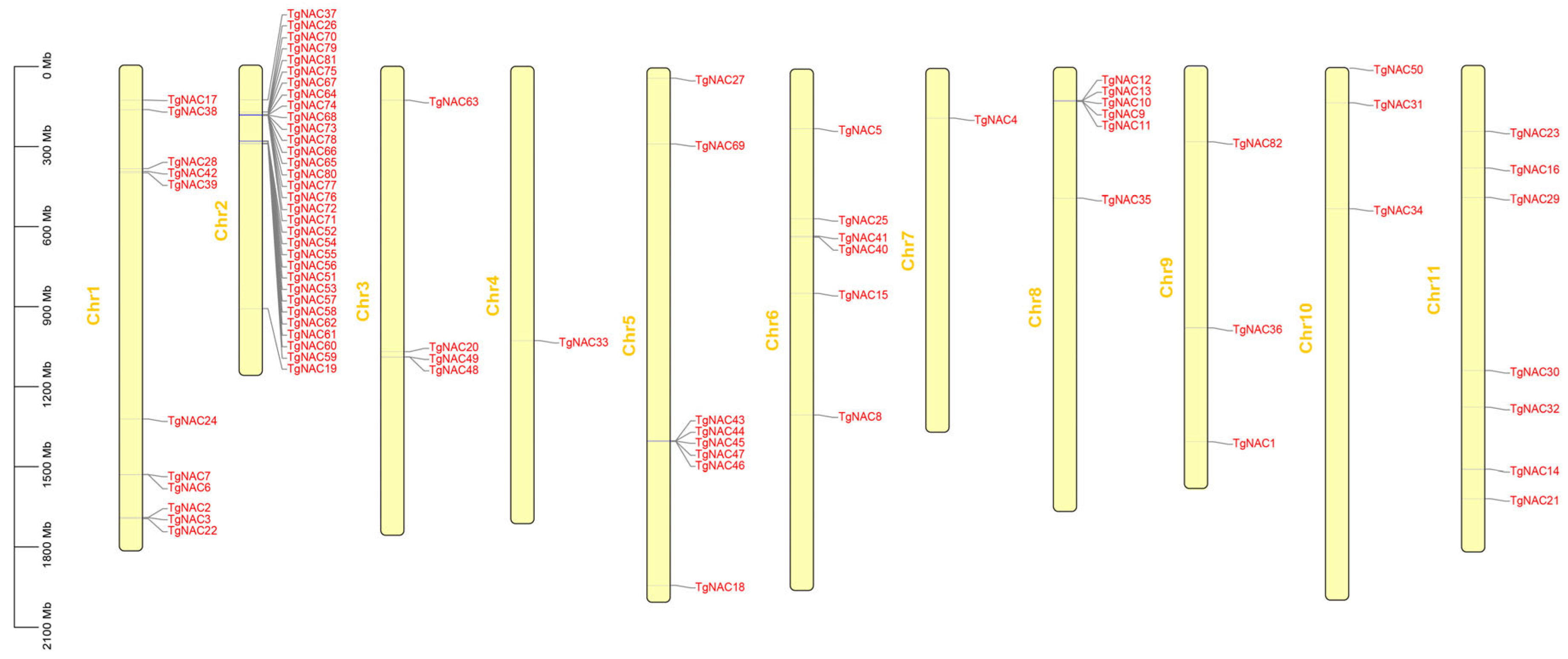
3.1. Identification and Structure of NAC Family in T. grandis
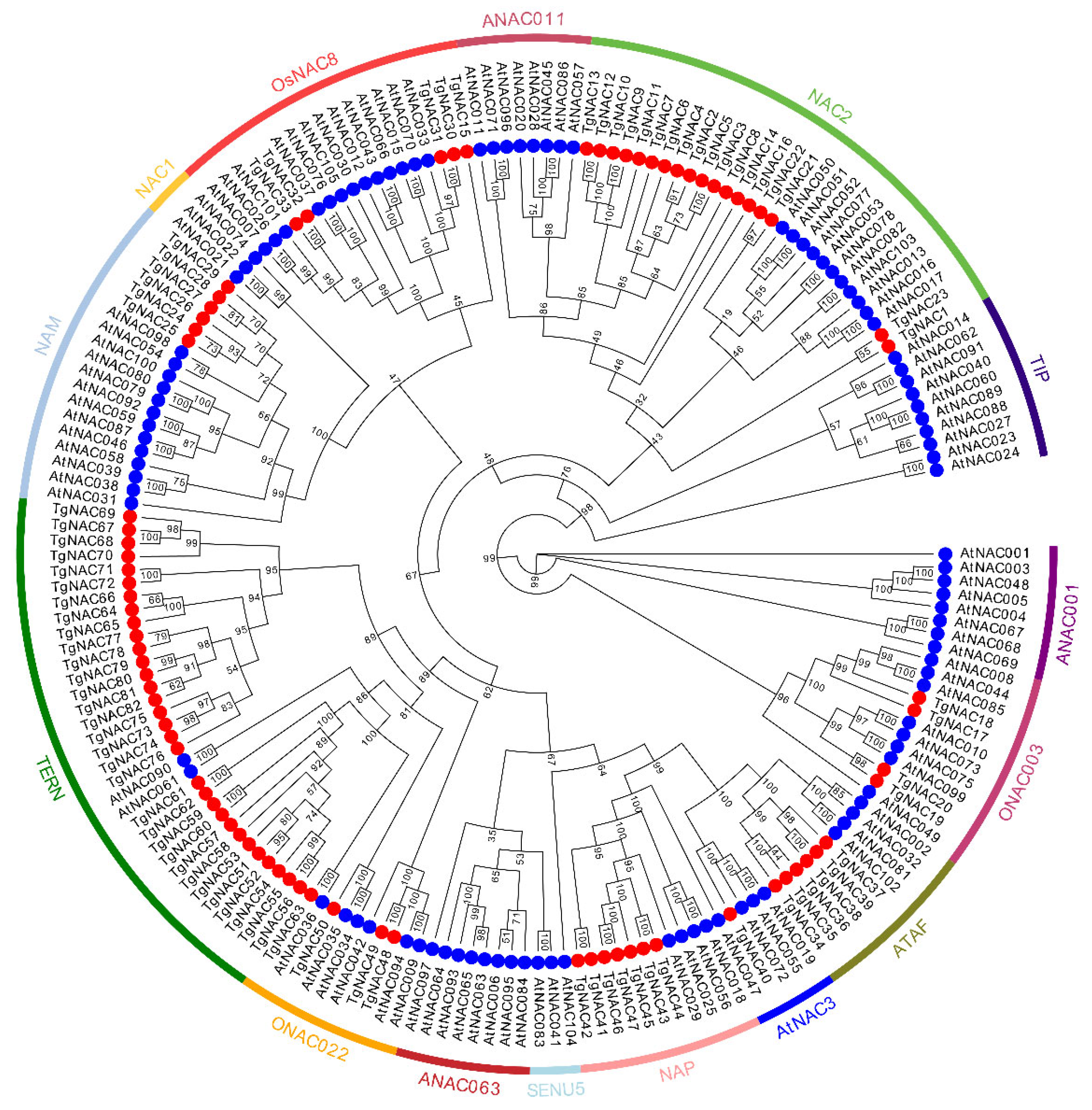
3.2. Phylogenetic Analysis of NAC Family
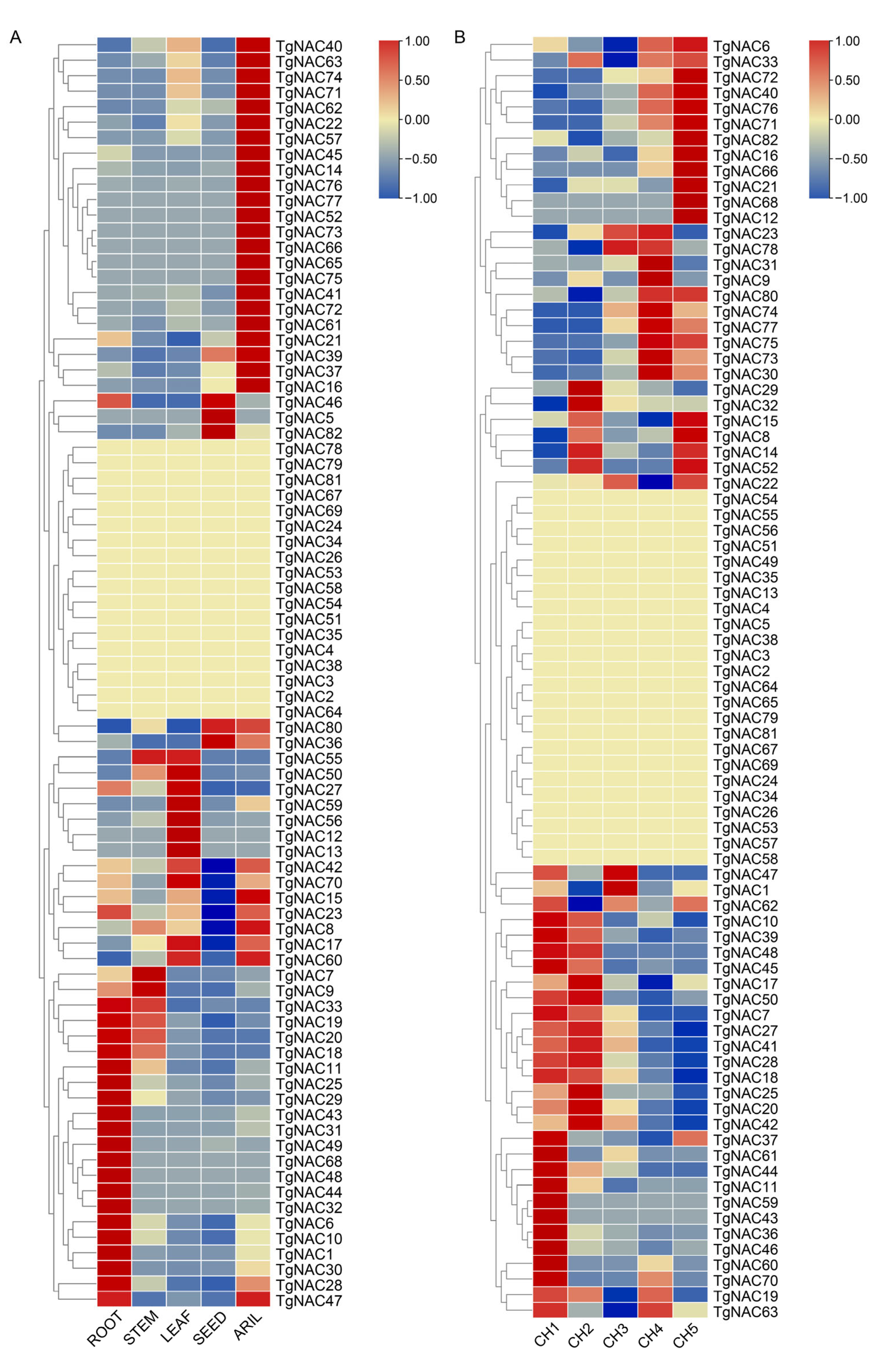
3.3. Tissue-Specific Expression Analysis of NAC Family in T. grandis
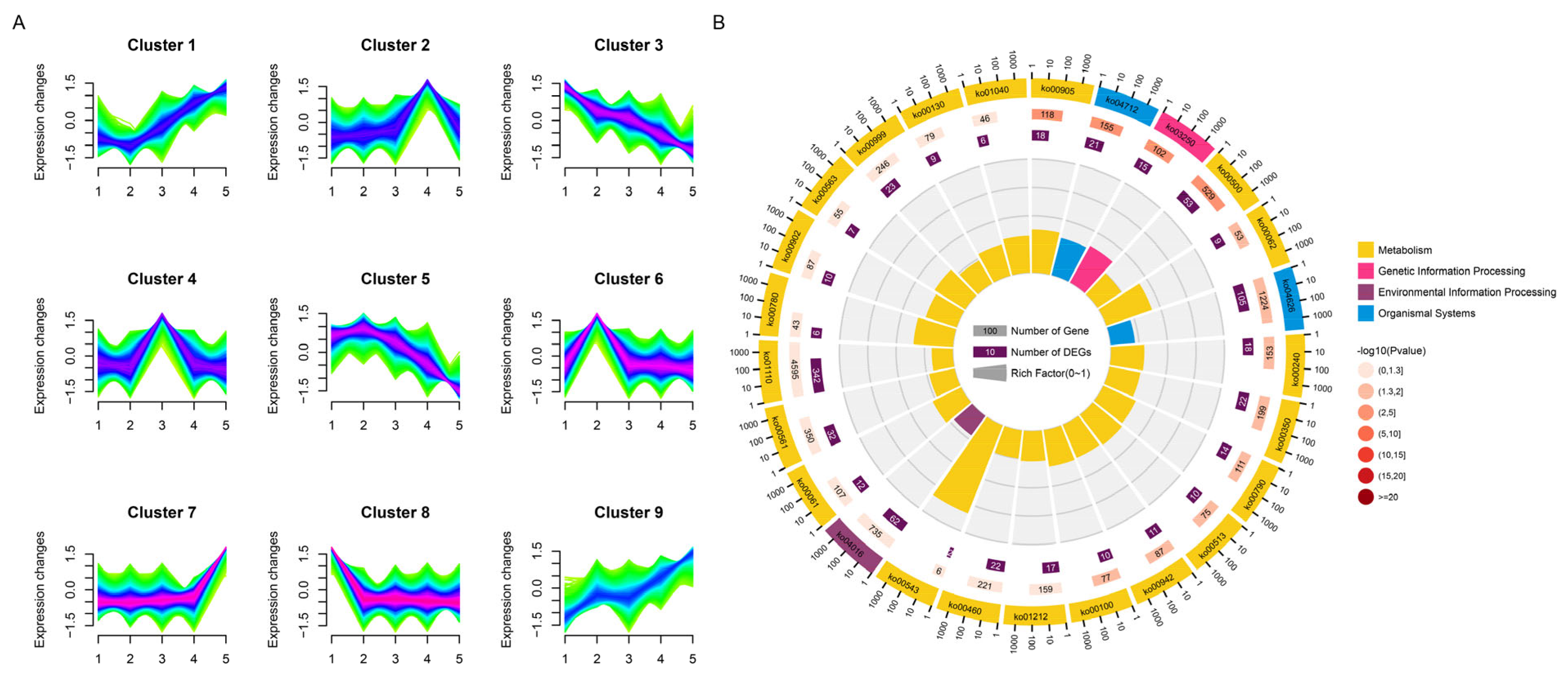
3.4. Expression Analysis of NAC Family During Female Cone Development in T. grandis
3.5. Cis-Element Analysis of NAC Family in T. grandis
3.6. Functional Characterization of TgNAC72 During Female Cone Development
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Han, K.; Zhao, Y.; Sun, Y.; Li, Y. NACs, generalist in plant life. Plant Biotechnol. J. 2023, 21, 2433–2457. [Google Scholar] [CrossRef]
- Aida, M.; Ishida, T.; Fukaki, H.; Fujisawa, H.; Tasaka, M. Genes involved in organ separation in Arabidopsis: An analysis of the cup-shaped cotyledon mutant. Plant Cell 1997, 9, 841–857. [Google Scholar] [CrossRef]
- Ooka, H.; Satoh, K.; Doi, K.; Nagata, T.; Otomo, Y.; Murakami, K.; Matsubara, K.; Osato, N.; Kawai, J.; Carninci, P.; et al. Comprehensive analysis of NAC family genes in Oryza sativa and Arabidopsis thaliana. DNA Res. 2003, 10, 239–247. [Google Scholar] [CrossRef]
- Guerin, C.; Roche, J.; Allard, V.; Ravel, C.; Mouzeyar, S.; Bouzidi, M.F. Genome-wide analysis, expansion and expression of the NAC family under drought and heat stresses in bread wheat (T. aestivum L.). PLoS ONE 2019, 14, e0213390. [Google Scholar] [CrossRef]
- Xiong, H.; He, H.; Chang, Y.; Miao, B.; Liu, Z.; Wang, Q.; Dong, F.; Xiong, L. Multiple roles of NAC transcription factors in plant development and stress responses. J. Integr. Plant Biol. 2025, 67, 510–538. [Google Scholar] [CrossRef]
- Fu, C.; Liu, M. Genome-wide identification and molecular evolution of NAC gene family in Dendrobium nobile. Front. Plant Sci. 2023, 14, 1232804. [Google Scholar] [CrossRef] [PubMed]
- Olsen, A.N.; Ernst, H.A.; Leggio, L.L.; Skriver, K. NAC transcription factors: Structurally distinct, functionally diverse. Trends Plant Sci. 2005, 10, 79–87. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.-S.; Li, H.-L.; Grierson, D.; Fu, D.-Q. NAC transcription factor family regulation of fruit ripening and quality: A review. Cells 2022, 11, 525. [Google Scholar] [CrossRef]
- Fuertes-Aguilar, J.; Matilla, A.J. Transcriptional control of seed life: New insights into the role of the NAC family. Int. J. Mol. Sci. 2024, 25, 5369. [Google Scholar] [CrossRef]
- Forlani, S.; Mizzotti, C.; Masiero, S. The NAC side of the fruit: Tuning of fruit development and maturation. BMC Plant Biol. 2021, 21, 238. [Google Scholar] [CrossRef]
- Kunieda, T.; Mitsuda, N.; Ohme-Takagi, M.; Takeda, S.; Aida, M.; Tasaka, M.; Kondo, M.; Nishimura, M.; Hara-Nishimura, I. NAC family proteins NARS1/NAC2 and NARS2/NAM in the outer integument regulate embryogenesis in Arabidopsis. Plant Cell 2008, 20, 2631–2642. [Google Scholar] [CrossRef]
- Huysmans, M.; Buono, R.A.; Skorzinski, N.; Radio, M.C.; De Winter, F.; Parizot, B.; Mertens, J.; Karimi, M.; Fendrych, M.; Nowack, M.K. NAC transcription factors ANAC087 and ANAC046 control distinct aspects of programmed cell death in the Arabidopsis columella and lateral root cap. Plant Cell 2018, 30, 2197–2213. [Google Scholar] [CrossRef]
- Shan, W.; Kuang, J.-F.; Wei, W.; Fan, Z.-Q.; Deng, W.; Li, Z.-G.; Bouzayen, M.; Pirrello, J.; Lu, W.-J.; Chen, J.-Y. MaXB3 modulates MaNAC2, MaACS1, and MaACO1 stability to repress ethylene biosynthesis during banana fruit ripening. Plant Physiol. 2020, 184, 1153–1171. [Google Scholar] [CrossRef]
- Sun, Q.; Li, Y.; Gong, D.; Hu, A.; Zhong, W.; Zhao, H.; Ning, Q.; Tan, Z.; Liang, K.; Mu, L.; et al. A NAC-EXPANSIN module enhances maize kernel size by controlling nucellus elimination. Nat. Commun. 2022, 13, 5708. [Google Scholar] [CrossRef]
- Huang, C.; Zhao, J.; Huang, Q.; Peng, L.; Huang, Z.; Li, W.; Sun, S.; He, Y.; Wang, Z. OsNAC3 regulates seed germination involving abscisic acid pathway and cell elongation in rice. New Phytol. 2024, 241, 650–664. [Google Scholar] [CrossRef]
- Pimenta, M.R.; Silva, P.A.; Mendes, G.C.; Alves, J.R.; Caetano, H.D.N.; Machado, J.P.B.; Brustolini, O.J.B.; Carpinetti, P.A.; Melo, B.P.; Silva, J.C.F.; et al. The stress-induced soybean NAC transcription factor GmNAC81 plays a positive role in developmentally programmed leaf senescence. Plant Cell Physiol. 2016, 57, 1098–1114. [Google Scholar] [CrossRef]
- Puranik, S.; Sahu, P.P.; Srivastava, P.S.; Prasad, M. NAC proteins: Regulation and role in stress tolerance. Trends Plant Sci. 2012, 17, 369–381. [Google Scholar] [CrossRef]
- Zhu, Y.; Yan, J.; Liu, W.; Liu, L.; Sheng, Y.; Sun, Y.; Li, Y.; Scheller, H.V.; Jiang, M.; Hou, X.; et al. Phosphorylation of a NAC Transcription Factor by a Calcium/Calmodulin-Dependent Protein Kinase Regulates Abscisic Acid-Induced Antioxidant Defense in Maize. Plant Physiol. 2016, 171, 1651–1664. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.; Han, T.; Xiang, Y.; Wang, C.; Zhang, A. The transcription factor ZmNAC84 increases maize salt tolerance by regulating ZmCAT1 expression. Crop J. 2024, 12, 1344–1356. [Google Scholar] [CrossRef]
- Xu, P.; Ma, W.; Feng, H.; Cai, W. The NAC056 transcription factor confers freezing tolerance by positively regulating expression of CBFs and NIA1 in Arabidopsis. Plant Commun. 2024, 5, 100923. [Google Scholar] [CrossRef] [PubMed]
- Mei, C.; Yang, J.; Mei, Q.; Jia, D.; Yan, P.; Feng, B.; Mamat, A.; Gong, X.; Guan, Q.; Mao, K.; et al. MdNAC104 positively regulates apple cold tolerance via CBF—Dependent and CBF—Independent pathways. Plant Biotechnol. J. 2023, 21, 2057–2073. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, S.; Liu, X.; Wang, R.; Chen, W.; Suo, J.; Yan, J.; Wu, J. Agrobacterium-mediated transient expression in Torreya grandis cones: A simple and rapid tool for gene expression and functional gene assay. Sci. Hortic. 2024, 338, 113664. [Google Scholar] [CrossRef]
- Yan, J.; Wang, M.; Zeng, H.; Yang, H.; Lv, K.; Zhou, Z.; Hou, Y.; Zhang, J.; Kong, N.; Wu, J. Ti3C2Tx MXene nanosheets protect Torreya grandis against root rot disease. Chem. Eng. J. 2024, 481, 148687. [Google Scholar] [CrossRef]
- Yan, J.; Zeng, H.; Chen, W.; Zheng, S.; Luo, J.; Jiang, H.; Yang, B.; Farag, M.A.; Lou, H.; Song, L.; et al. Effects of tree age on flavonoids and antioxidant activity in Torreya grandis nuts via integrated metabolome and transcriptome analyses. Food Front. 2023, 4, 358–367. [Google Scholar] [CrossRef]
- Yan, J.; Zeng, H.; Chen, W.; Luo, J.; Kong, C.; Lou, H.; Wu, J. New insights into the carotenoid biosynthesis in Torreya grandis kernels. Hortic. Plant J. 2023, 9, 1108–1118. [Google Scholar] [CrossRef]
- Wang, R.; Wang, S.; Zheng, J.; Yan, J.; Liu, Z.; Liu, Y.; Zhong, H.; Chen, W.; Wu, J.; Yan, J. TgMYBS1-TgSUS1 regulatory module mediates sucrose metabolism in Torreya grandis kernels. Int. J. Biol. Macromol. 2025, 318, 145150. [Google Scholar] [CrossRef]
- Suo, J.; Liu, Y.; Yan, J.; Li, Q.; Chen, W.; Liu, Z.; Zhang, Z.; Hu, Y.; Yu, W.; Yan, J. Sucrose promotes cone enlargement via the TgNGA1—TgWRKY47—TgEXPA2 module in Torreya grandis. New Phytol. 2024, 243, 1823–1839. [Google Scholar] [CrossRef]
- Chen, W.; Yan, J.; Guan, Y.; Lou, H.; Wu, J. Genome-wide identification of WOX gene family and its expression pattern in rapid expansion of Torreya grandis ovulate and staminate strobili. Sci. Hortic. 2024, 330, 113050. [Google Scholar] [CrossRef]
- Lou, H.; Song, L.; Li, X.; Zi, H.; Chen, W.; Gao, Y.; Zheng, S.; Fei, Z.; Sun, X.; Wu, J. The Torreya grandis genome illuminates the origin and evolution of gymnosperm-specific sciadonic acid biosynthesis. Nat. Commun. 2023, 14, 1315. [Google Scholar] [CrossRef]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME Suite. Nucleic Acids Res. 2015, 43, W39–W49. [Google Scholar] [CrossRef]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y.; et al. TBtools-II: A “one for all, all for one” bioinformatics platform for biological big-data mining. Mol. Plant 2023, 16, 1733–1742. [Google Scholar] [CrossRef] [PubMed]
- Tian, F.; Yang, D.-C.; Meng, Y.-Q.; Jin, J.; Gao, G. PlantRegMap: Charting functional regulatory maps in plants. Nucleic Acids Res. 2019, 48, D1104–D1113. [Google Scholar] [CrossRef] [PubMed]
- Rozewicki, J.; Li, S.; Amada, K.M.; Standley, D.M.; Katoh, K. MAFFT-DASH: Integrated protein sequence and structural alignment. Nucleic Acids Res. 2019, 47, W5–W10. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.-T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies. Mol. Biol. Evol. 2014, 32, 268–274. [Google Scholar] [CrossRef]
- Wang, S.; Wang, R.; Zhong, H.; Yan, J.; Liu, Z.; Liu, Y.; Chen, W.; Wu, J.; Yan, J. Sugars will eventually be exported transporter 18 mediates glucose metabolism in Torreya grandis kernels. Int. J. Biol. Macromol. 2025, 310, 143529. [Google Scholar] [CrossRef]
- Kim, T.; Alvarez, J.C.; Rana, D.; Preciado, J.; Liu, T.; Begcy, K. Evolution of NAC transcription factors from early land plants to domesticated crops. Plant Cell Physiol. 2025, 66, 566–580. [Google Scholar] [CrossRef]
- Zhang, Z.; Dong, J.; Ji, C.; Wu, Y.; Messing, J. NAC-type transcription factors regulate accumulation of starch and protein in maize seeds. Proc. Natl. Acad. Sci. USA 2019, 116, 11223–11228. [Google Scholar] [CrossRef]
- Cairns, J.R.K.; Esen, A. β-Glucosidases. Cell. Mol. Life Sci. 2010, 67, 3389–3405. [Google Scholar] [CrossRef]
- Sengupta, S.; Datta, M.; Datta, S. β-Glucosidase: Structure, function and industrial applications. In Glycoside Hydrolases; Elsevier: Amsterdam, The Netherlands, 2023; pp. 97–120. [Google Scholar]
- Mól, P.C.G.; Júnior, J.C.Q.; Veríssimo, L.A.A.; Boscolo, M.; Gomes, E.; Minim, L.A.; Da Silva, R. β-glucosidase: An overview on immobilization and some aspects of structure, function, applications and cost. Process Biochem. 2023, 130, 26–39. [Google Scholar] [CrossRef]
- Du, W.; Yang, J.; Li, Q.; Jiang, W.; Pang, Y. Medicago truncatula β—Glucosidase 17 contributes to drought and salt tolerance through antioxidant flavonoid accumulation. Plant Cell Environ. 2024, 47, 3076–3089. [Google Scholar] [CrossRef]
- Xu, Z.-Y.; Lee, K.H.; Dong, T.; Jeong, J.C.; Jin, J.B.; Kanno, Y.; Kim, D.H.; Kim, S.Y.; Seo, M.; Bressan, R.A.; et al. A vacuolar β-glucosidase homolog that possesses glucose-conjugated abscisic acid hydrolyzing activity plays an important role in osmotic stress responses in Arabidopsis. Plant Cell 2012, 24, 2184–2199. [Google Scholar] [CrossRef]
- Ren, R.; Li, D.; Zhen, C.; Chen, D.; Chen, X. Specific roles of Os4BGlu10, Os6BGlu24, and Os9BGlu33 in seed germination, root elongation, and drought tolerance in rice. Planta 2019, 249, 1851–1861. [Google Scholar] [CrossRef]
- Niu, F.; Bu, Y.; Tang, S.; Yang, X.; Zhang, L.; Song, X. Comprehensive analysis of glycoside hydrolase family 1 β-glucosidase genes in wheat reveals TaBGLU81 functions in pollen development and fertility conversion. Plant Growth Regul. 2024, 104, 215–231. [Google Scholar] [CrossRef]
- Dong, X.; Jiang, Y.; Hur, Y. Genome-wide analysis of glycoside hydrolase family 1 β-glucosidase genes in Brassica rapa and their potential role in pollen development. Int. J. Mol. Sci. 2019, 20, 1663. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, L.; Chen, C.; Liu, M.; Bi, W.; Li, S.; Zhang, X.; Han, T. Comprehensive Identification and Expression Profiling of the NAC Family During Female Cone Development in Torreya grandis. Horticulturae 2025, 11, 1229. https://doi.org/10.3390/horticulturae11101229
Wang L, Chen C, Liu M, Bi W, Li S, Zhang X, Han T. Comprehensive Identification and Expression Profiling of the NAC Family During Female Cone Development in Torreya grandis. Horticulturae. 2025; 11(10):1229. https://doi.org/10.3390/horticulturae11101229
Chicago/Turabian StyleWang, Long, Chang Chen, Meiying Liu, Wenfei Bi, Su Li, Xiong Zhang, and Tong Han. 2025. "Comprehensive Identification and Expression Profiling of the NAC Family During Female Cone Development in Torreya grandis" Horticulturae 11, no. 10: 1229. https://doi.org/10.3390/horticulturae11101229
APA StyleWang, L., Chen, C., Liu, M., Bi, W., Li, S., Zhang, X., & Han, T. (2025). Comprehensive Identification and Expression Profiling of the NAC Family During Female Cone Development in Torreya grandis. Horticulturae, 11(10), 1229. https://doi.org/10.3390/horticulturae11101229




