Genetic Diversity and Structure of a Critically Endangered Ornamental Species, Rhododendron farinosum, with Extremely Small Populations
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Material and Extraction of DNA
2.2. ddRAD Library Preparation and Sequencing
2.3. SNP Calling
2.4. Data Analysis
2.4.1. Analysis of Genetic Diversity and Structure
2.4.2. Demographic History
3. Results
3.1. ddRAD Sequencing and Data Processing
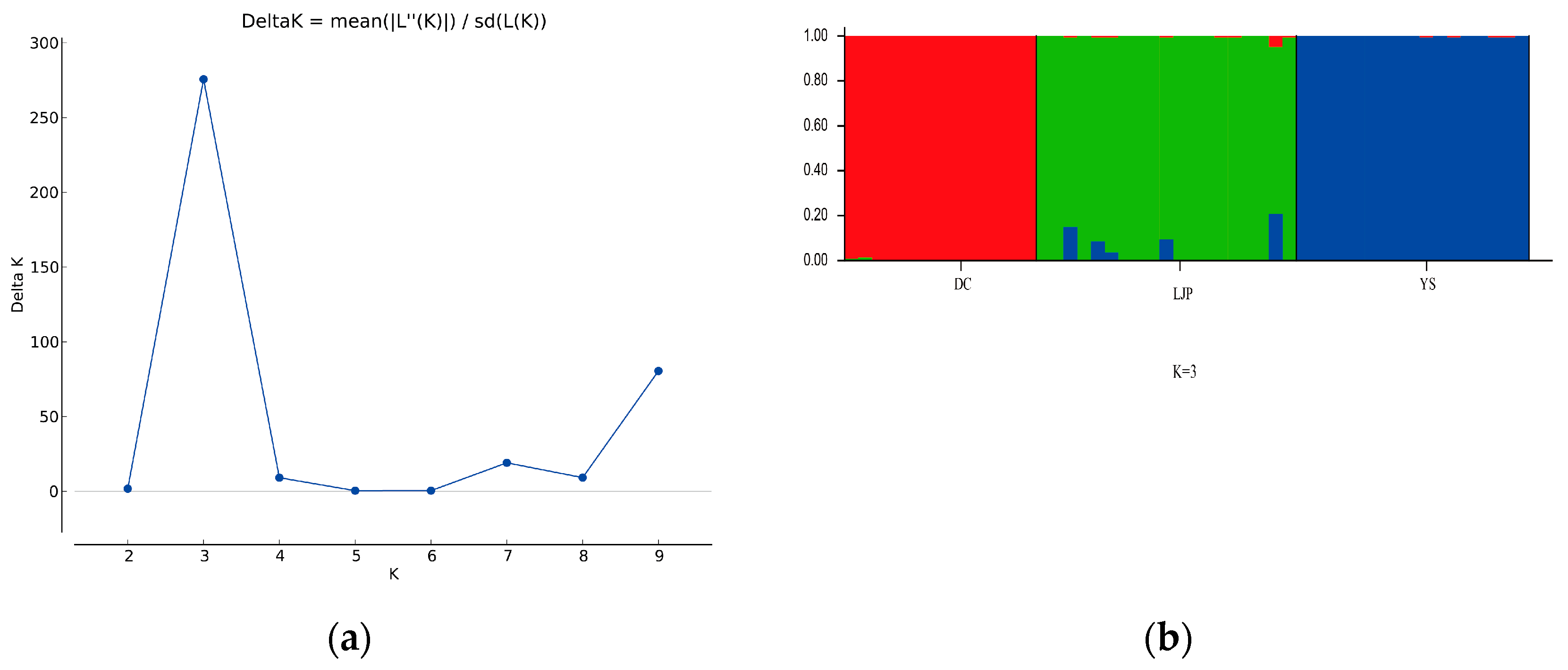
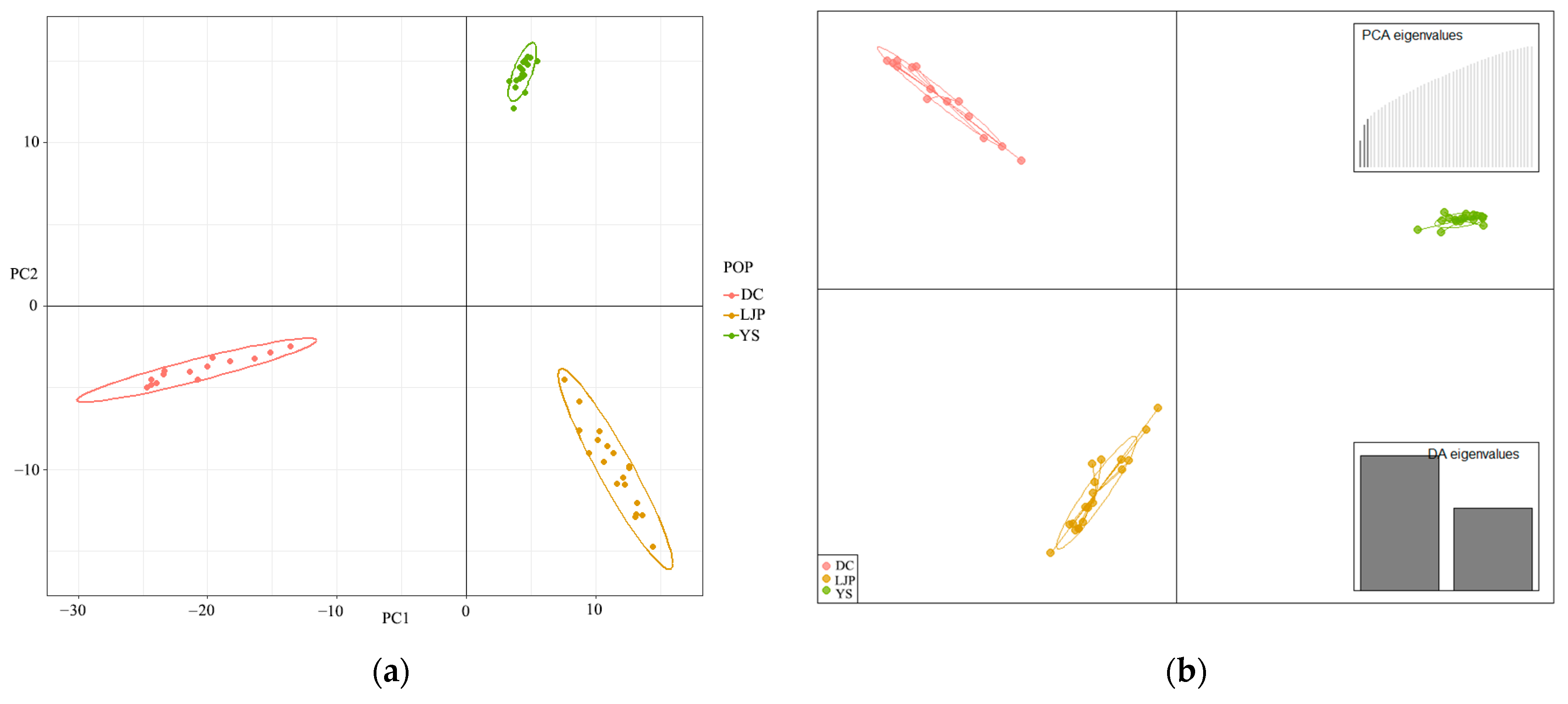
3.2. Genetic Diversity and Structure of R. farinosum
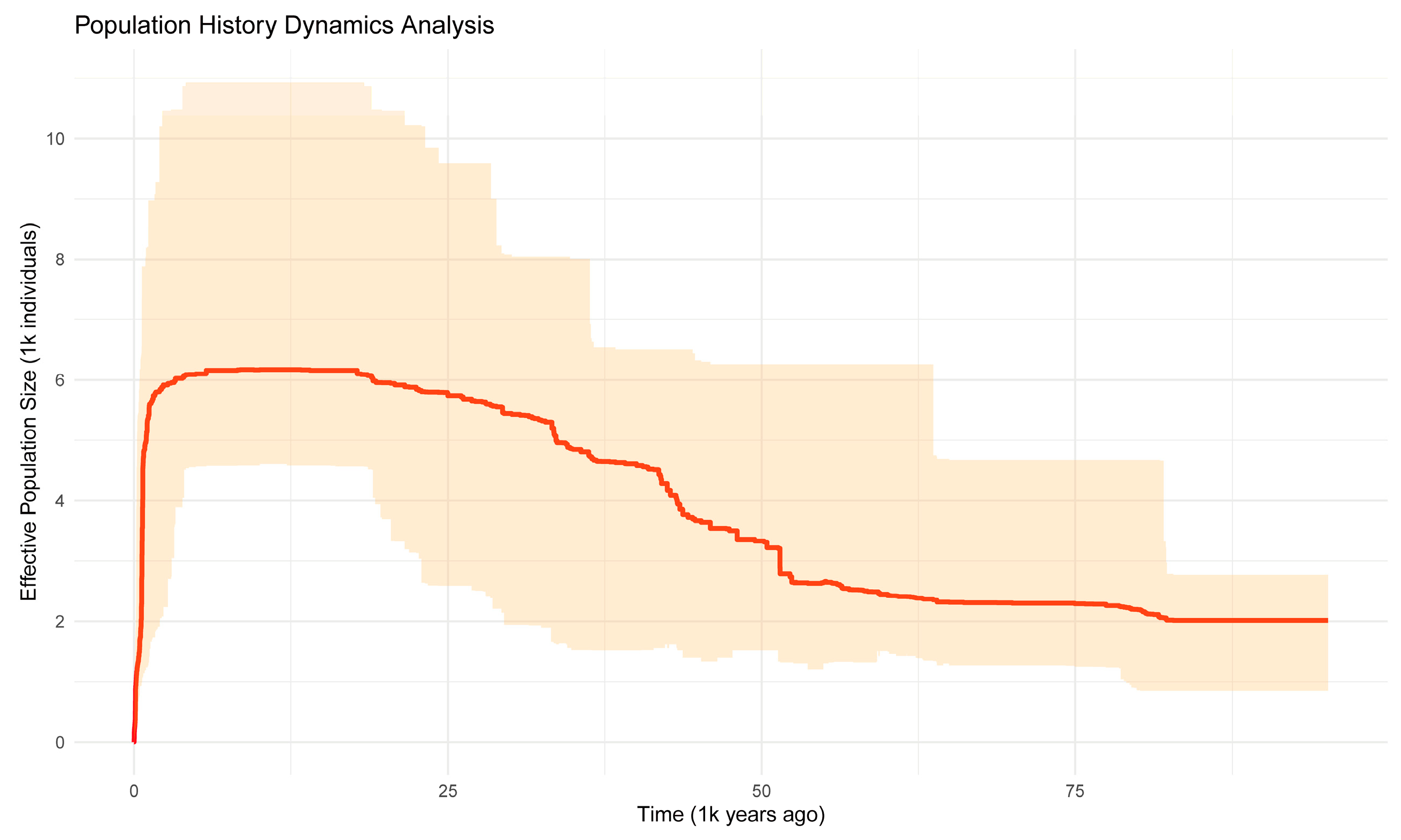
3.3. Population Demographic History of R. farinosum
4. Discussion
4.1. Genetic Diversity
4.2. Genetic Differentiation and Genetic Structure
4.3. Population Dynamic History of R. farinosum
4.4. Conservation and Management Strategies
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- De Kort, H.; Prunier, J.G.; Ducatez, S.; Honnay, O. Life history, climate, and biogeography interactively affect the worldwide genetic diversity of plant and animal populations. Nat. Commun. 2021, 12, 516–527. [Google Scholar] [CrossRef] [PubMed]
- Hamabata, T.; Kinoshita, G.; Kurita, K.; Cao, P.L.; Ito, M.; Murata, J.; Komaki, Y.; Isagi, Y.; Makino, T. Endangered island endemic plants have vulnerable genomes. Commun. Biol. 2019, 2, 244–253. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.L.; Qin, Y.Y.; Du, Z.; Yan, G.Q. Genetic diversity and differentiation of the endangered tree Elaeagnus mollis Diels (Elaeagnus L.) as revealed by simple sequence repeat (SSR) markers. Biochem. Syst. Ecol. 2012, 40, 25–33. [Google Scholar] [CrossRef]
- Hamrick, J.L.; Godt, M.J.W.; Sherman-Broyles, S.L. Factors influencing levels of genetic diversity in woody plant species. New For. 1992, 6, 95–124. [Google Scholar] [CrossRef]
- Liu, X.F.; Ma, Y.P.; Wan, Y.M.; Li, Z.H.; Ma, H. Genetic diversity of Phyllanthus emblica from two different climate type areas. Front. Plant Sci. 2020, 11, 580812. [Google Scholar] [CrossRef] [PubMed]
- He, S.; Yuan, C.J.; Zhang, P.L.; Wang, H.D.; Luo, D.L.; Dai, X.Y. Study on the characteristics of genetic diversity of different populations of Guizhou endemic plant Rhododendron pudingense based on microsatellite markers. BMC Plant Biol. 2024, 24, 77–88. [Google Scholar] [CrossRef]
- Ruang-areerate, P.; Sonthirod, C.; Sangsrakru, D.; Waiyamitra, P.; Maknual, C.; Wanthongchai, P.; Chomriang, P.; Pootakham, W.; Tangphatsornruang, S. Elucidating SNP-Based Population Structure and Genetic Diversity of Bruguiera gymnorhiza (L.) Savigny in Thailand. Forests 2023, 14, 693. [Google Scholar] [CrossRef]
- Wang, Y.P.; Li, H.H.; Yang, Z.J.; Liu, B.Y.; Liu, Y.J.; Zhong, Y.D. Genotyping-by-Sequencing Study of the Genetic Diversity and Population Structure of the Endangered Plant Tsoongiodendron odorum Chun in China. Forests 2024, 15, 910. [Google Scholar] [CrossRef]
- Li, Z.; Ran, Z.H.; Zhang, Y.; An, M.T. Genetic Diversity and Structure of Geodorum eulophioides, a Plant Species with Extremely Small Populations in China. Diversity 2023, 15, 990. [Google Scholar] [CrossRef]
- Zhu, X.L.; Tang, J.M.; Jiang, H.D.; Yang, Y.S.; Chen, Z.Y.; Zou, R.; Xu, A.Z.; Luo, Y.J.; Deng, Z.H.; Wei, X. Genomic evidence reveals high genetic diversity in a narrowly distributed species and natural hybridization risk with a widespread species in the genus Geodorum. BMC Plant Biol. 2023, 23, 317–328. [Google Scholar] [CrossRef] [PubMed]
- Garcia, C.B.; Silva, A.V.; Carvalho, I.A.S.; Nascimento, W.F.D.; Ramos, S.L.F.; Rodrigues, D.P.; Zucchi, M.I.; Costa, F.M.; Alves-Pereira, A.; Araújo Batista, C.E.D. Low Diversity and High Genetic Structure for Platonia insignis Mart., an Endangered Fruit Tree Species. Plants 2024, 13, 1033. [Google Scholar] [CrossRef] [PubMed]
- Peterson, B.K.; Weber, J.N.; Kay, E.H.; Fisher, H.S.; Hoekstra, H.E. Double digest RADseq: An inexpensive method for de novo SNP discovery and genotyping in model and non-model species. PLoS ONE 2012, 7, e37135. [Google Scholar] [CrossRef] [PubMed]
- Cai, M.; Wen, Y.; Uchiyama, K.; Onuma, Y.; Tsumura, Y. Population genetic diversity and structure of ancient tree populations of Cryptomeria japonica var. sinensis based on RAD-seq data. Forests 2020, 11, 1192. [Google Scholar] [CrossRef]
- Wang, X.; Huang, Y.; Long, C. Cross-amplification and characterization of microsatellite loci for the genus Rhododendron. HortScience 2010, 45, 1394–1397. [Google Scholar] [CrossRef]
- Liu, N.Y.; Zhang, L.; Zhou, Y.L.; Tu, M.L.; Wu, Z.Z.; Gui, D.P.; Ma, Y.P.; Wang, J.H.; Zhang, C.J. The Rhododendron Plant Genome Database (RPGD): A comprehensive online omics database for Rhododendron. BMC Genom. 2021, 22, 376–385. [Google Scholar] [CrossRef] [PubMed]
- Li, K.J.; Liu, X.F.; Zhang, J.H.; Zhou, X.L.; Yang, L.; Shen, S.K. Complexity responses of Rhododendron species to climate change in China reveal their urgent need for protection. For. Ecosyst. 2023, 10, 100124. [Google Scholar] [CrossRef]
- Gibbs, D.; Chamberlain, D.; Argent, G. The Red List of Rhododendrons; Botanic Gardens Conservation International: Richmond, UK, 2011. [Google Scholar]
- Doyle, J.J.; Doyle, J.L. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 1987, 19, 11–15. [Google Scholar]
- Catchen, J.; Hohenlohe, P.A.; Bassham, S.; Amores, A.; Cresko, W.A. Stacks: An analysis tool set for population genomics. Mol. Ecol. 2013, 22, 3124–3140. [Google Scholar] [CrossRef] [PubMed]
- Danecek, P.; Auton, A.; Abecasis, G.; Albers, C.A.; Banks, E.; DePristo, M.A.; Handsaker, R.E.; Lunter, G.; Marth, G.T.; Sherry, S.T. The variant call format and VCFtools. Bioinformatics 2011, 27, 2156–2158. [Google Scholar] [CrossRef]
- Tajima, F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 1989, 123, 585–595. [Google Scholar] [CrossRef]
- Lischer, H.E.L.; Excoffier, L. PGDSpider: An automated data conversion tool for connecting population genetics and genomics programs. Bioinformatics 2012, 28, 298–299. [Google Scholar] [CrossRef] [PubMed]
- Excoffier, L.; Lischer, H.E.L. Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 2010, 10, 564–567. [Google Scholar] [CrossRef] [PubMed]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef] [PubMed]
- Earl, D.A.; VonHoldt, B.M. STRUCTURE HARVESTER: A website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 2012, 4, 359–361. [Google Scholar] [CrossRef]
- Liu, X.; Fu, Y.X. Stairway Plot 2: Demographic history inference with folded SNP frequency spectra. Genome Biol. 2020, 21, 280–288. [Google Scholar] [CrossRef]
- Walsh, C.A.J.; Momigliano, P.; Boussarie, G.; Robbins, W.D.; Bonnin, L.; Fauvelot, C.; Kiszka, J.J.; Mouillot, D.; Vigliola, L.; Manel, S. Genomic insights into the historical and contemporary demographics of the grey reef shark. Heredity 2022, 128, 225–235. [Google Scholar] [CrossRef] [PubMed]
- Cross, J.R. Rhododendron Ponticum L. J. Ecol. 1975, 63, 345–364. [Google Scholar] [CrossRef]
- Yoichi, W.; Tamaki, I.; Sakaguchi, S.; Song, J.S.; Yamamoto, S.I.; Tomaru, N. Population demographic history of a temperate shrub, Rhododendron weyrichii (Ericaceae), on continental islands of Japan and South Korea. Ecol. Evol. 2016, 6, 8800–8810. [Google Scholar] [CrossRef]
- Liu, D.T.; Zhang, L.; Wang, J.H.; Ma, Y.P. Conservation genomics of a threatened Rhododendron: Contrasting patterns of population structure revealed from neutral and selected SNPs. Front. Genet. 2020, 11, 757–766. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.J.; Liu, X.F.; Liu, D.T.; Cao, Y.R.; Li, Z.H.; Ma, Y.P.; Ma, H. Genetic diversity and structure of Rhododendron meddianum, a plant species with extremely small populations. Plant Divers. 2021, 43, 472–479. [Google Scholar] [CrossRef] [PubMed]
- Wachowiak, W.; Salmela, M.J.; Ennos, R.A.; Iason, G.; Cavers, S. High genetic diversity at the extreme range edge: Nucleotide variation at nuclear loci in Scots pine (Pinus sylvestris L.) in Scotland. Heredity 2011, 106, 775–787. [Google Scholar] [CrossRef] [PubMed]
- Wright, S. Evolution and the Genetics of Populations: A Treatise in Four Volumes: Volume 4: Variability Within and Among Natural Populations; University of Chicago Press: Chicago, IL, USA, 1978. [Google Scholar]
- Laikre, L.; Allendorf, F.W.; Aroner, L.C.; Baker, C.S.; Gregovich, D.P.; Hansen, M.M.; Jackson, J.A.; Kendall, K.C.; McKelvey, K.; Neel, M.C. Neglect of genetic diversity in implementation of the convention of biological diversity. Conserv. Biol. 2010, 24, 86–88. [Google Scholar] [CrossRef] [PubMed]
- Mondon, A.; Owens, G.L.; Poverene, M.; Cantamutto, M.; Rieseberg, L.H. Gene flow in Argentinian sunflowers as revealed by genotyping-by-sequencing data. Evol. Appl. 2018, 11, 193–204. [Google Scholar] [CrossRef]
- Xiao, L.Q.; Ge, X.J.; Gong, X.; Hao, G.; Zheng, S.X. ISSR variation in the endemic and endangered plant Cycas guizhouensis (Cycadaceae). Ann. Bot. 2004, 94, 133–138. [Google Scholar] [CrossRef]
- Wu, Q.; Dong, S.; Zhao, Y.; Yang, L.; Qi, X.J.; Ren, Z.H.; Dong, S.B.; Cheng, J. Genetic diversity, population genetic structure and gene flow in the rare and endangered wild plant Cypripedium macranthos revealed by genotyping-by-sequencing. BMC Plant Biol. 2023, 23, 254–266. [Google Scholar] [CrossRef] [PubMed]
- Guilliams, C.M.; Hasenstab-Lehman, K.E. Conservation Genetics of the Endangered Lompoc Yerba Santa (Eriodictyon capitatum Eastw., Namaceae), including Phylogenomic Insights into the Evolution of Eriodictyon. Plants 2023, 13, 90. [Google Scholar] [CrossRef] [PubMed]
- Wu, F.Q.; Shen, S.K.; Zhang, X.J.; Wang, Y.H.; Sun, W.B. Genetic diversity and population structure of an extremely endangered species: The world’s largest Rhododendron. AoB Plants 2015, 7, plu082. [Google Scholar] [CrossRef] [PubMed]
- Feyissa, T.; Nybom, H.; Bartish, I.V.; Welander, M. Analysis of genetic diversity in the endangered tropical tree species Hagenia abyssinica using ISSR markers. Genet. Resour. Crop Evol. 2007, 54, 947–958. [Google Scholar] [CrossRef]
- Miao, Y.C.; Zhang, Z.J.; Su, J.R. Low genetic diversity in the endangered following a population bottleneck, a low effective population size and increased inbreeding. Silvae Genet. 2016, 65, 59–66. [Google Scholar] [CrossRef]
- López-A, D.M.; Bock, B.C.; Bedoya, G. Genetic structure in remnant populations of an endangered Andean Magnolia. Biotropica 2008, 40, 375–379. [Google Scholar] [CrossRef]
- Kramer, A.T.; Ison, J.L.; Ashley, M.V.; Howe, H.F. The paradox of forest fragmentation genetics. Conserv. Biol. 2008, 22, 878–885. [Google Scholar] [CrossRef]
- Nybom, H. Comparison of different nuclear DNA markers for estimating intraspecific genetic diversity in plants. Mol. Ecol. 2004, 13, 1143–1155. [Google Scholar] [CrossRef] [PubMed]
- Du, Y.L.; Fan, W.X.; Liu, Y.L. Characteristic and cause of debris flow in Dongchuan copper mining area. Northwest Geol. 2010, 43, 130–136. [Google Scholar]
- Zhang, X.P.; Li, X.H.; Qiu, Y.X. Genetic diversity of the endangered species Kirengeshoma palmata (Saxifragaceae) in China. Biochem. Syst. Ecol. 2006, 34, 38–47. [Google Scholar] [CrossRef]
- Hamrick, J.L.; Godt, M.J.W. Effects of life history traits on genetic diversity in plant species. Philos. Trans. Biol. Sci. 1996, 351, 1291–1298. [Google Scholar]
- Ennos, R.A. Estimating the relative rates of pollen and seed migration among plant populations. Heredity 1994, 72, 250–259. [Google Scholar] [CrossRef]
- Slatkin, M. Gene flow in natural populations. Annu. Rev. Ecol. Syst. 1985, 16, 393–430. [Google Scholar] [CrossRef]
- Ng, S.; Corlett, R.T. Comparative reproductive biology of the six species of Rhododendron (Ericaceae) in Hong Kong, South China. Can. J. Bot. 2000, 78, 221–229. [Google Scholar] [CrossRef]
- Wright, S. Isolation by distance. Genetics 1943, 28, 114–138. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.H.; Li, J.; Zhang, L.M.; He, Z.W.; Mei, Q.M.; Gong, X.; Jian, S.G. Population differentiation and demographic history of the Cycas taiwaniana Complex (Cycadaceae) endemic to South China as indicated by DNA sequences and microsatellite markers. Front. Genet. 2019, 10, 1238–1252. [Google Scholar] [CrossRef] [PubMed]
- Liang, L.Y. Impact of the Quaternary Ice Age on the flora and vegetation of China. Chin. Place Name 2020, 7, 51+53. [Google Scholar]
- Davis, M.B.; Shaw, R.G. Range shifts and adaptive responses to Quaternary climate change. Science 2001, 292, 673–679. [Google Scholar] [CrossRef] [PubMed]
- Kuang, M.S.; Li, J.Y.; Zhao, Y.; Chen, X.Q.; Zhang, Y.P.; Guo, T.X. A Study on the Quaternary Glacial Relics in the Gongwang Mountains in the Northeast Part of Yunnan Province. J. Glaciol. Geocryol. 1997, 19, 78–84. [Google Scholar]
- Hou, H.M.; Ye, H.; Wang, Z.; Wu, J.H.; Gao, Y.; Han, W.; Na, D.C.; Sun, G.L.; Wang, Y.L. Demographic history and genetic differentiation of an endemic and endangered Ulmus lamellosa (Ulmus). BMC Plant Biol. 2020, 20, 1–14. [Google Scholar] [CrossRef]
- Gilpin, M.E. Minimum viable populations: Processes of species extinction. Conserv. Biol. Sci. Scarcity Divers. 1986, 19–34. [Google Scholar]
- Ceballos, G.; Ehrlich, P.R.; Barnosky, A.D.; García, A.; Pringle, R.M.; Palmer, T.M. Accelerated modern human–induced species losses: Entering the sixth mass extinction. Sci. Adv. 2015, 1, e1400253. [Google Scholar] [CrossRef] [PubMed]
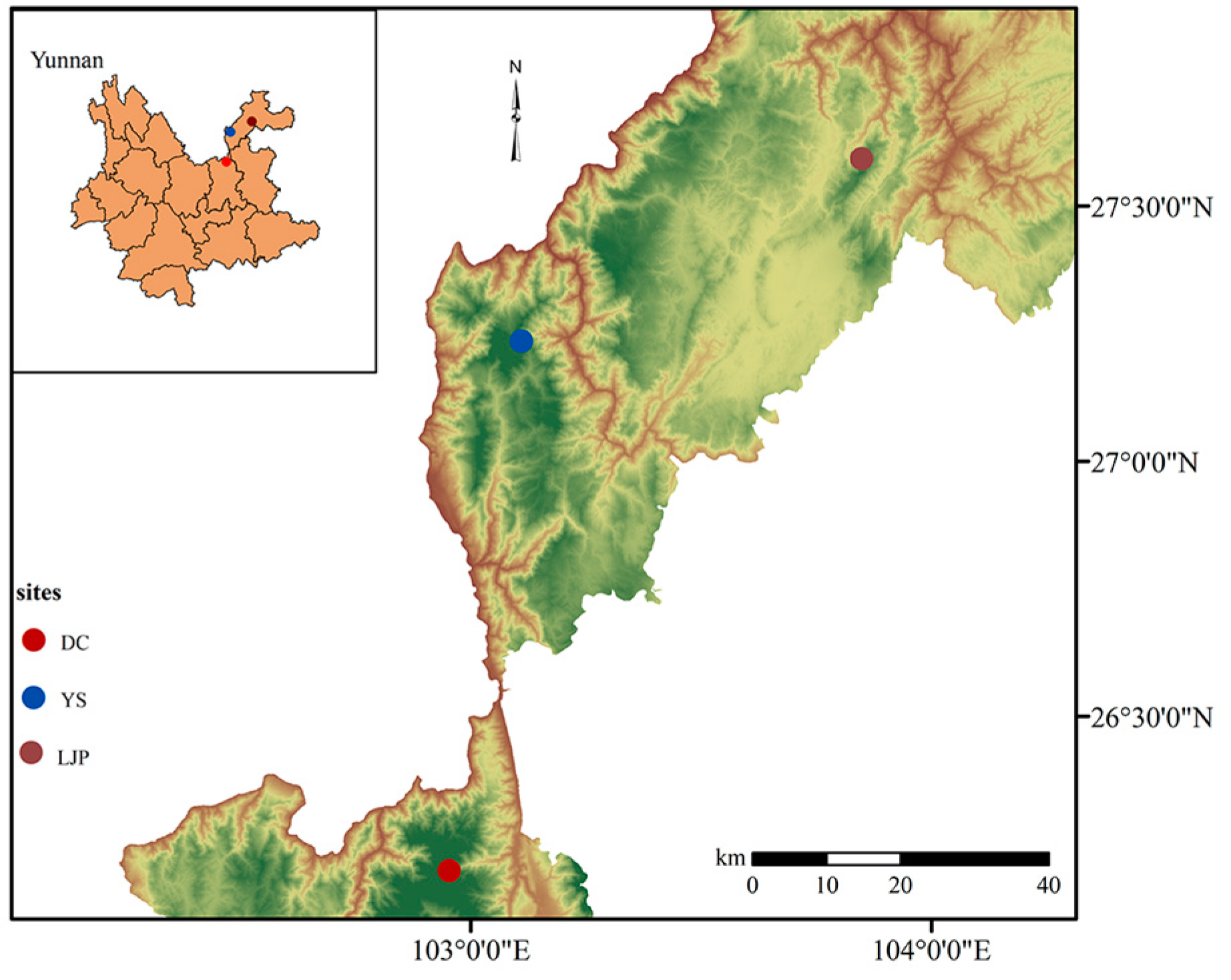

| Populations | Geographic Localities | Sample Size | Longitude (E) | Latitude (N) | Altitude (m) |
|---|---|---|---|---|---|
| DC | Lanniping | 14 | 102°57′6.53″ | 26°11′3.48″ | 3068 |
| YS | Yaoshan | 17 | 103°5′29.26″ | 27°13′25.5″ | 2757 |
| LJP | Lengjiaping | 19 | 103°50′3.19″ | 27°35′22.96″ | 2952 |
| Pop ID | Private | Ho | HE | π | FIS |
|---|---|---|---|---|---|
| DC | 1135 | 0.082 ± 0.0016 | 0.1544 ± 0.0020 | 0.1615 ± 0.0020 | 0.222 0± 0.0131 |
| LJP | 1464 | 0.1363 ± 0.0019 | 0.2026 ± 0.0020 | 0.2089 ± 0.0020 | 0.2155 ± 0.0162 |
| YS | 1229 | 0.1461 ± 0.0020 | 0.2069 ± 0.0020 | 0.2140 ± 0.0020 | 0.2014 ± 0.0160 |
| mean | 1276 | 0.1215 ± 0.0018 | 0.1880 ± 0.0020 | 0.1948 ± 0.0020 | 0.2130 ± 0.0151 |
| DC | LJP | YS | |
|---|---|---|---|
| DC | — | 0.2231 | 0.2074 |
| LJP | 165.8458 | — | 0.1383 |
| YS | 147.8450 | 24.5997 | — |
| Source of Variation | d.f. | Sum of Squares | Variance Components | Percentage Variation | p-Value |
|---|---|---|---|---|---|
| Among populations | 2 | 28,423.140 | 408.72680 Va | 37.17 | <0.01 |
| Within populations | 97 | 67,016.100 | 690.88763 Vb | 62.83 | <0.01 |
| Total | 99 | 95,439.240 | 1099.61442 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, F.; Fan, L.; Zhang, J.; Liu, W.; Li, W.; Ma, Y.; Ma, H. Genetic Diversity and Structure of a Critically Endangered Ornamental Species, Rhododendron farinosum, with Extremely Small Populations. Horticulturae 2025, 11, 51. https://doi.org/10.3390/horticulturae11010051
Li F, Fan L, Zhang J, Liu W, Li W, Ma Y, Ma H. Genetic Diversity and Structure of a Critically Endangered Ornamental Species, Rhododendron farinosum, with Extremely Small Populations. Horticulturae. 2025; 11(1):51. https://doi.org/10.3390/horticulturae11010051
Chicago/Turabian StyleLi, Fengjuan, Linyuan Fan, Jingli Zhang, Wen Liu, Wei Li, Yongpeng Ma, and Hong Ma. 2025. "Genetic Diversity and Structure of a Critically Endangered Ornamental Species, Rhododendron farinosum, with Extremely Small Populations" Horticulturae 11, no. 1: 51. https://doi.org/10.3390/horticulturae11010051
APA StyleLi, F., Fan, L., Zhang, J., Liu, W., Li, W., Ma, Y., & Ma, H. (2025). Genetic Diversity and Structure of a Critically Endangered Ornamental Species, Rhododendron farinosum, with Extremely Small Populations. Horticulturae, 11(1), 51. https://doi.org/10.3390/horticulturae11010051





