Advancements in Molecular Mechanism Research on Bolting Traits in Vegetable Crops
Abstract
1. Introduction
2. Bolting and Flowering Factors
2.1. Preconditions
2.2. Bolting and Flowering Pathways
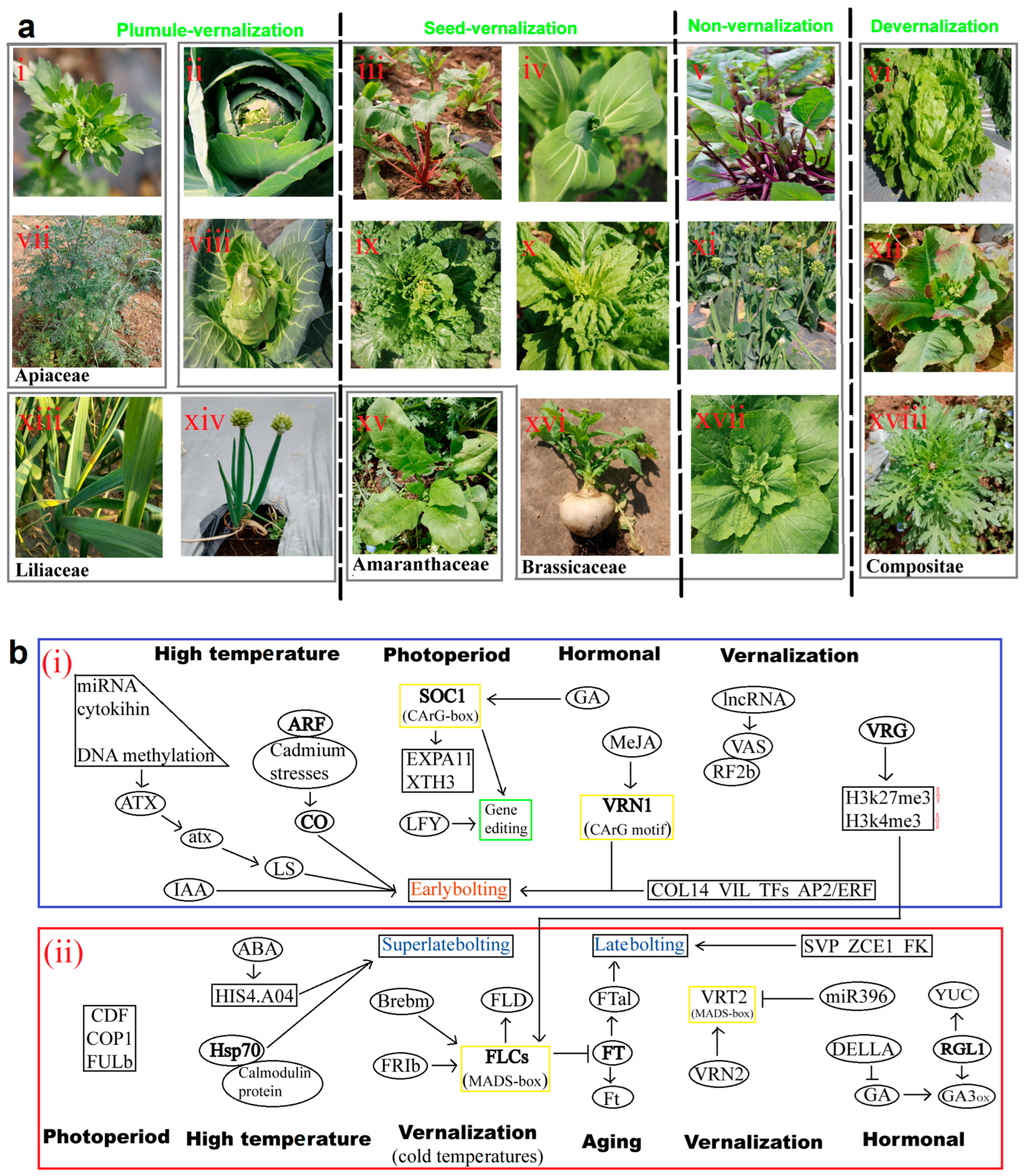
2.3. Vernalization and Devernalization Pathway
3. Physiological and Biochemical Changes before and after Bolting
3.1. Carbohydrates
3.2. Soluble Proteins
3.3. Endogenous Hormones
3.4. Enzyme Activity
3.5. Other Factors
4. Molecular Mechanism of Bolting
4.1. Molecular Markers
4.2. Mining and Expression of Regulatory Genes
4.3. Protein and Protein Interaction Regulation
5. Genome and Transcriptome Analysis
5.1. Genome and Transcriptome
5.2. RNA (miRNAs/lncRNAs/circRNAs/ceRNA)
5.3. Functional Verification and Genetic Transformation
6. Identification of Bolting and Breeding
6.1. Investigating Epigenetics
6.2. Identification of the Bolting Properties
6.3. Hereditability and Hereditary Effects
6.4. Breeding Methods
7. Discussion
7.1. Studying Blooming in Other Crops
7.2. Advantages and Disadvantages of Early and Late Bolting
8. The Concluding Overview
Author Contributions
Funding
Conflicts of Interest
References
- Coustham, V.; Li, P.J.; Strange, A.; Lister, C.; Song, J.; Dean, C. Quantitative modulation of polycomb silencing under lies natural variation in vernalization. Science 2012, 337, 584–587. [Google Scholar] [CrossRef] [PubMed]
- Elers, B.; Wiebe, H.J. Flower formation of Chinese cabbage. I. Response to vernalization and photoperiods. Sci. Hortic. 1984, 22, 219–231. [Google Scholar] [CrossRef]
- Yang, P.; Yang, H.J.; Yang, J.C.; Xu, C.; Zhang, K.H.; Ma, X.C. Identification and evaluation of bolting characters of turnip (Brassicarapa L.) germplasm resources. Mol. Plant Breed. 2022, 20, 1363–1372. [Google Scholar]
- Yui, S.; Yoshikawa, H. Bolting resistant breeding of Chinese cabbage. 1. flower induction of late bolting variety without chilling treatment. Euphytica 1991, 52, 171–176. [Google Scholar] [CrossRef]
- Bohuon, E.J.R.; Ramsay, L.D.; Craft, J.A.; Arthur, D.F.; Marshall, D.F.; Lydiate, D.J. The association of flowering time quantitative trait loci with duplicated regions and candidate loci in Brassica oleracea. Genetics 1998, 50, 393–401. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.I.; Wang, J.G.; Poon, S.Y.; Su, C.L.; Wang, S.S.; Chiou, T.J. Differential regulation of Flowering Locus C expression by vernalization in cabbage and Arabidopsis. Plant Physiol. 2005, 137, 1037–1048. [Google Scholar] [CrossRef] [PubMed]
- Nishioka, M.; Tamura, K.; Hayashi, M.; Fujimori, Y.; Ohkawa, Y.; Kuginuki, Y. Mapping of QTLs for bolting time in Brassica rapa (syn. campestris) under different environmental conditions. Breed Sci. 2005, 55, 127–133. [Google Scholar] [CrossRef]
- Moe, R.; Guttormsen, G. Effect of photoperiod and temperature on bolting in Chinese cabbage. Sci. Hortic. 1985, 27, 49–54. [Google Scholar] [CrossRef]
- Axelsson, T.; Shavorskaya, O.; Lagercrantz, U. Multiple flowering time QTLs with in several Brassica species could be the result of duplicated copies of oneance stralgene. Genome 2001, 44, 856–864. [Google Scholar] [CrossRef]
- Thomas, T.H. Relationship between bolting-resistance and seed dormancy of different celery cultivars. Sci. Hortic. 1978, 9, 311–316. [Google Scholar] [CrossRef]
- Jiang, Q.; Wang, F.; Li, M.Y.; Tan, H.W.; Ma, J.; Xiong, A.S. High-through put analysis of small RNAs and characterization of novel microRNAs affected by abiotic stress in alocal celery cultivar. Sci. Hortic. 2014, 169, 36–43. [Google Scholar] [CrossRef]
- Jia, X.L.; Li, M.Y.; Jiang, Q.; Xu, Z.S.; Wang, F.; Xiong, A.S. High-throughput sequencing of small RNAs and anatomical characteristics associated with leaf development in celery. Sci. Rep. 2015, 5, e11093. [Google Scholar] [CrossRef] [PubMed]
- Li, M.Y.; Hou, X.L.; Wang, F.; Tan, G.F.; Xu, Z.S.; Xiong, A.S. Advances in the research of celery, an important Apiaceae vegetable crop. Crit. Rev. Biotechnol. 2018, 38, 172–183. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.Y.; Chen, Z.J.; Lv, S.S.; Ning, K.; Ji, X.L.; Liu, X.Y.; Wang, Q.; Liu, R.Y.; Fan, S.X.; Zhang, X.L. MADS-box genes and gibberel lins regulate bolting in lettuce (Lactuca sativa L.). Front. Plant Sci. 2016, 7, e1889. [Google Scholar] [CrossRef] [PubMed]
- Abolghasemi, R.; Haghighi, M.; Etemadi, N.; Wang, S.; Soorni, A. Transcriptom earchitecture reveals genetic net works of bolting regulation in spinach. BMC Plant Biol. 2021, 21, 179. [Google Scholar] [CrossRef] [PubMed]
- Alessandro, M.S.; Galmarini, C.R. Inheritance of vernalization requirement in carrot. J. Am. Soc. Hor. Sci. 2007, 132, 525–529. [Google Scholar] [CrossRef]
- Ma, J.; Xu, Z.S.; Tan, G.F.; Wang, F.; Xiong, A.S. Distinct transcription profile of genes involved in carotenoid biosynthesis among six different color carrot (Daucus carota L.) cultivars. Acta Biochim. Biophys. Sin. 2017, 49, 817–826. [Google Scholar] [CrossRef]
- Tan, G.F.; Wang, F.; Zhang, X.Y.; Xiong, A.S. Different lengths, copies and expression levels of the mitochondrial atp6 gene in male sterile and fertile lines of carrot (Daucus carota L). Mitochondrial DNA 2018, 29, 446–454. [Google Scholar] [CrossRef] [PubMed]
- Que, F.; Hou, X.L.; Wang, G.L.; Xu, Z.S.; Tan, G.F.; Li, T.; Xiong, A.S. Advances in research on the carrot, an important root vegetable in the Apiaceae family. Hortic. Res. 2019, 6, 860–874. [Google Scholar] [CrossRef]
- Wang, X.J.; Luo, Q.; Li, T.; Meng, P.H.; Pu, Y.T.; Liu, J.X.; Tan, G.F. Origin, evolution, breeding, and omics of Apiaceae: Afamily of vegetables and medicinal plants. Hortic. Res. 2022, 9, uhac076. [Google Scholar] [CrossRef]
- Sheen, T.F. Effect of day and night temperature variation and of high temperature on devernalization in radish. Acta Hortic. 2000, 5, 157–162. [Google Scholar] [CrossRef]
- Curtis, G.J.; Hornesy, K.G. Graft-transmissible inductionofel ongation and flowering in scions of sugar-beet red for resistance to bolting. Nature 1964, 202, 1238. [Google Scholar] [CrossRef]
- Gupta, A.J.; Mahajan, V.; Lawande, K.E. Genotypic response to bolting tolerance in short day onion (Allium cepa L.). Veg. Sci. 2018, 45, 92–96. [Google Scholar]
- Wu, C.N.; Wang, M.Y.; Dong, Y.X.; Cheng, Z.H.; Meng, H.W. Growth, bolting and yield of garlic (Allium sativum L.) in response to clove chilling treatment. Sci. Hortic. 2015, 194, 43–52. [Google Scholar] [CrossRef]
- Ellis, R.H.; Summerfield, R.J.; Roberts, E.H. Effects of temperature, photoperiod and seed vernalization on flowering in fababean viciafaba. Ann. Bot. 1988, 61, 17–27. [Google Scholar] [CrossRef]
- Fujime, Y.; Mesuda, Y.; Okuda, N.; Varanyanond, W. Promotive effects of seed vernalization on flowering and pod setting of some broad bean cultivars. Acta Hortic. 2000, 533, 75–82. [Google Scholar] [CrossRef]
- Filek, W.; Kościelniak, J.; Grzesiak, S. The effect of seed vernalization and irradiation on growth and photosynthesis of field bean plants (Vicia faba L. minor) and on nitrogenase activity of root nodules. J. Agron. Crop. Sci. 2010, 185, 229–236. [Google Scholar] [CrossRef]
- Jiang, X.M.; Wu, M.X.; Yu, X.H.; Liu, S.C.; Xu, W. Effect of vernalization on nucleic acid content of Brassica oleracea L. var. italica Plenck. China Veg. 2011, 10, 41–44. [Google Scholar]
- Yu, R.F.; Su, T.B.; Yu, S.C.; Zhang, F.L.; Yu, Y.J.; Zhang, D.S. Effects of plumule-vernalization and seeding-vernalization on bolting and flowering times of different Chinese cabbage varieties. China Veg. 2016, 5, 27–32. [Google Scholar]
- Dai, Y.; Li, G.L.; Gao, X.Y.; Wang, S.X.; Li, Z.; Song, C. Identification of long noncoding RNAs involved in plumule-vernalization of Chinese cabbage. Front. Plant Sci. 2023, 14, e1147494. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.M.; Zhao, R.Q.; Yu, X.H. Effect of devernalization on nucleicacid content of cabbage. Acta Physiol. Plant 2009, 45, 673–676. [Google Scholar] [CrossRef]
- Yu, X.H.; Li, W.P.; Jiang, X.M.; Liu, D.; Zhang, Y. Changes of endogenous hormone during devernalization in cabbage. China Veg. 2010, 20, 38–41. [Google Scholar] [CrossRef]
- Akter, A.; Miyazaki, J.; Shea, D.J.; Nishida, N.; Takada, S.; Miyaji, N. Gene expression analysis in response to vernalization in Chinese cabbage (Brassica rapa L.). Hortic. J. 2020, 89, 268–277. [Google Scholar] [CrossRef]
- Chen, G.P.; Hu, G.L.; Hu, Z.L.; Li, Y.; Gu, F. Cloning and expression of vernalization-related genes in purple flowering stalk (Brassica compestrisssp. chinensis var. pupurea Hort). LSR 2010, 14, 27–33. [Google Scholar] [CrossRef]
- Ma, C.F.; Dai, S.L. Advances in photo receptor-mediated signaling transduction in flowering time regulation. Chin. Bull. Bot. 2019, 54, 9–22, (In Chinese with English Title and Abstract). [Google Scholar] [CrossRef]
- Rosental, L.; Still, D.W.; You, Y.; Hayes, R.J.; Simko, L. Mapping and identification of genetic loci affecting earliness of bolting and flowering in lettuce. Theor. Appl. Genet. 2021, 134, 3319–3337. [Google Scholar] [CrossRef]
- Kim, D.H. Current understanding of flowering path ways in plants: Focusing on the vernalization pathway in Arabidopsis and several vegetable crop plants. Hortic. Environ. Biot. 2020, 61, 133–143. [Google Scholar] [CrossRef]
- Burn, J.E.; Bagnall, D.J.; Metzger, J.D.; Dennis, E.S.; Peacock, W.J. DNA methylation, vernalization, and the initiation of flowering. Proc. Natl. Acad. Sci. USA 1993, 90, 287–291. [Google Scholar] [CrossRef]
- Fornara, F.; Panigrahi, K.C.S.; Gissot, L.; Sauerbrunn, N.; Rühl, M.; Jarillo, J.A. Arabidopsis DOF transcription factors act redundantly to reduce CONSTANS expression and are essential for a photoperiodic flowering response. Dev. Cell 2009, 17, 75–86. [Google Scholar] [CrossRef]
- Bendix, C.; Marshall, C.; Harmon, F. Circadian clock genes universally control key agricultural traits. Mol. Plant 2015, 8, 1135–1152. [Google Scholar] [CrossRef]
- Wang, Q.; Zuo, Z.C.; Wang, X.; Gu, L.F.; Yoshizumi, T.; Yang, Z.H. Photo activation and inactivation of Arabidopsis cryptochrome2. Science 2016, 354, 343–347. [Google Scholar] [CrossRef]
- Itabashi, E.; Shea, D.J.; Fukino, N.; Fujimoto, R.; Okazaki, K.; Kakizaki, T. Comparison of cold responses for orthologs of cabbage vernalization-relatedgenes. Hortic. J. 2019, 88, 462–470. [Google Scholar] [CrossRef]
- Mao, J.H.; Mao, S.M.; Zhuang, F.Y.; Ou, C.G.; Zhao, Z.W.; Bao, S.Y. Heredity and environmental regulation of prematurebolting in carrot. Acta Agric. Boreal-Occident Sin. 2013, 28, 67–72. [Google Scholar]
- Beharav, A.; Hellier, B. Bolting and flowering response of Lactuca georgica, a wild lettuce relative, to low temperatures. Am. J. Plant Sci. 2020, 11, 2139–2154. [Google Scholar] [CrossRef]
- Michael, B.; Shemesh-Mayer, T.; Kimhi, E.; Gershberg, S.; Forer, C.; Avila, I. Temporal and spatial effect of low pre-planting temperatures on plant architecture and flowering in bolting garlic. Sci. Hortic. 2018, 242, 69–75. [Google Scholar] [CrossRef]
- Mayer, E.S.; Ben-Michael, T.; Kimhi, S.; Forer, I.; Rabinowitch, H.D.; Kamenetsky, R. Effects of different temperature regimes on flower development, microsporogenesis and fertility in bolting garlic (Allium sativum). Funct. Plant Biol. 2015, 42, 514–526. [Google Scholar] [CrossRef]
- Bao, S.Y.; Ou, C.G.; Zhuang, F.Y.; Chen, J.F.; Zhao, Z.W. Study of premature bolting of carrot in spring cultivation. China Veg. 2010, 6, 38–42. [Google Scholar]
- Pressman, E.; Negbi, M. The effect of day length on the response of celery to vernalization. J. Exp. Bot. 1980, 31, 1291–1296. [Google Scholar] [CrossRef]
- Huang, Z.J.; Liu, S.Q.; Zhang, Z.K.; Chen, Y.C.; Zhang, Y.; Ma, L. Effect of prolonged photoperiod on bolting of garlic. Acta Agric. Boreal-Occident Sin. 2010, 20, 100–103, (In Chinese with English Title and Abstract). [Google Scholar]
- Wang, S.L.; Luo, C.; Sun, L.; Ning, K.; Chen, Z.J.; Yang, J.J. LsRGL1 controls the bolting and flowering times of lettuce by modulating the gibberellin pathway. Plant Sci. 2022, 316, e111175. [Google Scholar] [CrossRef]
- Wang, Y.D.; Huang, X.; Huang, X.M.; Su, W.; Hao, Y.W.; Liu, H.C. BcSOC1 promotes bolting and stem elongation in flowering Chinese cabbage. Int. J. Mol. Sci. 2022, 23, 3459. [Google Scholar] [CrossRef]
- Mero, C.E.; Honma, S. A method for evaluating bolting-resistancein Brassica species. Sci. Hortic. 1984, 24, 13–19. [Google Scholar] [CrossRef]
- Bernier, G.; Havelange, A.; Houssa, C.; Petitjean, A.; Lejeune, P. Physiological signals that induce flowering. Plant Cell 1993, 5, 1147–1155. [Google Scholar] [CrossRef]
- He, L.; Yao, N.; Pei, Y.; Yu, T.; Zu, G.D.; Zhang, W.P. Physiological and biochemical changes and spatial and temporal expressions of flowering genes in radish (Raphanus sativus L.) duringbolting. Mol. Plant Breed. 2020, 18, 6837–6843. [Google Scholar]
- Wang, B.L.; Deng, J.Y.; Zeng, G.W. Changes in carbohydrate content of stem apices and leaves during floral bud differentiation in radish (Raphanus sativus L.). Acta Hortic. Sin. 2004, 31, 375–377. [Google Scholar]
- Wang, Z.Z.; Hou, R.X.; Li, X.F.; Zhu, H.F.; Zhu, Y.Y.; Hou, X.L. Analysis of carbon and nitrogen indexes among different in bred lines during bolting in pak-choi. Plant Physiol. J. 2011, 47, 893–898. [Google Scholar] [CrossRef]
- Dai, Y.; Sun, X.; Wang, C.G.; Li, F.; Zhang, S.F.; Zhang, H. Geneco-expression net work analysis reveals key pathways and hubgenes in Chinese cabbage (Brassica rapa L.) during vernalization. BMC Genom. 2021, 22, 236. [Google Scholar] [CrossRef]
- Wang, S.Y.; Yu, F.W.; Zhang, W.; Yu, L.; Li, J.B. Change of anti-oxidative indexes and protein during bolting process of cabbage (Brassica oleracea L. var. capitata. L.). Acta Agric. Boreal-Occident Sin. 2023, 32, 101–108, (In Chinese with English Title and Abstract). [Google Scholar] [CrossRef]
- Li, M.L.; Wang, Q.M.; Zhu, Z.J.; Zeng, G.W. Studies on the changes of DNA methylation level‚ GA content and protein innon-heading Chinese cabbage during vernalization. Acta Hortic. Sin. 2002, 29, 353–357, (In Chinese with English Title and Abstract). [Google Scholar] [CrossRef]
- Oka, M.; Tasaka, Y.; Iwabuchi, M.; Mino, M. Elevated sensitivity to gibberellins by vernalization in the vegetative rosette plants of Eustoma grandiflorum and Arabidopsis thaliana. Plant Sci. 2001, 160, 1237–1245. [Google Scholar] [CrossRef]
- Cheng, F.; Zhang, S.N.; Sun, C.H.; Li, S.J.; Chen, G.F. Study on index of morphology and physiology for spring Chinese cabbage. Acta Hortic. Sin. 1999, 26, 120–122, (In Chinese with English Title and Abstract). [Google Scholar] [CrossRef]
- Wu, C.N.; Wang, M.Y.; Cheng, Z.H.; Meng, H.W. Response of garlic (Allium sativum L.) bolting and bulbing to temperature and photoperiod treatments. Biol. Open. 2016, 5, 507–518. [Google Scholar] [CrossRef]
- Liu, X.Y.; Lv, S.S.; Liu, R.; Fan, S.X.; Liu, C.J.; Liu, R.Y. Transcriptomic analysis reveals the roles of gibberellin-regulated genes and transcription factors in regulating bolting in lettuce (Lactuca sativa L.). PLoS ONE 2018, 13, e0191518. [Google Scholar] [CrossRef]
- Yang, Y.G.; Zhang, H.S.; Li, Y.L.; Wang, X.W.; Yu, J.H.; Wang, X.L. Endogenous hormone content of fleshy root in relation to early bolting in summer cultivated carrot on plateau. Acta Hortic. Sin. 2010, 37, 1102–1108. [Google Scholar] [CrossRef]
- Wu, C.; Wang, M.; Dong, Y.; Cheng, Z.; Meng, H. Effect of plant age and vernalizationon bolting, plant growth and enzyme activity of garlic (Allium sativum L.). Sci. Hortic. 2016, 201, 295–305. [Google Scholar] [CrossRef]
- Guo, H.Y.; Liu, Y.J.; Yuan, S.H.; Yue, J.R.; Li, Y.M.; Liao, X.Z. Superoxide dismutase promotes early flowering in Triticum aestivum L. Agric. Commum. 2023, 1, e100007. [Google Scholar] [CrossRef]
- Finnegan, E.J.; Genger, R.K.; Kovac, K.; Peacock, W.J.; Dennis, E.S. DNA methylation and the promotion of flowering by vernalization. Proc. Natl. Acad. Sci. USA 1988, 95, 5824–5829. [Google Scholar] [CrossRef]
- Yang, Z.F.; Yan, H.D.; Wang, J.P.; Nie, G.; Feng, G.Y.; Xu, X.H. DNA hyper methylation promotes the flowering of orchard grass during vernalization. Plant Physiol. 2022, 190, 1490–1505. [Google Scholar] [CrossRef]
- He, Y.H.; Michaels, S.D.; Amasino, R.M. Regulation of flowering time by histone acetylation in Arabidopsis. Science 2003, 302, 1751–1754. Available online: https://www.researchgate.net/publication/9030793 (accessed on 5 December 2003). [CrossRef]
- Osborn, T.C.; Kole, C.; Parkin, I.A.P.; Kuiper, M.; Lydiate, D.J.; Trick, M. Comparison of flowering time genes in Brassica rapa, B. napus and Arabidopsis thaliana. Genetics 1997, 146, 1123–1129. [Google Scholar] [CrossRef]
- Li, F.; Kitashiba, H.; Inaba, K.; Nishiio, T. A Brassica rapa linkage map of EST-based SNP markers for identification of candidate genes controlling flowering time and leaf morphological traits. DNA Res. 2009, 16, 311–323. [Google Scholar] [CrossRef]
- Yang, S.J.; Wang, Z.Y.; Zhao, Y.Y.; Wei, X.C.; Yuan, Y.X.; Zhang, X.W. Development of KASP marker for bolting related gene BrFLC1 in Chinese cabbage (Brassica rapa L. ssp. pekinensis). J. Nucl. Agric. Sci. 2020, 34, 265–272. [Google Scholar] [CrossRef]
- Yang, X.; Yu, Y.J.; Zhang, F.L.; Zou, Z.R.; Zhao, X.Y.; Zhang, D.S. Link age map construction and quantitative trait loci analysis for bolting based on a double haploid population of Brassica rapa. J. Integr. Plant Biol. 2007, 49, 664–671. [Google Scholar] [CrossRef]
- Li, J.L.; Wang, C.; Zhang, X.X.; Jiang, K.X.; Zhao, L.J. A CAPS marker derived from the SCAR marker linked to later bolting gene in Brassica oleracea var. capitata. Mol. Plant Breed. 2020, 18, 1529–1534. [Google Scholar] [CrossRef]
- Kakizaki, T.; Kato, T.; Fukino, N.; Ishida, M.; Hatakeyama, K.; Matsumoto, S. Identification of quantitative trait loci controlling late bolting in Chinese cabbage (Brassica rapa L.) parental line Nou 6 gou. Breed. Sci. 2011, 61, 151–159. [Google Scholar] [CrossRef]
- Xu, W.L.; Wang, S.F.; Mu, J.H.; Wang, C.H.; Liu, X.X. Identification of AFLP and SCAR molecular markers linked to bolting trait in radish. Mol. Plant Breed. 2009, 7, 743–749. [Google Scholar] [CrossRef]
- Liu, Y.T.; Zhang, L.; Ji, X.H.; Lv, X.X.; Yang, Y.G.; Feng, H. QTL mapping of vernalization requirement in Brassica rapa. Agric. Biotechnol. 2015, 4, 1–5. [Google Scholar] [CrossRef]
- Gan, C.X.; Cui, L.; Pang, W.X.; Wang, A.H.; Yu, X.Q.; Deng, X.H. QTL mapping of bolting and flowering traits based on high density genetic map of radish. Acta Hortic. Sin. 2021, 48, 1273–1281. [Google Scholar] [CrossRef]
- Yuan, Y.X.; Sun, R.F.; Zhang, X.W.; Wu, J.; Xu, D.H.; Zhang, H. A caps marker linked to bolting related gene BrFLC1 in Brassica rapa. Acta Hortic. Sin. 2008, 35, 1635–1640. [Google Scholar]
- Zhang, X.M.; Liu, B.; Hu, Y.Y.; Liu, J.; Wang, X.W.; Wu, J. Development of dCAPs marker for BrFLC5 related with flowering time in Brassica rapa. Acta Hortic. Sin. 2014, 41, 2035–2042. [Google Scholar]
- Wang, W.H.; Wang, J.L.; Li, B.Y.; Wei, Q.Z.; Hu, T.H.; Hu, H.J. Genetic and QTL mapping analysis of bolting time in cabbage (Brassica oleracea). Acta Hortic. Sin. 2022, 47, 974–982. [Google Scholar] [CrossRef]
- Meng, Q.; Liu, Z.Y.; Feng, C.D.; Zhang, H.L.; Xu, Z.S.; Wang, X.W. Quantitative trait locus mapping and identification of candidate genes controlling bolting in spinach (Spinacia oleracea L.). Front. Plant Sci. 2022, 13, e850810. [Google Scholar] [CrossRef] [PubMed]
- Sheldon, C.C.; Burn, J.; Perez, P.P.; Metzger, J.; Edwards, J.A.; Peacock, W.J. The FLFMADS boxgene: A repressor of flowering in Arabidopsis regulated by vernalization and methylation. Plant Cell 1999, 11, 445–458. [Google Scholar] [CrossRef] [PubMed]
- Michaels, S.D.; Amasino, M.R. Flowering Locus C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell 1999, 11, 949–956. [Google Scholar] [CrossRef] [PubMed]
- Sheldon, C.C.; Conn, A.B.; Dennis, E.S.; Peacock, W.J. Different regulatory regions are required for the vernalization-induced repression of Flowering Locus C and for the epigenetic maintenance of repression. Plant Cell 2002, 14, 2527–2537. [Google Scholar] [CrossRef] [PubMed]
- Xiao, X.F.; Cao, B.H.; Wang, Y.; Chen, G.J.; Lei, J.J. Study on flower bud differentiation and cloning and expression of BrcuFLC in Brassica campestris L. ssp. Chinese (L.) makino var utilis. Acta Hortic. Sin. 2008, 35, 827–832. [Google Scholar] [CrossRef]
- Basualdo, J.; Díaz, M.L.; Cuppari, S.; Cardone, S.; Soresi, D.; Camargo, G.P.; Carrera, A.; Jung, C. Allelic variation and differential expression of VRN-A1 in durum wheat genotypes varying in the vernalization response. Plant Breed. 2015, 134, 520–528. [Google Scholar] [CrossRef]
- Zitzewitz, J.V.; Szucs, P.; Dubcovsky, J.; Yan, L.L.; Francia, E.; Pecchioni, N. Molecular and structural characterization of barley vernalization genes. Plant Mol. Biol. 2005, 59, 449–467. [Google Scholar] [CrossRef]
- Putterill, J.; Laurie, R.; Macknight, R. It’s time to flower: The genetic control of flowering time. Bioessays 2004, 26, 363–373. [Google Scholar] [CrossRef]
- Fornara, F.; Montaigu, A.D.; Coupland, G. Snap Shot: Control of flowering in Arabidopsis. Cell 2010, 141, 550. [Google Scholar] [CrossRef]
- Guo, D.; Wong, W.S.; Xu, W.Z.; Sun, F.F.; Qing, D.J.; Li, N. Cis-cinnamic acid-enhanced 1 gene plays a role in regulation of Arabidopsis bolting. Plant Mol. Biol. 2011, 75, 481–495. [Google Scholar] [CrossRef] [PubMed]
- Huang, B.Y.; Qian, P.P.; Gao, N.; Shen, J.; Hou, S.W. Fackel interacts with gibberellic acid signaling and vernalization to mediate flowering in Arabidopsis. Planta 2017, 245, 939–950. [Google Scholar] [CrossRef]
- Gu, A.X.; Zhang, Y.K.; Zhao, J.J.; Wang, Y.H.; Chen, X.P.; Shen, S.X. Effects of the added cabbage chromosome 4 on bolting time and BrFLCs expression of Chinese cabbage. Acta Hortic. Sin. 2015, 42, 2405–2411. [Google Scholar] [CrossRef]
- Feng, G.Y.; Xu, L.; Wang, J.P.; Nie, G.; Bushman, B.S.; Xie, W.G. Integration of small RNAs and transcriptome sequencing uncovers acomplex regulatory network during vernalization and heading stages of orchard grass (Dactylis glomerata L.). BMC Genom. 2018, 19, 727. [Google Scholar] [CrossRef]
- Shim, Y.; Lim, C.; Seong, G.; Choi, Y.; Kang, K.; Paek, N.C. The AP2/ERF transcription factor LATE FLOWERING SEMI-DWARF suppresses long-day-dependent repression of flowering. Plant Cell Environ. 2022, 45, 2446–2459. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Q.; Wang, Z.; Hu, G.M.; Yao, X.H. Genome-wide identification and characterization of AP2/ERF gene super family during flower development in Actinidia eriantha. BMC Genom. 2022, 23, 650. [Google Scholar] [CrossRef] [PubMed]
- Okazaki, K.; Sakamoto, K.; Kikuchi, R.; Saito, A.; Togashi, E.; Kuginuki, Y.; Hirai, S.M. Mapping and characterization of FLC homologs and QTL analysis of flowering time in Brassica oleracea. Theor. Appl. Genet. 2007, 114, 595–608. [Google Scholar] [CrossRef]
- Razi, H.; Howell, E.C.; Newbury, H.J.; Kearesy, M.J. Does sequence polymorphism of FLC paralogues underlie flowering time QTL in Brassica oleracea? Theor. Appl. Genet. 2008, 116, 179–192. [Google Scholar] [CrossRef]
- Zhao, R.Q.; Hu, Y.; Jiang, X.M.; Yu, X.H. Clone and expression analysis of BoVIN3 from Cabbage. Acta Hortic. Sin. 2012, 39, 1099–1106. [Google Scholar]
- Wang, Y.; Jiang, W.; Yan, K.; Zhou, W.W.; Wang, Z.M.; Wei, D.Y. Protein interactions of flowering in hibitors AGL18 and HDA9 with integrator factors in Brassica oleracea var.italic. Acta Hortic. Sin. 2018, 45, 2383–2394. [Google Scholar] [CrossRef]
- Jiang, W.; Zhou, W.W.; Li, C.C.; Yan, K.; Wang, Y.; Wang, Z.M. Interactions of flowering promoting factor AGL19 with integrator factors AGL24 and SOC1 in Brassica oleracea var. italica. Acta Hortic. Sin. 2017, 44, 1905–1913. [Google Scholar] [CrossRef]
- Michaels, S.D.; Ditta, G.; Gustafson-Brown, C.; Pelaz, S.; Yanofsky, M.; Amasino, R.M. AGL24 acts as a promoter of flowering in Arabidopsis and is positively regulated by vernalization. Plant J. 2003, 33, 867–874. [Google Scholar] [CrossRef] [PubMed]
- Xi, X.; Wei, K.Y.; Gao, B.Z.; Liu, J.H.; Liang, J.L.; Cheng, F. BrFLC5: A weak regulator of flowering time in Brassica rapa. Theor. Appl. Genet. 2018, 131, 2107–2116. [Google Scholar] [CrossRef] [PubMed]
- Xin, X.Y.; Su, T.B.; Li, P.R.; Wang, W.H.; Zhao, X.Y.; Yu, Y.J. A histone H4 gene prevents drought-induced boltingin Chinese cabbage by attenuating the expression of flowering genes. J. Exp. Bot. 2020, 72, 623–635. [Google Scholar] [CrossRef] [PubMed]
- Wei, X.C.; Rahim, M.A.; Zhao, Y.Y.; Yang, S.J.; Wang, Z.Y.; Su, H.N. Inheritance and genetic mapping of late-bolting to early-bolting gene, BrEb-1, in Chinese cabbage (Brassica rapa L.). Agronomy 2022, 12, 1048. [Google Scholar] [CrossRef]
- Dong, X.S.; Yi, H.; Han, C.T.; Nou, I.S.; Swaraz, A.M.; Hur, Y. Genome-wide analysis of genes associated with bolting in heading type chinese cabbage. Euphytica 2016, 212, 65–82. [Google Scholar] [CrossRef]
- Hong, J.K.; Park, S.R.; Suh, E.J.; Park, J.; Lee, Y.H. Effects of over expression of Brassica Rapa Short Vegetative Phase gene on flowering time. Kor. J. Breed. Sci. 2020, 52, 244–251. [Google Scholar] [CrossRef]
- Li, Z.G.; Zhao, L.X.; Cui, C.S.; Kai, G.Y.; Zhang, L.D.; Sun, X.F. Molecular cloning and characterization of an anti-bolting related gene (BrpFLC) from Brassica rapa ssp. Pekinensis. Plant Sci. 2005, 168, 407–413. [Google Scholar] [CrossRef]
- Huang, J.S.; Li, J.Y.; Yu, X.L.; Sun, B.J.; Cao, J.S. Cloning and expression analysis of vernalization-related gene BcFLC in Brassica campestris. Acta Hortic. Sin. 2007, 34, 1169–1176. [Google Scholar] [CrossRef]
- Rong, Z.L.; Hou, X.L.; Shi, G.J.; Xiao, D.; Wang, Q.; Zhang, L. Cloning and expression analysis of late bolting BcFLC3 gene from Brassica campestris ssp. Chinensis. Acta Bot Boreal-Occident Sin. 2010, 12, 2373–2378, (In Chinese with English Titleand Abstract). [Google Scholar]
- Guan, H.L.; Huang, X.M.; Zhu, Y.N.; Xie, B.X.; Liu, H.C.; Song, S.W. Identification of DELLA genes and key stage for GA sensitivity in bolting and flowering of flowering Chinese cabbage. Int. J. Mol. Sci. 2021, 22, 12092. [Google Scholar] [CrossRef] [PubMed]
- Jung, W.Y.; Park, H.J.; Lee, A.; Lee, S.S.; Kim, Y.S.; Cho, H.S. Identification of flowering-related genes responsible for differences in bolting time between two radish in bred lines. Front. Plant Sci. 2016, 7, e1844. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.B.; Zhang, Y.J.; Zhang, L.B. A naturally occurring insertion in the RsFLC2 gene associated with late-bolting trait in radish (Raphanus sativus L.). Mol. Breed. 2018, 38, e137. [Google Scholar] [CrossRef]
- Mao, J.H.; Zhuang, F.Y.; Ou, C.G.; Zhao, Z.W.; Wang, H.; Ma, Z.G. Expression analysis of FLC homologues responding to low temperature and photoperiod in carrot. Acta Hortic. Sin. 2013, 40, 2453–2462. [Google Scholar]
- Alessandro, M.S.; Galmarini, C.R.; Iorizzo, M.; Simon, P.W. Molecular mapping of vernalization requirement and fertility restoration genes in carrot. Theor. Appl. Genet. 2013, 126, 415–423. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.J.; Ou, C.G.; Zhao, Z.W.; Sun, T.T.; Zhuang, F.Y. Function analysis of carrot SOC1 homologues responding to photoperiod. Acta Hortic. Sin. 2016, 43, 1099–1106. [Google Scholar] [CrossRef]
- Li, Y.F.; Zhu, J.Q.; Feng, Y.X.; Li, Z.F.; Ren, Z.; Liu, C.J. LsARF3 mediates thermally induced bolting through promoting the expression of LsCO in lettuce (Lactuca sativa L.). Front. Plant Sci. 2022, 13, e958833. [Google Scholar] [CrossRef] [PubMed]
- Hu, M.M.; Qi, Z.Y.; Ren, Z.; Tong, J.; Wang, B.J.; Wu, Z.H. Genome-wide analysis of auxin response factors in lettuce (Lactuca sativa L.) reveals the positive roles of LsARF8a in thermally induced bolting. Int. J. Mol. Sci. 2023, 23, 13509. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.J.; Han, Y.Y.; Ning, K.; Ding, Y.Y.; Zhao, W.S.; Yan, S.S. Inflorescence development and the role of LsFT in regulating bolting in lettuce (Lactuca sativa L.). Front. Plant Sci. 2017, 8, e2248. [Google Scholar] [CrossRef]
- Fu, D.; Dunbar, M.; Dubcovsky, J. Wheat VIN3-like PHD finger genes are up-regulated by vernalization. Mol. Genet. Genomics 2007, 277, 301–313. [Google Scholar] [CrossRef]
- Schmitz, R.J.; Amasino, R.M. Vernalization: A model for investigating epigenetics and eukaryotic gene regulation in plants. Biochim. Et Biophys. Acta 2007, 1769, 269–275. [Google Scholar] [CrossRef] [PubMed]
- Kamiya, Y.; Shiraki, S.; Fujiwara, K.; Akter, M.A.; Akter, A.; Fujimoto, R. The role of epigenetic transcriptional regulation in Brassica vegetables: A potential resource for epigenetic breeding. In Smart Plant Breeding for Vegetable Crops in Post-Genomics Era; Springer Nature Singapore Pte Ltd.: Singapore, 2023; pp. 1–24. [Google Scholar] [CrossRef]
- Zhang, S.J.; Qiu, Y.; Song, J.P.; Wang, H.P.; Zhang, X.H.; Shen, D. Identification for the tolerance to bolting of raddish germplasm resources. J. Plant. Genet. Res. 2014, 15, 262–269, (In Chinese with English Title and Abstract). [Google Scholar]
- Liu, C.X.; Sui, M.; Liu, C.X.; Cao, H.X.; Chang, T.L.; Yin, C.L. Back cross breeding of late bolting and detection of first intron PCR amplification of FLC2 gene in radish. Acta Agric. Boreal-Occident Sin. 2019, 34, 83–92, (In Chinese with English Title and Abstract). [Google Scholar] [CrossRef]
- Zhang, S.X.; Fan, W.Q.; Zheng, S.Y.; Wang, C.N.; Hua, D.P.; Zhang, H. Identification of bolting tolerance and genetic relationship analysis of green radish germplasm resources. Mol. Plant Breed. 2022, 20, 6566–6576, (In Chinese with English Title and Abstract). [Google Scholar]
- Zhuo, Z.C.; Wan, E.M.; Zhang, L.G.; Zhang, M.K.; Hui, M.X. Major gene plus poly gene inheritance analysis of bolting trait in heading Chinese cabbage. Acta Bot. Boreal-Occident Sin. 2009, 29, 867–873. [Google Scholar] [CrossRef]
- Ji, X.H.; Zhang, L.; Lu, X.X.; Liu, Y.T.; Wang, Y.G.; Feng, H. Inheritance analysis of bolting associated traits using mixed major gene plus polygenemodel in Brassica rapa. Agric. Biotechnol. 2013, 2, 26–30. [Google Scholar] [CrossRef]
- Li, X.F.; Zhu, H.F.; Zhu, Y.Y.; Hou, R.X. Combining ability and heritability analysis of bolting character of non-heading Chines cabbage. Plant Div. Res. 2015, 37, 788–792. [Google Scholar]
- Li, M.; Liu, Y.M.; Fang, Z.H.; Yang, L.M.; Zhuang, M.; Zhang, Y.Y. Combining ability and heritability analysis of bolting and flowering characters in cabbage (Brassica oleracea var. capitata). Acta Agric. Boreal-Occident Sin. 2009, 24, 86–89, (In Chinese with English Title and Abstract). [Google Scholar]
- Li, X.F.; Zhu, H.F.; Zhu, Y.Y.; Hou, R.X.; Zhai, W. Inheritance of major gene plus polygene underlying bolting and flowering traits in pak-choi. J. Nucl. Agric. Sci. 2016, 12, 2318–2325, (In Chinese with English Title and Abstract). [Google Scholar] [CrossRef]
- Motoki, K.; Kinoshita, Y.; Hosokawa, M. Non-vernalization flowering and seed set of cabbage induced by grafting onto radish roots tocks. Front. Plant Sci. 2019, 9, e1967. [Google Scholar] [CrossRef]
- Ipek, M.; Ipek, A.; Senalik, D.; Simon, P.W. Characterization of anunusualcy to plasmic chimera detected in bolting garlic clones. J. Am. Soc. Hortic. Sci. 2007, 132, 664–669. [Google Scholar] [CrossRef]
- Li, X.F.; Zhu, H.F.; Zhu, Y.Y.; Hou, R.X. Development of a new late-bolting germplasm and breeding a new cultivar yanchun of pak-choi by using space mutation. J. Nucl. Agric. Sci. 2018, 32, 1249–1255. [Google Scholar] [CrossRef]
- Abuyusuf, M.; Nath, U.K.; Kim, H.T.; Biswas, M.K.; Park, J.I.; Nou, I.S. Intronic sequence variations in a gene with peroxidase domain alter bolting time in cabbage (Brassica oleracea var. capitata). Plant Mol. Biol. Rep. 2018, 36, 725–737. [Google Scholar] [CrossRef]
- Fu, W.; Huang, S.S.; Liu, Z.Y.; Gao, Y.; Zhang, M.D.; Wang, N. Fine mapping of Brebm6, a geneconferring the early-bolting phenotype in Chinese cabbage (Brassica rapa ssp. pekinensis). Veg. Res. 2021, 1, e7. [Google Scholar] [CrossRef]
- Li, Q.F.; Peng, A.; Yang, J.Q.; Zheng, S.D.; Li, Z.P.; Mu, Y.H. A 215-bp in delatintron I of BoFLC2 affects flowering time in Brassica oleracea var. capitata during vernalization. Theor. Appl. Genet. 2022, 135, 2785–2797. [Google Scholar] [CrossRef]
- Tang, Q.W.; Kuang, H.H.; Yu, C.C.; An, G.H.; Tao, R.; Zhang, W.Y. Non-vernalization requirement in Chinese kale caused by loss of BoFLC and low expressions of its paralogs. Theor. Appl. Genet. 2022, 135, 473–483. [Google Scholar] [CrossRef]
- Shin, Y.H.; Lee, H.M.; Park, Y.D. CRISPR/Cas9-mediated editing of Agamous-like genes results in a late-bolting phenotype in Chinese cabbage (Brassica rapa ssp. pekinensis). Int. J. Mol. Sci. 2022, 23, 15009. [Google Scholar] [CrossRef]
- Shin, Y.H.; Park, Y.D. CRISPR/Cas9-mediated mutagenesis of BrLEAFY delays the bolting time in Chinese cabbage (Brassica rapa L. ssp. pekinensis). Int. J. Mol. Sci. 2023, 24, 541. [Google Scholar] [CrossRef]
- Choi, S.H.; Ahn, W.S.; Jie, E.Y.; Cho, H.S.; Kim, S.W. Development of late-bolting plants by CRISPR/Cas9-mediated genome editing from mesophyll protoplasts of lettuce. Plant Cell Rep. 2022, 41, 1627–1630. [Google Scholar] [CrossRef]
- Fan, S.T.; Li, Y.; Luo, X.Y.; Zong, Y.; Liu, B.L.; Li, J.M. Bioinformatics analysis of keygenes FT and SOC1 inradix Angelica sinensi. Mol. Plant Breed. 2022, 20, 3562–3569, (In Chinese with English Title and Abstract). [Google Scholar]
- Kitamoto, N.; Yui, S.; Nishikawa, K.; Takahata, Y.; Yokoi, S. A naturally occurring long insertion in the first intron in the Brassica rapa FLC2 gene causes delayed bolting. Euphytica 2014, 196, 213–223. [Google Scholar] [CrossRef]
- Chia, T.Y.P.; Müller, A.; Jung, C.; Mutasa-Göttgens, E.S. Sugar beet contains a large Constans-Like gene family including a CO homologue that is independent of the early-bolting (B) genelocus. J. Exp. Bot. 2008, 59, 2735–2748. [Google Scholar] [CrossRef] [PubMed]
- Szűcs, P.; Karsai, I.; Zitzewitz, J.V.; Meszaros, K.; Cooper, L.L.D.; Gu, Y.Q. Positional relationships between photoperiod response QTL and photoreceptor and vernalization genes in barley. Theor. Appl. Genet. 2006, 112, 1277–1285. [Google Scholar] [CrossRef]
- Liu, S.N.; Ma, T.F.; Ma, L.Y.; Lin, X.C. Ectopic expression of PvSOC1, a homolog of SOC1 from Phyllostachys violascens, promotes flowering in Arabidopsis andrice. Acta Physiol. Plant 2016, 38, 166. [Google Scholar] [CrossRef]
- Xu, S.J.; Dong, Q.; Deng, M.; Lin, D.X.; Xiao, J.; Cheng, P.L. The vernalization-induced long non-coding RNA VAS functions with the transcription factor TaRF2b to promote TaVRN1 expression for flowering in hexaploid wheat. Mol. Plant 2021, 14, 1525–1538. [Google Scholar] [CrossRef]
- Dios, E.A.D.; Delaye, L.; Simpson, J. Transcriptome analysis of bolting in A. tequilana reveals roles for florigen, MADS, fructans and gibberellins. BMC Genomics 2019, 20, 473. [Google Scholar] [CrossRef]
- Wen, Z.Z.; Guo, W.Z.; Li, J.C.; Lin, H.S.; He, C.M.; Liu, Y.Q. Comparative transcriptomic analysis of vernalization and cytokinin induced floral transition in Dendrobium nobile. Sci. Rep. 2017, 7, e45748. [Google Scholar] [CrossRef]
- Yan, X.; Wang, L.J.; Zhao, Y.Q.; Jia, G.X. Expression patterns of key genes in the photoperiod and vernalization flowering pathways in Lilium longiflorum with different bulbsizes. Int. J. Mol. Sci. 2022, 15, 8341. [Google Scholar] [CrossRef]
- Sharma, B.; Batz, T.A.; Kaundal, R.; Kramer, E.M.; Sanders, U.R.; Mellano, V.J. Developmental and molecular changes underlying the vernalization-induced transition of lowering in Aquilegia coerulea (James). Genes 2019, 10, 734. [Google Scholar] [CrossRef]
- Jaudal, M.; Yeoh, C.C.; Zhang, L.L.; Stockum, C.; Mysore, K.S.; Ratet, P. Retroelement insertions at the medicago FTa1 locus in spring mutants eliminate vernalisation but not long-day requirements for early flowering. Plant J. 2013, 76, 580–591. [Google Scholar] [CrossRef]
- Shojaei, E.; Mirzaie-Asl, A.; Mahmoudi, S.B.; Nazeri, S. Identification of sugar beet flowering genes based on Arabidopsis homologous genes. J. Agric. Sci. Technol. 2017, 19, 719–729. [Google Scholar]
- Kane, N.A.; Agharbaoui, Z.; Diallo, A.O.; Adam, H.; Tominaga, Y.; Ouellet, F. TaVRT2 represses transcription of the wheat vernalization gene TaVRN1. Plant J. 2007, 51, 670–680. [Google Scholar] [CrossRef]
- Takada, S.; Akter, A.; Itabashi, E.; Nishida, N.; Shea, D.J.; Miyaji, N. The role of Frigida and Flowering Locus C genes in flowering time of Brassica rapa leafy vegetables. Sci. Rep. 2019, 9, e13843. [Google Scholar] [CrossRef]
- Liang, N.G.; Cheng, D.Y.; Zhao, L.; Lu, H.D.; Xu, L.; Bi, Y.H. Identification of the genes encoding B3 domain-containing proteins related to vernalization of Beta vulgaris. Genes 2022, 13, 2217. [Google Scholar] [CrossRef] [PubMed]
- Hinckley, W.E.; Brusslan, J.A. Gene expression changes occurring at bolting time are associatedwith leaf senescence in Arabidopsis. Plant Direct. 2020, 4, e00279. [Google Scholar] [CrossRef] [PubMed]
| Families | Species | Gene | Characteristic of Genes, Expression, and Regulation | References |
|---|---|---|---|---|
| Brassicaceae | Cabbage (B. oleracea L.) | BoFLC2 | Negative regulation | [97,98] |
| BoVIN3 | Stems; gene length was 1680 bp; amino acid was 560 aa; isoelectric point was 6.56 | [99] | ||
| BoFLC(1,2,3) | Conserved with the MADS-box domain | [42] | ||
| AGL18, HDA9 | Gene length was 777 and 1281 bp, conserved region was MIKC/HDAC; amino acids were 258 and 426 aa; negative regulation | [100] | ||
| AGL19, SOC1, AGL24 | Amino acids were 221, 221, and 214 bp, conserved region was MIKC; AGL19 and SOC1 were SOC1/TM3 subfamily; positive regulation | [101,102] | ||
| Chinese cabbage (B. rapa L.) | BrFLC5 | Weak positive regulator | [103] | |
| BrFLC2 | A candidate gene for a vernalization | [99] | ||
| BrFLC, BrMAF, BrVIP, BrVRN | Negative regulation, response to vernalization; BrVRN and BrVIP genes showed different expression patterns | [33] | ||
| BrHIS4.A04 | A histone H4 gene; prevents premature bolting by weakening the expression of flowering genes under drought conditions through the ABA signaling pathway | [104] | ||
| BrEb-1 | Positive regulation; an incomplete dominant gene, mapped from 20,070,000 to 25,290,000 bp (5.22 Mb) and harbored on chromosome A07 | [105] | ||
| BrPIF4, BrPIF5, BrFLCs, BrFRL, BrMAF1s, BrCOLs | BrPIF4, BrPIF5, and BrCOLs were predominantly expressed in core tissues, promoting bolting, BrFLCs, BrFRL, and BrMAF1s were predominantly expressed in core tissue | [106] | ||
| BrSVP | Negative regulation; repressed the expression of the floral integrator genes AGL20, AGL24, and FT during vernalization | [107] | ||
| B. rapa ssp. pekinensis | BrpFLC | Negative regulation; gene length was 851 bp and contained a 591 bp ORF; amino acid length was 197 aa; conserved region was MADS-box; belongs to a multi-gene family | [108] | |
| B. campestris ssp. chinensis | BcFLC-1, BcFLC-2, BcFLC3 | Negative regulation, responded to low-temperature vernalization in leaves; gene length was 1017 bp; conserved region was MADS-box; amino acid length was 197 aa; isoelectric point was 9.36; multi-copy | [109,110] | |
| Chinese cabbage | BcSOC1 | Positive regulation; promoted stem elongation and bolting in flowering Chinese cabbage | [51] | |
| DELLA | Negative regulation of GA signal transduction; its proteins contained VHYNP-, DELLA-, SAW-, and VHIID-conserved domains; tissue-specific expression | [111] | ||
| Radish (R. Sativus L.) | BrcuFRI, BrcuFLC | BrcuFRI in stems and leaves, while BrcuFLC in roots; Gen Bank accession number was EU700362/EF138603 | [86] | |
| RsFLC, RsSOC1 | RsFLC was Ft genes; negative regulation;RsSOC1 positive negative | [112] | ||
| RsFLC2 | A late-bolting gene was detected in a 1.1 cM on chromosome R02; contains a 1627 bp insertion; weakened gene repression | [113] | ||
| FLC1.1, FLC1.2, VRN1, VRN2, SOC1 | FLC1.1 and FLC1.2 induced positive regulation while VRN1, VRN2, and SOC1 induced negative regulation | [54] | ||
| Carrot (D. carota L.) | DcFLC1, DcFLC2, DcFLC3 | DcFLC1 and DcFLC3 responded to low temperature in late bolting while DcFLC2 responded to light; amino acid length was 209, 212, and 219 aa; conserved region was MADS-box/K-box | [114] | |
| Vrn1, Rf1 | Early-flowering gene, mapped to chromosomes 2 and 9 with flanking markers from 0.70 to 4.38 cM and 0.46 to 1.12 cM | [115] | ||
| DcSOC1-1DcSOC1-2 | DcSOC1-1 promoted by long day early-bolting; amino acids were 217 and 211 aa, conserved region was MADS-box/K-box, sub cellular location was SOC1/TM3 subfamily | [116] | ||
| Lettuce (L. sativa L.) | LsARF3 | Response to high temperature; activate the expression of LsCO | [117] | |
| LsARF8a | 24 LsARFs in the lettuce genome; have been classified into three clusters; respond to heat | [118] | ||
| LsRGL1 | One of the DELLA-encoding genes; negatively regulates the GA pathway; interacts with LsGA3ox and the LsYUC4 promoter region | [50] | ||
| LsFT | Over expression of it recovered the late-flowering phenotype of ft-2 mutant and it was promoted by heat treatment; knockdown of it by RNA interference dramatically delayed bolting | [119] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tan, G.-F.; Luo, Q.; Zhu, S.-H.; Zhong, X.-L.; Meng, P.-H.; Li, M.-Y.; Chen, Z.-F.; Xiong, A.-S. Advancements in Molecular Mechanism Research on Bolting Traits in Vegetable Crops. Horticulturae 2024, 10, 670. https://doi.org/10.3390/horticulturae10070670
Tan G-F, Luo Q, Zhu S-H, Zhong X-L, Meng P-H, Li M-Y, Chen Z-F, Xiong A-S. Advancements in Molecular Mechanism Research on Bolting Traits in Vegetable Crops. Horticulturae. 2024; 10(7):670. https://doi.org/10.3390/horticulturae10070670
Chicago/Turabian StyleTan, Guo-Fei, Qing Luo, Shun-Hua Zhu, Xiu-Lai Zhong, Ping-Hong Meng, Meng-Yao Li, Zhi-Feng Chen, and Ai-Sheng Xiong. 2024. "Advancements in Molecular Mechanism Research on Bolting Traits in Vegetable Crops" Horticulturae 10, no. 7: 670. https://doi.org/10.3390/horticulturae10070670
APA StyleTan, G.-F., Luo, Q., Zhu, S.-H., Zhong, X.-L., Meng, P.-H., Li, M.-Y., Chen, Z.-F., & Xiong, A.-S. (2024). Advancements in Molecular Mechanism Research on Bolting Traits in Vegetable Crops. Horticulturae, 10(7), 670. https://doi.org/10.3390/horticulturae10070670








