Genetic Diversity Analysis of Capsicum frutescens Based on Simplified Genome Sequencing Technology
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Methods
2.2.1. DNA Extraction and GBS Library Construction
2.2.2. Filtering of GBS Genotyping Sequencing Data
2.2.3. Genome-Wide SNP Screening and Typing Analysis
2.2.4. Capsaicin Content Extraction and Assay
2.3. Data Processing
3. Results
3.1. Genome Sequencing Quality Assessment
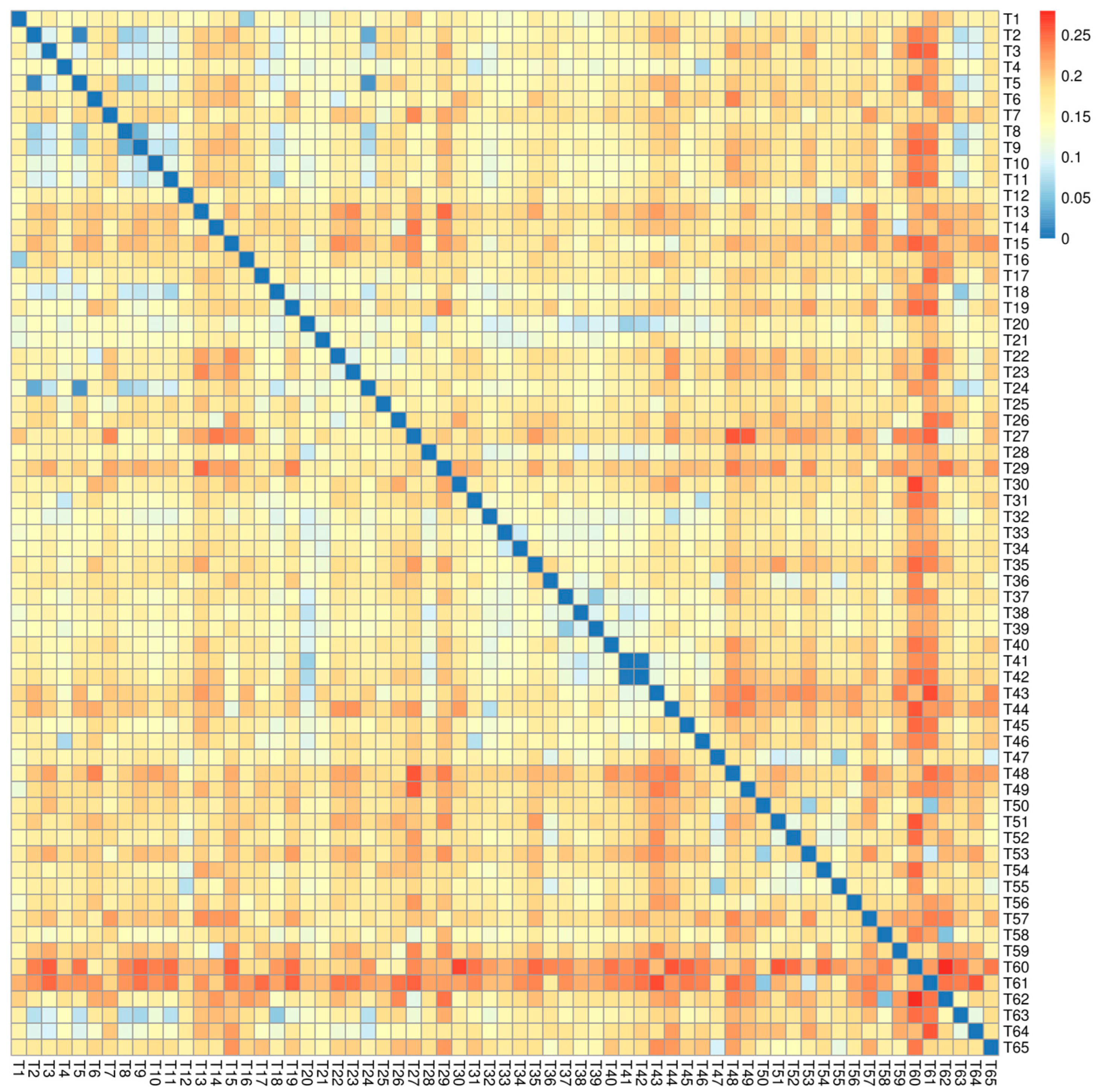
3.2. SNP Variation in 65 Pepper Germplasm Resources
3.3. Population Genetic Structure Analysis
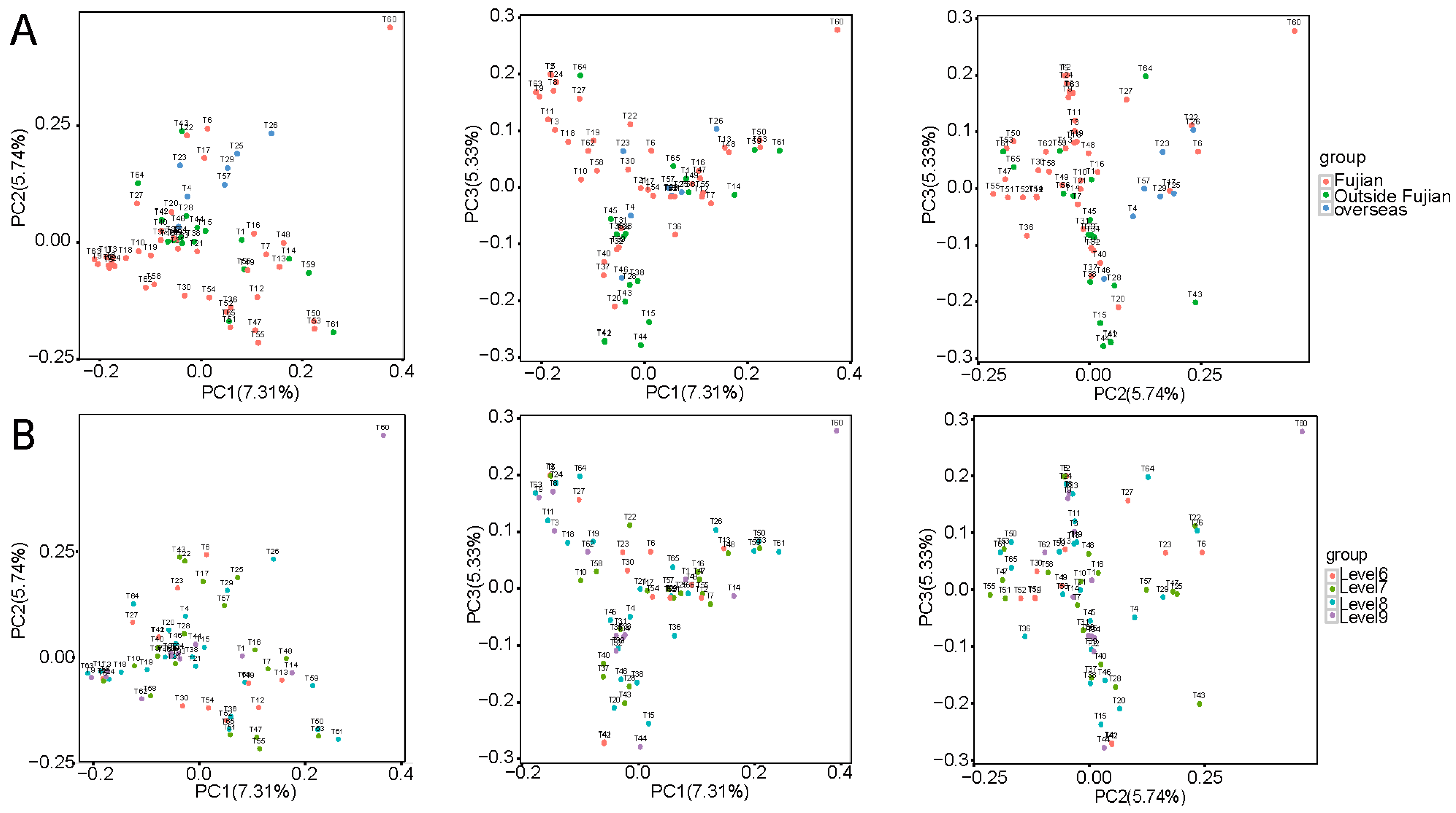
3.3.1. Population Principal Component Analysis
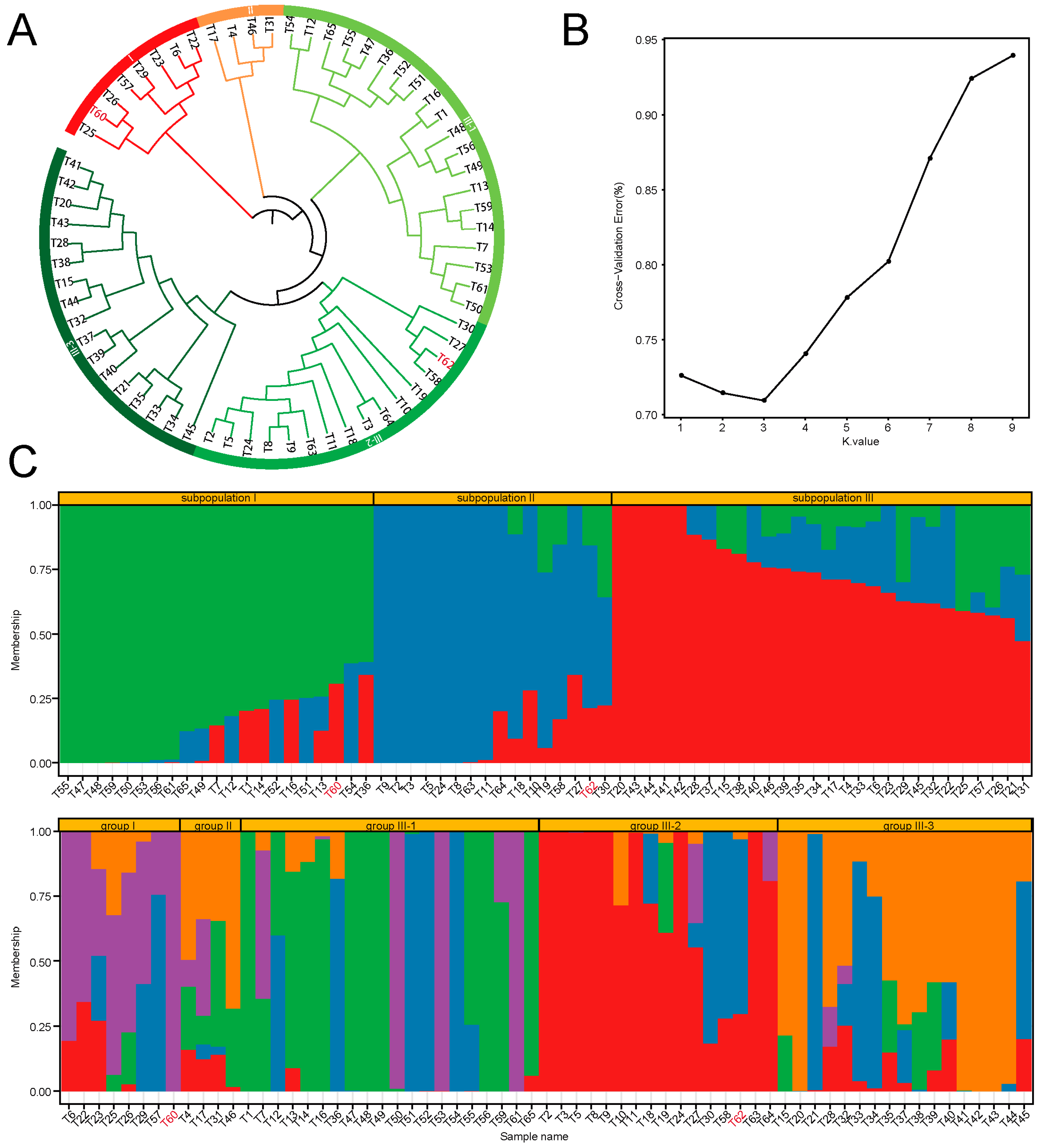
3.3.2. Group Structure Analysis
4. Discussion
4.1. Genetic Structure of C. frutescens Resources Can Be Significantly Differentiated Using GBS
4.2. Use of GBS Allows Easy and Rapid Exploitation of Pepper Hybrid Dominance
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lozada, D.N.; Bosland, P.W.; Barchenger, D.W.; Haghshenas-Jaryani, M.; Sanogo, S.; Walker, S. Chile Pepper (Capsicum) Breeding and Improvement in the “Multi-Omics” Era. Front. Plant Sci. 2022, 13, 879182. [Google Scholar] [CrossRef] [PubMed]
- Fu, G.; Yu, S.; Wu, K.; Yang, M.; Altaf, M.A.; Wu, Z.; Deng, Q.; Lu, X.; Fu, H.; Wang, Z.; et al. Genome-wide association study and candidate gene identification for agronomic traits in 182 upward-growing fruits of C. frutescens and C. annuum. Sci. Rep. 2024, 14, 14691. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Zhang, K.; Yu, H.; Chen, S.; Xu, D.; Zhao, H.; Zhang, Z.; Yang, Y.; Gu, X.; Liu, X.; et al. Pepper variome reveals the history and key loci associated with fruit domestication and diversification. Mol. Plant 2022, 15, 1744–1758. [Google Scholar] [CrossRef]
- Liu, F.; Zhao, J.; Sun, H.; Xiong, C.; Sun, X.; Wang, X.; Wang, Z.; Jarret, R.; Wang, J.; Tang, B.; et al. Genomes of cultivated and wild Capsicum species provide insights into pepper domestication and population differentiation. Nat. Commun. 2023, 14, 5487. [Google Scholar] [CrossRef]
- Ye, Z.; Shang, Z.; Li, M.; Zhang, X.; Ren, H.; Hu, X.; Yi, J. Effect of ripening and variety on the physiochemical quality and flavor of fermented Chinese chili pepper (Paojiao). Food Chem. 2021, 368, 130797. [Google Scholar] [CrossRef]
- Batiha, G.E.-S.; Alqahtani, A.; Ojo, O.A.; Shaheen, H.M.; Wasef, L.; Elzeiny, M.; Ismail, M.; Shalaby, M.; Murata, T.; Zaragoza-Bastida, A.; et al. Biological Properties, Bioactive Constituents, and Pharmacokinetics of Some Capsicum spp. and Capsaicinoids. Int. J. Mol. Sci. 2020, 21, 5179. [Google Scholar] [CrossRef]
- Jeong, H.-B.; Jang, S.-J.; Kang, M.-Y.; Kim, S.; Kwon, J.-K.; Kang, B.-C. Candidate Gene Analysis Reveals That the Fruit Color Locus C1 Corresponds to PRR2 in Pepper (Capsicum frutescens). Front. Plant Sci. 2020, 11, 399. [Google Scholar] [CrossRef] [PubMed]
- Manikharda, M.; Makoto, T.; Mika, A.; Kaoru, Y.; Fumio, H.; Kensaku, T.; Koji, W. Influence of Fruit Ripening on Color, Organic Acid Contents, Capsaicinoids, Aroma Compounds, and Antioxidant Capacity of Shimatogarashi (Capsicum frutescens). J. Oleo Sci. 2017, 67, 113–123. [Google Scholar] [CrossRef] [PubMed]
- Ziv, C.; Lers, A.; Fallik, E.; Paran, I. Genetic and biotechnological tools to identify breeding targets for improving postharvest quality and extending shelf life of peppers. Curr. Opin. Biotechnol. 2022, 78, 102794. [Google Scholar] [CrossRef]
- Guo, G.; Pan, B.; Yi, X.; Khan, A.; Zhu, X.; Ge, W.; Liu, J.; Diao, W.; Wang, S. Genetic diversity between local landraces and current breeding lines of pepper in China. Sci. Rep. 2023, 13, 4058. [Google Scholar] [CrossRef]
- Carvalho, S.I.C.; Bianchetti, L.B.; Ragassi, C.F.; Ribeiro, C.S.C.; Reifschneider, F.J.B.; Buso, G.S.C.; Faleiro, F.G. Genetic variability of a Brazilian Capsicum frutescens germplasm collection using morphological characteristics and SSR markers. Genet. Mol. Res. 2017, 16, gmr16039689. [Google Scholar] [CrossRef] [PubMed]
- Lima, M.F.; Carvalho, S.I.C.; Ragassi, C.F.; Bianchetti, L.B.; Faleiro, F.G.; Reifschneider, F.J.B. Characterization of a pepper collection (Capsicum frutescens L.) from Brazil. Genet. Mol. Res. 2017, 16, gmr16039704. [Google Scholar] [CrossRef] [PubMed]
- Thul, S.T.; Darokar, M.P.; Shasany, A.K.; Khanuja, S.P.S. Molecular profiling for genetic variability in Capsicum species based on ISSR and RAPD markers. Mol. Biotechnol. 2011, 51, 137–147. [Google Scholar] [CrossRef] [PubMed]
- Elshire, R.J.; Glaubitz, J.C.; Sun, Q.; Poland, J.A.; Kawamoto, K.; Buckler, E.S.; Mitchell, S.E. A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS ONE 2011, 6, e19379. [Google Scholar] [CrossRef] [PubMed]
- Riangwong, K.; Wanchana, S.; Aesomnuk, W.; Saensuk, C.; Nubankoh, P.; Ruanjaichon, V.; Kraithong, T.; Toojinda, T.; Vanavichit, A.; Arikit, S. Mining and validation of novel genotyping-by-sequencing (GBS)-based simple sequence repeats (SSRs) and their application for the estimation of the genetic diversity and population structure of coconuts (Cocos nucifera L.) in Thailand. Hortic. Res. 2020, 7, 156. [Google Scholar] [CrossRef]
- Melo, A.T.O.; Bartaula, R.; Hale, I. GBS-SNP-CROP: A reference-optional pipeline for SNP discovery and plant germplasm characterization using variable length, paired-end genotyping-by-sequencing data. BMC Bioinform. 2016, 17, 29. [Google Scholar] [CrossRef]
- Gürcan, K.; Teber, S.; Ercisli, S.; Yilmaz, K.U. Genotyping by Sequencing (GBS) in Apricots and Genetic Diversity Assessment with GBS-Derived Single-Nucleotide Polymorphisms (SNPs). Biochem. Genet. 2016, 54, 854–885. [Google Scholar] [CrossRef]
- Armin, S.; Jacqueline, B.; David, E. Revolution in Genotyping Platforms for Crop Improvement. Adv. Biochem. Eng. Biotechnol. 2018, 164, 37–52. [Google Scholar] [CrossRef]
- Larsen, B.; Gardner, K.; Pedersen, C.; Ørgaard, M.; Migicovsky, Z.; Myles, S.; Toldam-Andersen, T.B. Population structure, relatedness and ploidy levels in an apple gene bank revealed through genotyping-by-sequencing. PLoS ONE 2018, 13, e0201889. [Google Scholar] [CrossRef]
- Pavan, S.; Delvento, C.; Nazzicari, N.; Ferrari, B.; D’agostino, N.; Taranto, F.; Lotti, C.; Ricciardi, L.; Annicchiarico, P. Merging genotyping-by-sequencing data from two ex situ collections provides insights on the pea evolutionary history. Hortic. Res. 2022, 9, uhab062. [Google Scholar] [CrossRef]
- Sun, Y.; Li, X.; Ma, Z.; Chen, S. Quantitative trait locus mapping of fruit aroma compounds in cucumber (Cucumber sativus L.) based on a recombinant inbred line population. Hortic. Res. 2022, 9, uhac151. [Google Scholar] [CrossRef] [PubMed]
- Stansell, Z.; Björkman, T. From landrace to modern hybrid broccoli: The genomic and morphological domestication syndrome within a diverse B. oleracea collection. Hortic. Res. 2020, 7, 159. [Google Scholar] [CrossRef] [PubMed]
- Ding, X.Y.; Jin Song, X.U.; Huang, H.; Zhang, X.K. Unraveling waterlogging tolerance-related traits with QTL analysis in reciprocal intervarietal introgression lines using genotyping by sequencing in rapeseed (Brassica napus L.). J. Integr. Agric. 2020, 19, 1974–1983. [Google Scholar] [CrossRef]
- Pereira-Dias, L.; Vilanova, S.; Fita, A.; Prohens, J.; Rodríguez-Burruezo, A. Genetic diversity, population structure, and relationships in a collection of pepper (Capsicum spp.) landraces from the Spanish centre of diversity revealed by genotyping-by-sequencing (GBS). Hortic. Res. 2019, 6, 54. [Google Scholar] [CrossRef]
- Lozada, D.N.; Bhatta, M.; Coon, D.; Bosland, P.W. Single nucleotide polymorphisms reveal genetic diversity in New Mexican chile peppers (Capsicum spp.). BMC Genom. 2021, 22, 356. [Google Scholar] [CrossRef]
- Solomon, A.M.; Han, K.; Lee, J.-H.; Lee, H.-Y.; Jang, S.; Kang, B.-C. Genetic diversity and population structure of Ethiopian Capsicum germplasms. PLoS ONE 2019, 14, e0216886. [Google Scholar] [CrossRef]
- GB/T 21266-2007; Determination of Total Capsaicinoid Content and Representation of Pungency Degree in Capsicum and Its Products. Standards Press of China: Beijing, China, 2007.
- DB43/T276-2006; Method of Chillies and Its Products Pungency Degree Sensory Test. Standards Press of China: Beijing, China, 2006.
- Wang, Y.; Zhang, X.; Yang, J.; Chen, B.; Zhang, J.; Li, W.; Du, H.; Geng, S. Optimized Pepper Target SNP-Seq Applied in Population Structure and Genetic Diversity Analysis of 496 Pepper (Capsicum spp.) Lines. Genes 2024, 15, 214. [Google Scholar] [CrossRef]
- Lopez-Moreno, H.; Basurto-Garduño, A.C.; Torres-Meraz, M.A.; Diaz-Valenzuela, E.; Arellano-Arciniega, S.; Zalapa, J.; Sawers, R.J.H.; Cibrián-Jaramillo, A.; Diaz-Garcia, L. Genetic analysis and QTL mapping of domestication-related traits in chili pepper (Capsicum annuum L.). Front. Genet. 2023, 14, 1101401. [Google Scholar] [CrossRef]
- McCoy, J.; Martínez-Ainsworth, N.; Bernau, V.; Scheppler, H.; Hedblom, G.; Adhikari, A.; McCormick, A.; Kantar, M.; McHale, L.; Jardón-Barbolla, L.; et al. Population structure in diverse pepper (Capsicum spp.) accessions. BMC Res. Notes 2023, 16, 20. [Google Scholar] [CrossRef]
- Zou, Z.; Zou, X. Geographical and Ecological Differences in Pepper Cultivation and Consumption in China. Front. Nutr. 2021, 8, 718517. [Google Scholar] [CrossRef]
- Wang, L.; Zhong, Y.; Liu, J.; Ma, R.; Miao, Y.; Chen, W.; Zheng, J.; Pang, X.; Wan, H. Pigment Biosynthesis and Molecular Genetics of Fruit Color in Pepper. Plants 2023, 12, 2156. [Google Scholar] [CrossRef] [PubMed]
- Berry, H.M.; Rickett, D.V.; Baxter, C.J.; Enfissi, E.M.A.; Fraser, P.D. Carotenoid biosynthesis and sequestration in red chilli pepper fruit and its impact on colour intensity traits. J. Exp. Bot. 2019, 70, 2637–2650. [Google Scholar] [CrossRef] [PubMed]
- Rosa-Martínez, E.; García-Martínez, M.D.; Adalid-Martínez, A.M.; Pereira-Dias, L.; Casanova, C.; Soler, E.; Figàs, M.R.; Raigón, M.D.; Plazas, M.; Soler, S.; et al. Fruit composition profile of pepper, tomato and eggplant varieties grown under uniform conditions. Food Res. Int. 2021, 147, 110531. [Google Scholar] [CrossRef]
- Zhang, J.; Wang, C.; Wang, J.; Yang, Y.; Han, K.; Bakpa, E.P.; Li, J.; Lyu, J.; Yu, J.; Xie, J. Comprehensive fruit quality assessment and identification of aroma-active compounds in green pepper (Capsicum annuum L.). Front. Nutr. 2023, 9, 1027605. [Google Scholar] [CrossRef] [PubMed]
- Naves, E.R.; Scossa, F.; Araújo, W.L.; Nunes-Nesi, A.; Fernie, A.R.; Zsögön, A. Heterosis for capsacinoids accumulation in chili pepper hybrids is dependent on parent-of-origin effect. Sci. Rep. 2022, 12, 14450. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Zhang, Z.; Chen, W.; Li, X.; Zhou, S.; Liang, C.; Li, X.; Yang, B.; Zou, X.; Liu, F.; et al. Integration of mRNA and miRNA profiling reveals the heterosis of three hybrid combinations of Capsicum annuum varieties. GM Crop. Food 2021, 12, 224–241. [Google Scholar] [CrossRef]
- Costa, M.P.S.D.; Rêgo, M.M.D.; da Silva, A.P.G.; Rêgo, E.R.D.; Barroso, P.A. Characterization and genetic diversity of pepper (Capsicum spp.) parents and interspecific hybrids. Genet. Mol. Res. 2016, 15, gmr.15027652. [Google Scholar] [CrossRef]
- Liu, W.; He, G.; Deng, X.W. Toward understanding and utilizing crop heterosis in the age of biotechnology. iScience 2024, 27, 108901. [Google Scholar] [CrossRef]
- Paril, J.; Reif, J.; Fournier-Level, A.; Pourkheirandish, M. Heterosis in crop improvement. Plant J. 2023, 117, 23–32. [Google Scholar] [CrossRef]


| Population | Long Lantern-Shaped | Long Ram’s Horn-Shaped | Long Finger-Shaped | Short Ram’s Horn-Shaped |
|---|---|---|---|---|
| Long lantern-shaped | - | 0.2729 | 0.2839 | 0.2837 |
| Long ram’s horn-shaped | 0.1375 | - | 0.2709 | 0.2717 |
| Long finger-shaped | 0.1458 | 0.0747 | - | 0.2785 |
| Short ram’s horn-shaped | 0.1530 | 0.0849 | 0.0825 | - |
| Population | Level 6 | Level 7 | Level 8 | Level 9 |
|---|---|---|---|---|
| Level 6 | - | 0.2898 | 0.2985 | 0.2731 |
| Level 7 | 0.0588 | - | 0.2891 | 0.2721 |
| Level 8 | 0.0752 | 0.0672 | - | 0.2832 |
| Level 9 | 0.0918 | 0.1048 | 0.1247 | - |
| Population | Dark Red | Orange-Yellow | Orange-Red | Light Red |
|---|---|---|---|---|
| Dark red | - | 0.2979 | 0.2752 | 0.2761 |
| Orange-yellow | 0.1530 | - | 0.3086 | 0.2984 |
| Orange-red | 0.1693 | 0.1901 | - | 0.2938 |
| Light red | 0.0623 | 0.0862 | 0.1077 | - |
| Population | Yellow-White | Yellow-Green | Green | Light Green |
|---|---|---|---|---|
| Yellow-white | - | 0.2908 | 0.3333 | 0.3039 |
| Yellowish green | 0.1285 | - | 0.3115 | 0.2802 |
| Green | 0.1609 | 0.1121 | - | 0.2881 |
| Light green | 0.1673 | 0.1005 | 0.0854 | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, L.; Qiu, Y.; Lin, S.; Zhang, R.; Wang, L.; Li, Y.; Cao, Y. Genetic Diversity Analysis of Capsicum frutescens Based on Simplified Genome Sequencing Technology. Horticulturae 2024, 10, 1004. https://doi.org/10.3390/horticulturae10091004
Wu L, Qiu Y, Lin S, Zhang R, Wang L, Li Y, Cao Y. Genetic Diversity Analysis of Capsicum frutescens Based on Simplified Genome Sequencing Technology. Horticulturae. 2024; 10(9):1004. https://doi.org/10.3390/horticulturae10091004
Chicago/Turabian StyleWu, Lidong, Yinhui Qiu, Shuting Lin, Rui Zhang, Lihao Wang, Yongqing Li, and Yacong Cao. 2024. "Genetic Diversity Analysis of Capsicum frutescens Based on Simplified Genome Sequencing Technology" Horticulturae 10, no. 9: 1004. https://doi.org/10.3390/horticulturae10091004
APA StyleWu, L., Qiu, Y., Lin, S., Zhang, R., Wang, L., Li, Y., & Cao, Y. (2024). Genetic Diversity Analysis of Capsicum frutescens Based on Simplified Genome Sequencing Technology. Horticulturae, 10(9), 1004. https://doi.org/10.3390/horticulturae10091004






