Genome-Wide Identification, Characterization, and Expression Profile of SWEETs Gene Family in Grapevine (Vitis vinifera L.)
Abstract
1. Introduction
| Type | Species | Number of SWEET Members | Reference |
|---|---|---|---|
| Monocotyledons | Oryza sativa | 21 | [23] |
| Sorghum bicolor | 23 | [24] | |
| Musa acuminata | 25 | [25] | |
| Ananas comosus | 39 | [12] | |
| Bletilla striata | 17 | [26] | |
| Saccharum spontaneum | 22 | [27] | |
| Zea mays | 24 | [28] | |
| Prunus mume | 17 | [29] | |
| Triticum aestivuml | 59 | [30] | |
| Hordeum vulgare | 23 | [31] | |
| Dicotyledons | Arabidopsis thaliana | 17 | [6] |
| Manihot esculenta | 23 | [32] | |
| Citrus sinensis | 16 | [33] | |
| Eucalyptus grandis | 52 | [34] | |
| Glycine max | 52 | [35] | |
| Solanum lycopersicum | 29 | [20] | |
| Solanum tuberosum | 35 | [36] | |
| Pyrus bretschneideri | 18 | [37] | |
| Gossypium hirsutum | 55 | [38] | |
| Malus domestica | 25 | [13] | |
| Camellia sinensis | 13 | [39] | |
| Litchi | 16 | [40] | |
| Medicago truncatula | 25 | [41] | |
| Brassica oleracea | 30 | [42] | |
| Juglans regia | 25 | [43] | |
| Citrullus lanatus | 22 | [44] | |
| Punica granatum | 20 | [45] | |
| Dimocarpus longan | 20 | [46] | |
| Beta vulgaris | 16 | [47] | |
| Rosa rugosa | 25 | [48] | |
| Prunus salicina | 15 | [49] | |
| Capsicum annuum | 33 | [50] | |
| Medicago polymorpha | 23 | [51] | |
| Ziziphus jujuba | 19 | [52] | |
| Betula platyphylla | 13 | [53] | |
| Potentilla anserina | 23 | [54] |
2. Materials and Methods
2.1. Identification and Annotation of SWEET Genes in the Grapevine Genome
2.2. Phylogenetic and Conserved Domain Alignment Analysis
2.3. Analysis of Exon–Intron Structure and Conserved Motifs
2.4. Chromosomal Localization and Synteny Analysis
2.5. Analysis of Expression Profiles in Various Organs and Different Berry Developmental Stages
2.6. Analysis of Expression Profiles in Different Abiotic Stress Conditions
- Leaves from one-year-old potted grapevine plants with cold-resistant (V. amurensis S‘huangyou’) and cold-sensitive (V. vinifera cultivar R‘ed Globe’) varieties after cold stress (0 °C) for 3, 12, 48, and 72 h [74].
- Leaves of two-year-old potted cutting seedlings from the drought-resistant (V. yeshanensis ’Yanshan-1’) and the drought-sensitive (V. riparia’ He’an’) varieties, after drought stress for 0, 8, 16, and 24 days [75].
- Two-year-old potted grapevine rootstocks, including the salt-tolerant varieties 3309C (V. riparia × V. rupestris), 520A (V. berlandieri × V. riparia), and 1103P (V. berlandieri × V. rupestris), as well as the salt-sensitive varieties 5BB (V. berlandieri × V. riparia), 101–14 (V. riparia × V. rupestris), and Beta (V. riparia × V. labrusca), were irrigated with a 130 mmol L−1 NaCl solution for two consecutive days to induce salt stress [76].
2.7. Expression Profiles to Biotic Stress
2.8. RNA Extracted and qRT–PCR Analysis
3. Results
3.1. Identification and Characterization of Grape SWEET Genes
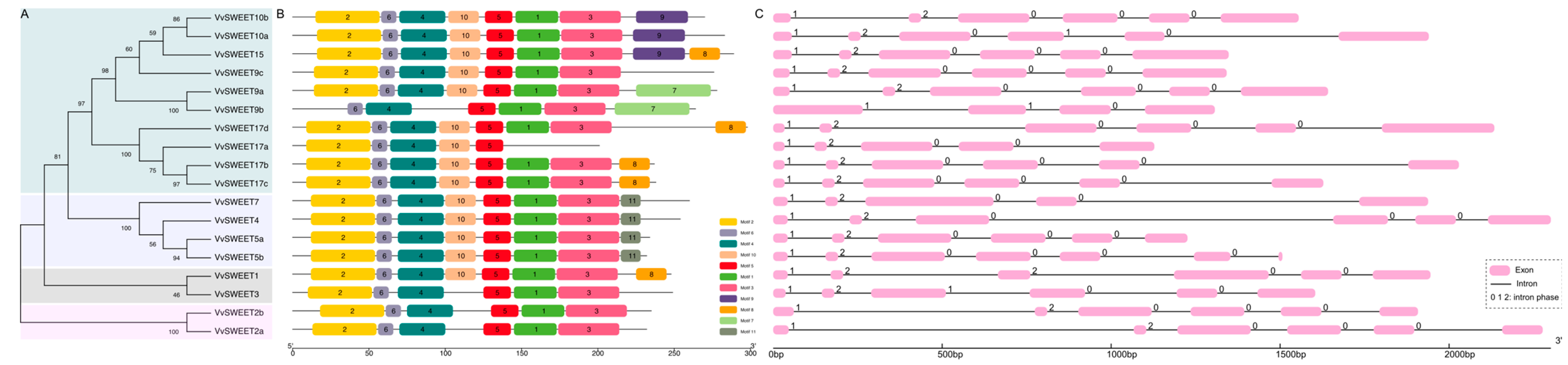
3.2. Phylogenetic Analysis of the Grape SWEET Proteins
3.3. Analysis of Conserved Protein Motifs and Exon–Intron Structure of Grape SWEET Genes
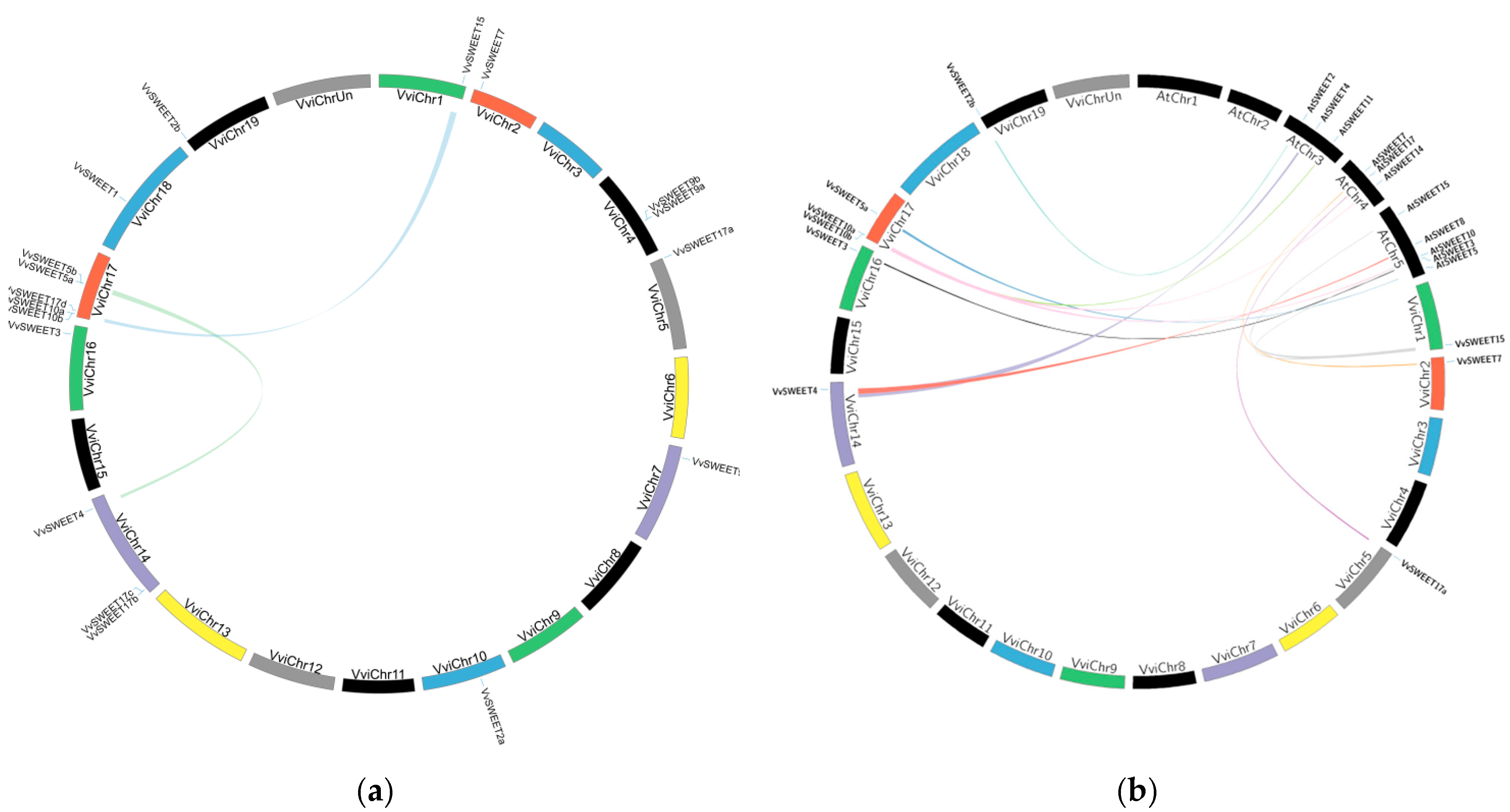
3.4. Chromosomal Distribution and Synteny Analysis among Grape SWEET Genes
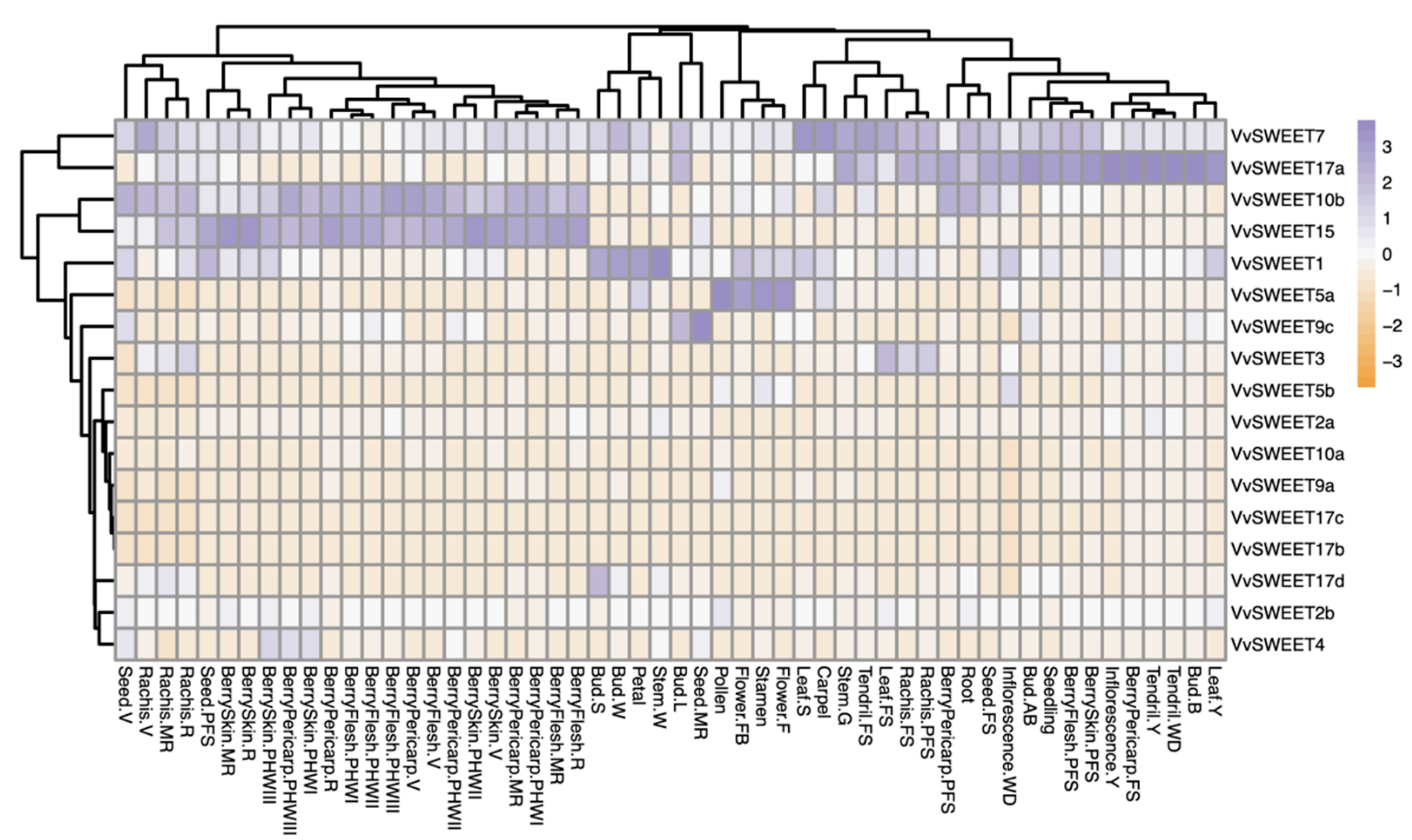
3.5. Expression Analysis of Grape SWEET Genes in Different Tissues
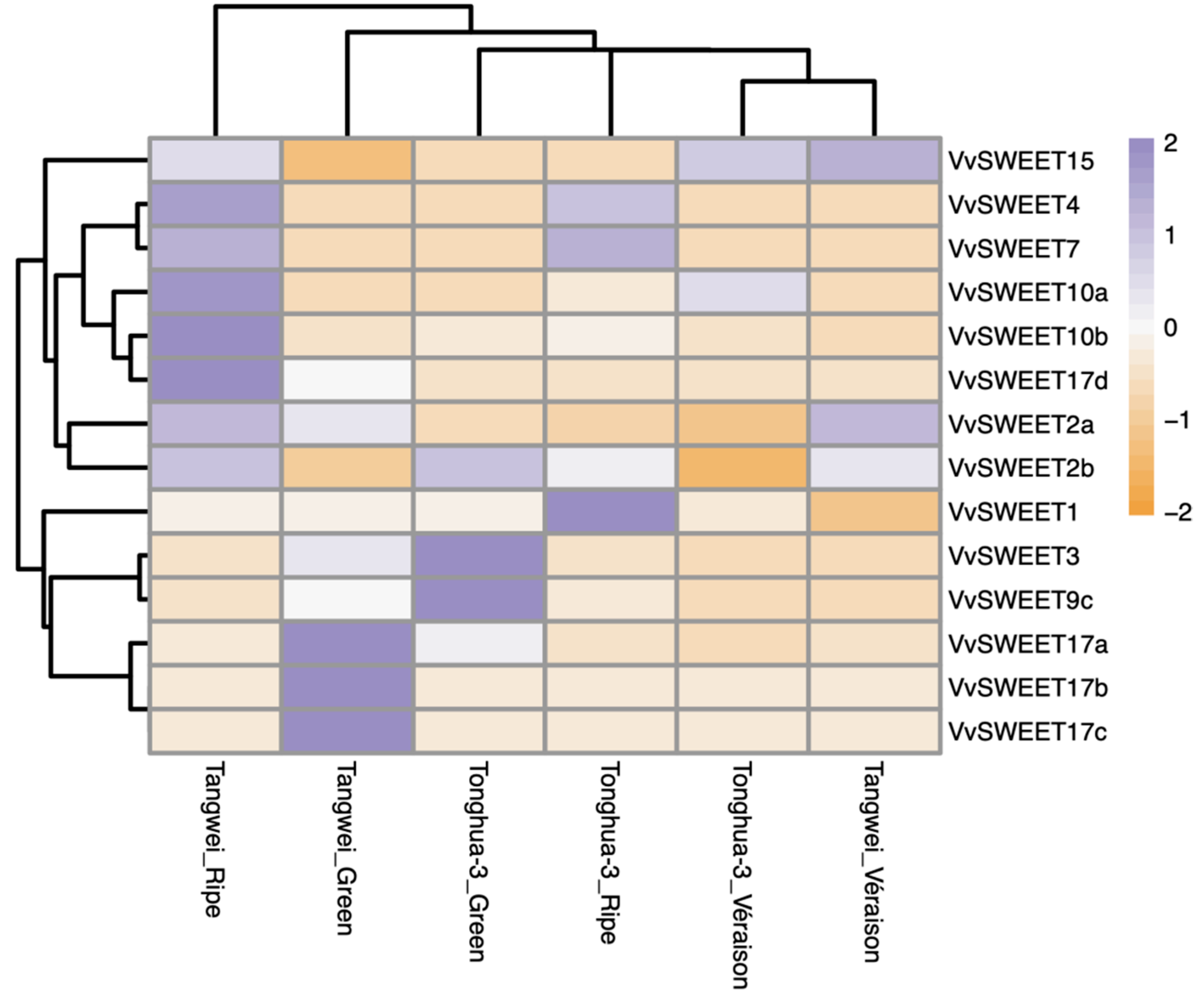
3.6. Expression Analysis of Grape SWEET Genes during Fruit Developmental Stages
3.7. Expression Patterns of Grape SWEET Genes under Abiotic Stress Conditions
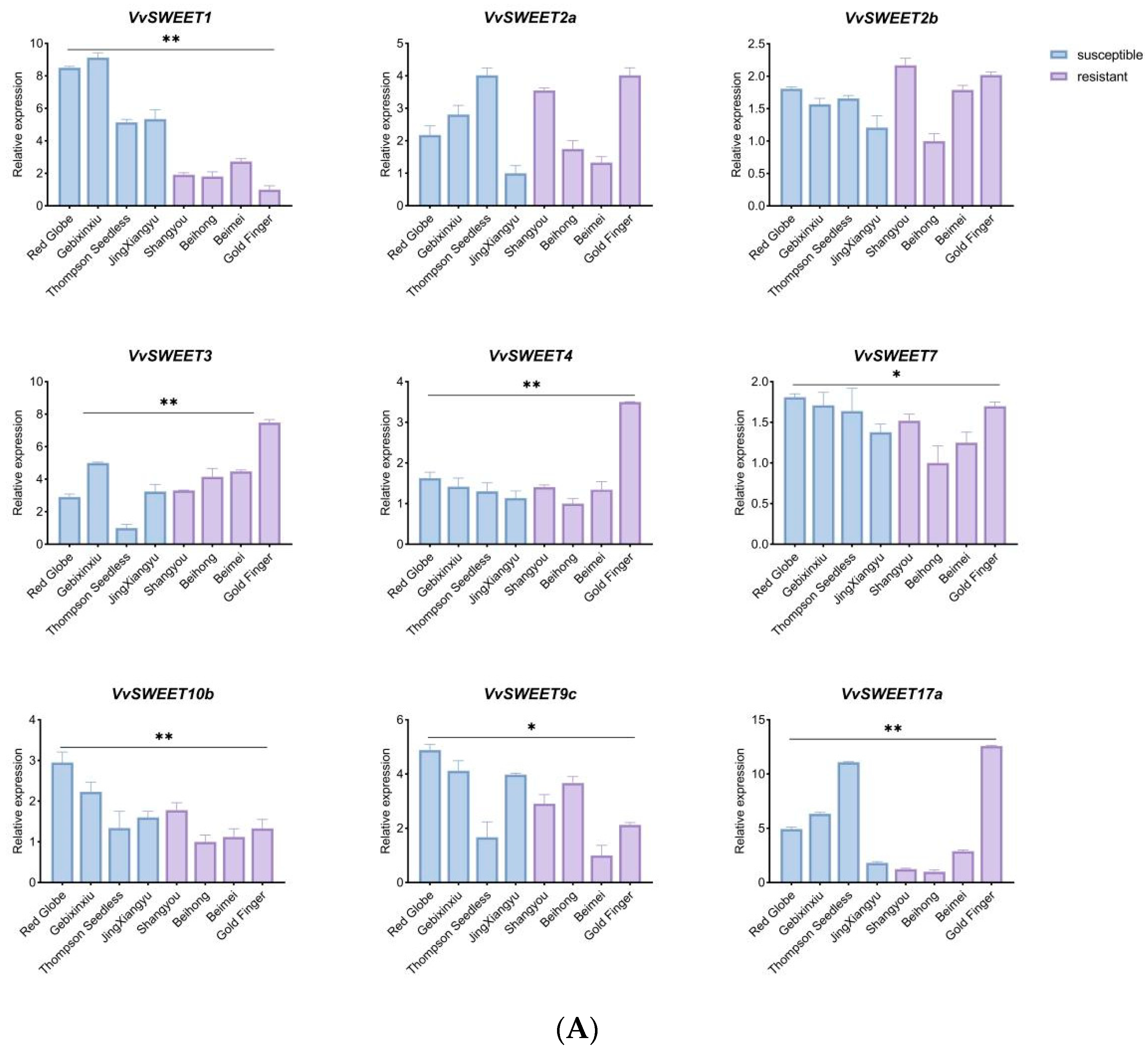
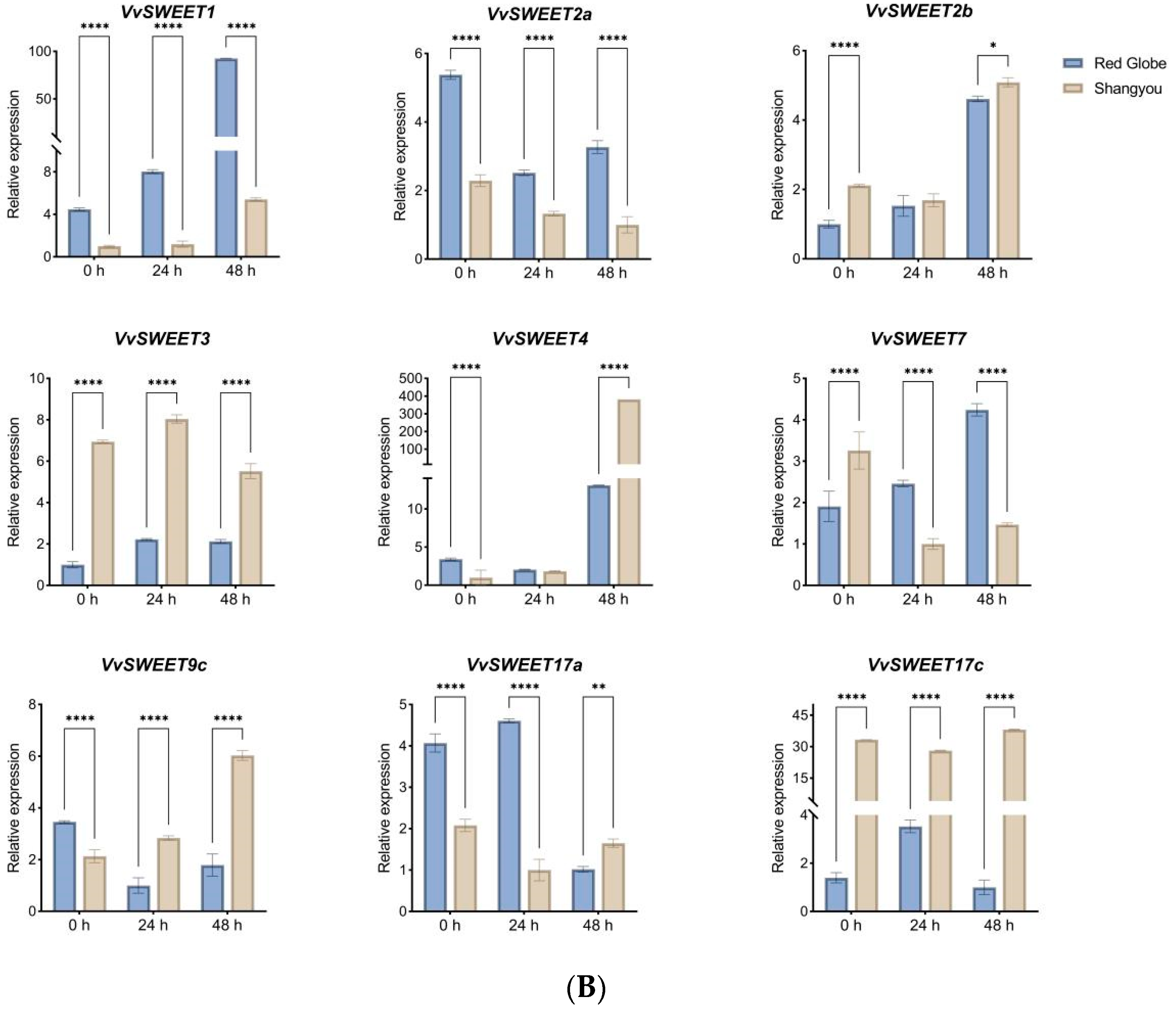
3.8. qRT-PCR Analysis of VvSWEET Genes in Relation to Grape Botrytis cinerea Infection
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Yamada, K.; Osakabe, Y. Sugar compartmentation as an environmental stress adaptation strategy in plants. Semin. Cell Dev. Biol. 2017, 83, 106–114. [Google Scholar] [CrossRef]
- Pommerrenig, B.; Ludewig, F.; Cvetkovic, J.; Trentmann, O.; Klemens, P.A.W.; Neuhaus, H.E. In Concert: Orchestrated Changes in Carbohydrate Homeostasis Are Critical for Plant Abiotic Stress Tolerance. Plant Cell Physiol. 2018, 59, 1290–1299. [Google Scholar] [CrossRef] [PubMed]
- Lalonde, S.; Wipf, D.; Frommer, W.B. Transport mechanisms for organic forms of carbon and nitrogen between source and sink. Annu. Rev. Plant Biol. 2004, 55, 341–372. [Google Scholar] [CrossRef] [PubMed]
- Slewinski, T.L. Diverse functional roles of monosaccharide transporters and their homologs in vascular plants: A physiological perspective. Mol. Plant 2011, 4, 641–662. [Google Scholar] [CrossRef] [PubMed]
- Kühn, C.; Grof, C.P. Sucrose transporters of higher plants. Curr. Opin. Plant Biol. 2010, 13, 288–298. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.Q.; Hou, B.H.; Lalonde, S.; Takanaga, H.; Hartung, M.L.; Qu, X.Q.; Guo, W.J.; Kim, J.G.; Underwood, W.; Chaudhuri, B.; et al. Sugar transporters for intercellular exchange and nutrition of pathogens. Nature 2010, 468, 527–532. [Google Scholar] [CrossRef]
- Chen, L.Q.; Qu, X.Q.; Hou, B.H.; Sosso, D.; Osorio, S.; Fernie, A.R.; Frommer, W.B. Sucrose efflux mediated by SWEET proteins as a key step for phloem transport. Science 2012, 335, 207–211. [Google Scholar] [CrossRef] [PubMed]
- Eom, J.S.; Chen, L.Q.; Sosso, D.; Julius, B.T.; Lin, I.W.; Qu, X.Q.; Braun, D.M.; Frommer, W.B. SWEETs, transporters for intracellular and intercellular sugar translocation. Curr. Opin. Plant Biol. 2015, 25, 53–62. [Google Scholar] [CrossRef] [PubMed]
- Kanno, Y.; Oikawa, T.; Chiba, Y.; Ishimaru, Y.; Shimizu, T.; Sano, N.; Koshiba, T.; Kamiya, Y.; Ueda, M.; Seo, M. AtSWEET13 and AtSWEET14 regulate gibberellin-mediated physiological processes. Nat. Commun. 2016, 7, 13245–13256. [Google Scholar] [CrossRef]
- Sosso, D.; Luo, D.; Li, Q.B.; Sasse, J.; Yang, J.; Gendrot, G.; Suzuki, M.; Koch, K.E.; McCarty, D.R.; Chourey, P.S.; et al. Seed filling in domesticated maize and rice depends on SWEET-mediated hexose transport. Nat. Genet. 2015, 47, 1489–1493. [Google Scholar] [CrossRef]
- Wu, L.-B.; Eom, J.-S.; Isoda, R.; Li, C.; Char, S.N.; Luo, D.; Schepler-Luu, V.; Nakamura, M.; Yang, B.; Frommer, W.B. OsSWEET11b, a potential sixth leaf blight susceptibility gene involved in sugar transport-dependent male fertility. New Phytol. 2022, 234, 975–989. [Google Scholar] [CrossRef] [PubMed]
- Guo, C.Y.; Li, H.Y.; Xia, X.Y.; Liu, X.Y.; Yang, L. Functional and evolution characterization of SWEET sugar transporters in Ananas comosus. Biochem. Biophys. Res. Commun. 2018, 496, 407–414. [Google Scholar] [CrossRef] [PubMed]
- Zhen, Q.L.; Fang, T.; Peng, Q.; Liao, L.; Zhao, L.; Owiti, A.; Han, Y.P. Developing gene-tagged molecular markers for evaluation of genetic association of apple SWEET genes with fruit sugar accumulation. Hortic. Res. 2018, 5, 14. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Liu, L.; Huang, W.; Yuan, M.; Zhou, F.; Li, X.; Lin, Y. Overexpression of OsSWEET5 in rice causes growth retardation and precocious senescence. PLoS ONE 2014, 9, e94210. [Google Scholar] [CrossRef] [PubMed]
- Klemens, P.A.W.; Patzke, K.; Krapp, A.; Chardon, F.; Neuhaus, H.E. SWEET16 and SWEET17, two novel vacuolar sugar carriers with impact on cellular sugar homeostasis and plant traits. Biochem. Cell Biol. 2014, 92, 589. [Google Scholar]
- Yue, C.; Cao, H.; Wang, L.; Zhou, Y.; Huang, Y.; Hao, X.; Wang, Y.; Wang, B.; Yang, Y.; Wang, X. Effects of cold acclimation on sugar metabolism and sugar-related gene expression in tea plant during the winter season. Plant Mol. Biol. 2015, 88, 591–608. [Google Scholar] [CrossRef] [PubMed]
- Seo, P.J.; Park, J.M.; Kang, S.K.; Kim, S.G.; Park, C.M. An Arabidopsis senescence-associated protein SAG29 regulates cell viability under high salinity. Planta 2011, 233, 189–200. [Google Scholar] [CrossRef] [PubMed]
- Julius, B.T.; Leach, K.A.; Tran, T.M.; Mertz, R.A.; Braun, D.M. Sugar Transporters in Plants: New Insights and Discoveries. Plant Cell Physiol. 2017, 58, 1442–1460. [Google Scholar] [CrossRef] [PubMed]
- Durand, M.; Porcheron, B.; Hennion, N.; Maurousset, L.; Lemoine, R. Water deficit enhances C export to the roots in Arabidopsis thaliana plants with contribution of sucrose transporters in both shoot and roots. Plant Physiol. 2016, 170, 1460–1479. [Google Scholar] [CrossRef]
- Feng, C.Y.; Han, J.X.; Han, X.X.; Jiang, J. Genome-wide identification, phylogeny, and expression analysis of the SWEET gene family in tomato. Gene 2015, 573, 261–272. [Google Scholar] [CrossRef]
- Streubel, J.; Pesce, C.; Hutin, M.; Koebnik, R.; Boch, J.; Szurek, B. Five phylogenetically close rice SWEET genes confer TAL effector-mediated susceptibility to Xanthomonas oryzae pv. oryzae. New Phytol. 2013, 200, 808–819. [Google Scholar] [CrossRef] [PubMed]
- Jaillon, O.; Aury, J.-M.; Noel, B.; Policriti, A.; Clepet, C.; Casagrande, A.; Choisne, N.; Aubourg, S.; Vitulo, N.; Jubin, C.; et al. The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 2007, 449, 463–467. [Google Scholar] [PubMed]
- Yuan, M.; Wang, S. Rice MtN3/saliva/SWEET family genes and their homologs in cellular organisms. Mol. Plant. 2013, 6, 665–674. [Google Scholar] [CrossRef] [PubMed]
- Mizuno, H.; Kasuga, S.; Kawahigashi, H. The sorghum SWEET gene family: Stem sucrose accumulation as revealed through transcriptome profiling. Biotechnol. Biofuels 2016, 9, 127. [Google Scholar] [CrossRef] [PubMed]
- Miao, H.; Sun, P.; Liu, Q.; Miao, Y.; Liu, J.; Zhang, K.; Hu, W.; Zhang, J.; Wang, J.; Wang, Z.; et al. Genome-wide analyses of SWEET family proteins reveal involvement in fruit development and abiotic/biotic stress responses in banana. Sci Rep 2017, 7, 3536. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Hao, L.; Wang, N.; Bai, X.L.; Zhang, Y.M. Transcriptome-wide identification and expression profiling of the SWEET family in Bletilla striata and regulation analysis with non-coding RNAs. Ind. Crop. Prod. 2023, 201, 116876. [Google Scholar] [CrossRef]
- Hu, W.; Hua, X.; Zhang, Q.; Wang, J.; Shen, Q.; Zhang, X.; Wang, K.; Yu, Q.; Lin, Y.R.; Ming, R.; et al. New insights into the evolution and functional divergence of the SWEET family in Saccharum based on comparative genomics. BMC Plant Biol. 2018, 18, 270. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.L.; Zhou, L.; Li, T.F.; Ruan, Y.Y.; Zhang, A.; Dong, X.M.; Zhu, Y.S.; Li, C.; Fan, J.J. Genome-Wide Investigation and Characterization of SWEET Gene Family with Focus on Their Evolution and Expression during Hormone and Abiotic Stress Response in Maize. Genes 2022, 13, 1682. [Google Scholar] [CrossRef]
- Wen, Z.Y.; Li, M.Y.; Meng, J.; Li, P.; Cheng, T.R.; Zhang, Q.X.; Sun, L.D. Genome-wide identification of the SWEET gene family mediating the cold stress response in Prunus mume. PeerJ 2022, 10, e13273. [Google Scholar] [CrossRef]
- Gao, Y.; Wang, Z.Y.; Kumar, V.; Xu, X.F.; Yuan, D.P.; Zhu, X.F.; Li, T.Y.; Jia, B.L.; Xuan, Y.H. Genome-wide identification of the SWEET gene family in wheat. Gene 2018, 642, 284–292. [Google Scholar] [CrossRef]
- Yue, W.H.; Cai, K.F.; Xia, X.; Liu, L.; Wang, J.M. Genome-wide identification, expression pattern and genetic variation analysis of SWEET gene family in barley reveal the artificial selection of HvSWEET1a during domestication and improvement. Front. Plant Sci. 2023, 14, 1137434. [Google Scholar] [CrossRef] [PubMed]
- Cohn, M.; Bart, R.S.; Shybut, M.; Dahlbeck, D.; Gomez, M.; Morbitzer, R.; Hou, B.H.; Frommer, W.B.; Lahaye, T.; Staskawicz, B.J. Xanthomonas axonopodis virulence is promoted by a transcription activator-like effector–mediated induction of a SWEET sugar transporter in cassava. Mol. Plant-Microbe Interact. 2014, 27, 1186–1198. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Q.; Tang, Z.; Xu, Q.; Deng, X.X. Isolation, phylogenetic relationship and expression profiling of sugar transporter genes in sweet orange (Citrus sinensis). Plant Cell Tissue Organ Cult. 2014, 119, 609–624. [Google Scholar] [CrossRef]
- Yin, Q.; Zhu, L.; Du, P.; Fan, C.; Wang, J.; Zhang, B.; Li, H. Comprehensive analysis of SWEET family genes in Eucalyptus (Eucalyptus grandis). Biotechnol. Biotechnol. Equip. 2020, 34, 595–604. [Google Scholar] [CrossRef]
- Patil, G.; Valliyodan, B.; Deshmukh, R.; Prince, S.; Nicander, B.; Zhao, M.Z.; Sonah, H.; Song, L.; Lin, L.; Chaudhary, J.; et al. Soybean (Glycine max) SWEET gene family: Insights through comparative genomics, transcriptome profiling and whole genome re-sequence analysis. BMC Genomics 2015, 16, 520. [Google Scholar] [CrossRef] [PubMed]
- Manck-Götzenberger, J.; Requena, N. Arbuscular mycorrhiza symbiosis induces a major transcriptional reprogramming of the potato SWEET sugar transporter family. Front. Plant Sci. 2016, 7, 487. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Qin, M.; Qiao, X.; Cheng, Y.; Li, X.; Zhang, H.; Wu, J. A new insight into the evolution and functional divergence of SWEET transporters in Chinese white pear (Pyrus bretschneideri). Plant Cell Physiol. 2017, 58, 839–850. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Ren, Z.Y.; Wang, Z.Y.; Sun, K.; Pei, X.Y.; Liu, Y.G.; He, K.L.; Zhang, F.; Song, C.X.; Zhou, X.J.; et al. Evolution and Stress Responses of Gossypium hirsutum SWEET Genes. Int. J. Mol. Sci. 2018, 19, 769. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Yao, L.; Hao, X.; Li, N.; Qian, W.; Yue, C.; Ding, C.; Zeng, J.; Yang, Y.; Wang, X. Tea plant SWEET transporters: Expression profiling, sugar transport, and the involvement of CsSWEET16 in modifying cold tolerance in Arabidopsis. Plant Mol. Biol. 2018, 96, 577–592. [Google Scholar] [CrossRef]
- Xie, H.H.; Wang, D.; Qin, Y.Q.; Ma, A.N.; Fu, J.X.; Qin, Y.H.; Hu, G.B.; Zhao, J.T. Genome-wide identification and expression analysis of SWEET gene family in Litchi chinensis reveal the involvement of LcSWEET2a/3b in early seed development. BMC Plant Biol. 2019, 19, 499. [Google Scholar] [CrossRef]
- Hu, B.; Wu, H.; Huang, W.F.; Song, J.B.; Zhou, Y.; Lin, Y.J. SWEET Gene Family in Medicago truncatula: Genome-Wide Identification, Expression and Substrate Specificity Analysis. Plants 2019, 8, 338. [Google Scholar] [CrossRef]
- Zhang, W.; Wang, S.; Yu, F.W.; Tang, J.; Shan, X.; Bao, K.; Yu, L.; Wang, H.; Fei, Z.J.; Li, J.B. Genome-wide characterization and expression profiling of SWEET genes in cabbage (Brassica oleracea var. capitata L.) reveal their roles in chilling and clubroot disease responses. BMC Genomics 2019, 20, 93. [Google Scholar] [CrossRef] [PubMed]
- Jiang, S.J.; Balan, B.; Assis, R.D.B.; Sagawa, C.H.D.; Wan, X.Q.; Han, S.; Wang, L.; Zhang, L.L.; Zaini, P.A.; Walawage, S.L.; et al. Genome-Wide Profiling and Phylogenetic Analysis of the SWEET Sugar Transporter Gene Family in Walnut and Their Lack of Responsiveness to Xanthomonas arboricola pv. juglandis Infection. Int. J. Mol. Sci. 2020, 21, 1251. [Google Scholar] [CrossRef]
- Xuan, C.Q.; Lan, G.P.; Si, F.F.; Zeng, Z.L.; Wang, C.X.; Yadav, V.; Wei, C.H.; Zhang, X. Systematic Genome-Wide Study and Expression Analysis of SWEET Gene Family: Sugar Transporter Family Contributes to Biotic and Abiotic Stimuli in Watermelon. Int. J. Mol. Sci. 2021, 22, 8407. [Google Scholar] [CrossRef]
- Zhang, X.H.; Wang, S.; Ren, Y.; Gan, C.Y.; Li, B.B.; Fan, Y.Y.W.; Zhao, X.Q.; Yuan, Z.H. Identification, Analysis and Gene Cloning of the SWEET Gene Family Provide Insights into Sugar Transport in Pomegranate (Punica granatum). Int. J. Mol. Sci. 2022, 23, 2471. [Google Scholar] [CrossRef]
- Fang, T.; Rao, Y.; Wang, M.Z.; Li, Y.; Liu, Y.J.; Xiong, P.P.; Zeng, L.H. Characterization of the SWEET Gene Family in Longan (Dimocarpus longan) and the Role of DlSWEET1 in Cold Tolerance. Int. J. Mol. Sci. 2022, 23, 8914. [Google Scholar] [CrossRef] [PubMed]
- La, H.V.; Chu, H.D.; Ha, Q.T.; Tran, T.T.H.; Tong, H.V.; Tran, T.V.; Le, Q.T.N.; Bui, H.T.T.; Cao, P.B. SWEET Gene Family in Sugar Beet (Beta vulgaris): Genome-Wide Survey, Phylogeny and Expression analysis. Pak. J. Biol. Sci. 2022, 25, 387–395. [Google Scholar]
- Song, X.S.; Kou, Y.P.; Duan, M.G.; Feng, B.; Yu, X.Y.; Jia, R.D.; Zhao, X.; Ge, H.; Yang, S.H. Genome-Wide Identification of the Rose SWEET Gene Family and Their Different Expression Profiles in Cold Response between Two Rose Species. Plants 2023, 12, 1474. [Google Scholar] [CrossRef] [PubMed]
- Jiang, C.C.; Zeng, S.M.; Yang, J.; Wang, X.A. Genome-Wide Identification and Expression Profiling Analysis of SWEET Family Genes Involved in Fruit Development in Plum (Prunus salicina Lindl). Genes 2023, 14, 1679. [Google Scholar] [CrossRef]
- Han, X.W.; Han, S.; Zhu, Y.X.; Liu, Y.Q.; Gao, S.H.; Yin, J.L.; Wang, F.; Yao, M.H. Genome-Wide Identification and Expression Analysis of the SWEET Gene Family in Capsicum annuum L. Int. J. Mol. Sci. 2023, 24, 17408. [Google Scholar] [CrossRef]
- Liu, N.; Wei, Z.; Min, X.; Yang, L.; Zhang, Y.; Li, J.; Yang, Y. Genome-Wide Identification and Expression Analysis of the SWEET Gene Family in Annual Alfalfa (Medicago polymorpha). Plants 2023, 12, 1948. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Zhao, X.; Luo, Z.; Wang, L.H.; Liu, M.J. Genome-wide identification and expression profile analysis of SWEET genes in Chinese jujube. PeerJ 2023, 11, e14704. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Ding, Y.T.; Yang, K.Y.; Wang, X.Y.; Gao, W.S.; Xie, Q.J.; Liu, Z.Y.; Gao, C.Q. An Insight of Betula platyphylla SWEET Gene Family through Genome-Wide Identification, Expression Profiling and Function Analysis of BpSWEET1c under Cold Stress. Int. J. Mol. Sci. 2023, 24, 13626. [Google Scholar] [CrossRef] [PubMed]
- Iqbal, J.; Zhang, W.H.; Fan, Y.D.; Dong, J.; Xie, Y.Y.; Li, R.H.; Yang, T.; Zhang, J.Z.; Che, D.D. Genome-Wide Bioinformatics Analysis of SWEET Gene Family and Expression Verification of Candidate PaSWEET Genes in Potentilla anserina. Plants 2024, 13, 406. [Google Scholar] [CrossRef] [PubMed]
- Canaguier, A.; Grimplet, J.; Di Gaspero, G.; Scalabrin, S.; Duchêne, E.; Choisne, N.; Mohellibi, N.; Guichard, C.; Rombauts, S.; Le Clainche, I.; et al. A new version of the grapevine reference genome assembly (12X. v2) and of its annotation (VCost. v3). Genom Data 2017, 14, 56–62. [Google Scholar] [CrossRef] [PubMed]
- El-Gebali, S.; Mistry, J.; Bateman, A.; Eddy, S.R.; Luciani, A.; Potter, S.C.; Qureshi, M.; Richardson, L.J.; Salazar, G.A.; Smart, A.; et al. The Pfam protein families database in 2019. Nucleic Acids Res. 2019, 47, D427–D432. [Google Scholar] [CrossRef] [PubMed]
- Eddy, S.R. Profile hidden Markov models. Bioinformatics 1998, 14, 755–763. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, L.; Ji, M.; Wu, Y.; Zhang, S.; Zhu, Y.; Yao, J.; Li, Z.; Gao, H.; Wang, X. Genome-wide identification and expression analysis of the B-box transcription factor gene family in grapevine (Vitis vinifera L.). BMC Genomics 2021, 22, 221. [Google Scholar] [CrossRef] [PubMed]
- Goodstein, D.M.; Shu, S.; Howson, R.; Neupane, R.; Hayes, R.D.; Fazo, J.; Mitros, T.; Dirks, W.; Hellsten, U.; Putnam, N.; et al. Phytozome: A comparative platform for green plant genomics. Nucleic Acids Res. 2012, 40, D1178–D1186. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. 20 years of the SMART protein domain annotation resource. Nucleic Acids Res. 2018, 46, D493–D496. [Google Scholar] [CrossRef]
- Marchler-Bauer, A.; Bo, Y.; Han, L.; He, J.; Lanczycki, C.J.; Lu, S.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; et al. CDD/SPARCLE: Functional classification of proteins via subfamily domain architectures. Nucleic Acids Res. 2017, 45, D200–D203. [Google Scholar] [CrossRef] [PubMed]
- Wilkins, M.R.; Gasteiger, E.; Bairoch, A.; Sanchez, J.C.; Williams, K.L.; Appel, R.D.; Hochstrasser, D.F. Protein identification and analysis tools on the ExPASy server. In The Proteomics Protocols Handbook; Walker, J.M., Ed.; Humana Press: Totowa, NJ, USA, 2005; pp. 571–607. [Google Scholar]
- Zhang, X.; Ma, J.; Yang, S.; Yao, W.; Zhang, N.; Hao, X.; Xu, W. Analysis of GATA transcription factors and their expression patterns under abiotic stress in grapevine (Vitis vinifera L.). BMC Plant Biol. 2023, 23, 611. [Google Scholar] [CrossRef]
- Crooks, G.E.; Hon, G.; Chandonia, J.M.; Brenner, S.E. WebLogo: A sequence logo generator. Genome Res. 2004, 14, 1188–1190. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Jin, J.; Guo, A.Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37 (Suppl. S2), W202–W208. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; Debarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.-h.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef]
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef]
- Cao, J.; Han, X.; Zhang, T.; Yang, Y.; Huang, J.; Hu, X. Genome-wide and molecular evolution analysis of the subtilase gene family in Vitis vinifera. BMC Genom. 2014, 15, 1116. [Google Scholar] [CrossRef]
- Fasoli, M.; Dal Santo, S.; Zenoni, S.; Tornielli, G.B.; Farina, L.; Zamboni, A.; Porceddu, A.; Venturini, L.; Bicego, M.; Murino, V.; et al. The grapevine expression atlas reveals a deep transcriptome shift driving the entire plant into a maturation program. Plant Cell 2012, 24, 3489–3505. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Wu, Y.; Zhang, Y.; Yin, X.; van Nocker, S.; Guo, J.; Li, Z.; Gao, M.; Song, J.; Wang, X. Identification of potential key genes in resveratrol biosynthesis via transcriptional analyses of berry development in grapevine (Vitis spp.) genotypes varying in trans-resveratrol content. Fruit Res. 2022, 2, 6. [Google Scholar] [CrossRef]
- Gu, B.; Zhang, B.; Ding, L.; Li, P.; Shen, L.; Zhang, J. Physiological Change and Transcriptome Analysis of Chinese Wild Vitis amurensis and Vitis vinifera in Response to Cold Stress. Plant Mol. Biol. Rep. 2020, 38, 478–490. [Google Scholar] [CrossRef]
- Cui, X.; Xue, J.; Zhang, B.; Chen, C.; Tang, Y.; Zhang, P.; Zhang, J. Physiological change and screening of diferentially expressed genes of wild Chinese Vitis yeshanensis and American Vitis riparia in response to drought stress. Sci. Hortic. 2020, 266, 109140. [Google Scholar] [CrossRef]
- Zhao, F.; Zheng, T.; Liu, Z.; Fu, W.; Fang, J. Transcriptomic Analysis Elaborates the Resistance Mechanism of Grapevine Rootstocks against Salt Stress. Plants 2022, 11, 1167. [Google Scholar] [CrossRef] [PubMed]
- Wan, R.; Hou, X.; Wang, X.; Qu, J.; Singer, S.D.; Wang, Y.; Wang, X. Resistance evaluation of Chinese wild Vitis genotypes against Botrytis cinerea and different responses of resistant and susceptible hosts to the infection. Front. Plant Sci. 2015, 6, 854. [Google Scholar] [CrossRef]
- Gabler, F.M.; Smilanick, J.L.; Mansour, M.; Ramming, D.W.; Mackey, B.E. Correlations of Morphological, Anatomical, and Chemical Features of Grape Berries with Resistance to Botrytis cinerea. Phytopathology 2003, 93, 1263–1273. [Google Scholar] [CrossRef]
- Rahman, M.U.; Hanif, M.; Wan, R.; Hou, X.; Ahmad, B.; Wang, X. Screening Vitis Genotypes for Responses to Botrytis cinerea and Evaluation of Antioxidant Enzymes, Reactive Oxygen Species and Jasmonic Acid in Resistant and Susceptible Hosts. Molecules 2019, 24, 5. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Grimplet, J.; Adam-Blondon, A.-F.; Bert, P.-F.; Bitz, O.; Cantu, D.; Davies, C.; Delrot, S.; Pezzotti, M.; Rombauts, S.; Cramer, G.R. The grapevine gene nomenclature system. BMC Genomics 2014, 15, 1077. [Google Scholar] [CrossRef]
- Cannon, S.B.; Mitra, A.; Baumgarten, A.; Young, N.D.; May, G. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 2004, 4, 10. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Duan, X.; Jiang, H.; Sun, Y.; Tang, Y.; Yuan, Z.; Guo, J.; Liang, W.; Chen, L.; Yin, J.; et al. Genome-wide analysis of basic/helix-loop-helix transcription factor family in rice and Arabidopsis. Plant Physiol. 2006, 141, 1167–1184. [Google Scholar] [CrossRef] [PubMed]
- Le Hir, R.; Spinner, L.; Klemens, P.A.W.; Chakraborti, D.; de Marco, F.; Vilaine, F.; Wolff, N.; Lemoine, R.; Porcheron, B.; Géry, C.; et al. Disruption of the Sugar Transporters AtSWEET11 and AtSWEET12 Affects Vascular Development and Freezing Tolerance in Arabidopsis. Mol. Plant 2015, 8, 1687–1690. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Zhang, Y.; Yang, C.; Tian, Z.; Li, J. AtSWEET4, a hexose facilitator, mediates sugar transport to axial sinks and affects plant development. Sci. Rep. 2016, 6, 24563. [Google Scholar] [CrossRef]
- Yao, L.; Ding, C.; Hao, X.; Zeng, J.; Yang, Y.; Wang, X.; Wang, L. CsSWEET1a and CsSWEET17 Mediate Growth and Freezing Tolerance by Promoting Sugar Transport across the Plasma Membrane. Plant Cell Physiol. 2020, 61, 1669–1682. [Google Scholar] [CrossRef]


| Gene Name | VCost. v3 ID | Gene Locus ID | Accession No | CDS (bp) | ORF (aa) | Chromosome | MW | pI | Instability Index | Aliphatic Index | GRAVY |
|---|---|---|---|---|---|---|---|---|---|---|---|
| VvSWEET1 | Vitvi18g01215.t01 | GSVIVT01010015001 | XP_002265836.1 | 747 | 248 | Chr18: 13476398—-13478625 (−) | 27,352.74 | 9.64 | 28.21 | 111.29 | 0.682 |
| VvSWEET2 | Vitvi10g00679.t01 | GSVIVT01021317001 | XP_002269484.1 | 699 | 232 | Chr10: 7519651—7522710 (+) | 25,777.62 | 8.84 | 53.21 | 115.99 | 0.839 |
| VvSWEET2b | Vitvi19g00024.t01 | GSVIVT01014088001 | XP_010644065.1 | 708 | 235 | Chr19: 278130—280739 (−) | 26,117.95 | 9.30 | 47.26 | 117.32 | 0.741 |
| VvSWEET3 | Vitvi16g01984.t01 | GSVIVT01028713001 | XP_002267886.1 | 750 | 249 | Chr16: 21023729—21025870 (+) | 28,036.27 | 9.13 | 39.81 | 116.14 | 0.535 |
| VvSWEET4 | Vitvi14g01783.t01 | GSVIVT01032489001 | XP_002274582.1 | 765 | 254 | Chr14: 27825326—27827944 (−) | 27,899.02 | 9.36 | 38.85 | 108.98 | 0.517 |
| VvSWEET5a | Vitvi17g00791.t01 | GSVIVT01007779001 | XP_002283068.1 | 705 | 234 | Chr17: 9249190—9250708 (+) | 26,160.48 | 9.43 | 29.69 | 124.06 | 0.693 |
| VvSWEET5b | Vitvi17g00793.t01 | GSVIVT01007777001 | XP_002279850.1 | 699 | 232 | Chr17: 9294434—9295940 (+) | 25,967.16 | 9.49 | 30.31 | 119.66 | 0.698 |
| VvSWEET7 | Vitvi02g00181.t01 | GSVIVT01019601001 | XP_002263697.1 | 783 | 260 | Chr02: 1670296—1672808 (−) | 28,845.49 | 9.62 | 43.99 | 116.54 | 0.695 |
| VvSWEET9a | Vitvi04g01075.t01 | GSVIVT01026399001 | XP_002267792.1 | 837 | 278 | Chr04: 15790881—15792999 (−) | 31,515.37 | 9.05 | 39.12 | 112.16 | 0.576 |
| VvSWEET9b | Vitvi04g01077.t01 | - | RVW45685.1 | 795 | 264 | Chr04: 15798938—15800244 (+) | 29,875.24 | 8.34 | 31.28 | 100.80 | 0.454 |
| VvSWEET9c | Vitvi07g00250.t01 | GSVIVT01010993001 | XP_002270131.1 | 831 | 276 | Chr07: 2749916—2751667 (−) | 31,004.89 | 9.30 | 34.08 | 106.05 | 0.535 |
| VvSWEET10a | Vitvi17g00069.t01 | GSVIVT01008597001 | XP_002280599.1 | 852 | 283 | Chr17: 678049—680301 (+) | 31,699.78 | 9.34 | 38.63 | 112.93 | 0.575 |
| VvSWEET10b | Vitvi17g00070.t01 | GSVIVT01008595001 | XP_002284244.1 | 813 | 270 | Chr17: 682658—684513 (+) | 30,608.80 | 8.90 | 34.63 | 127.70 | 0.836 |
| VvSWEET15 | Vitvi01g01719.t01 | GSVIVT01000938001 | NP_001384792.1 | 864 | 289 | Chr01: 23092093—23093857 (−) | 32,149.03 | 9.08 | 35.66 | 110.00 | 0.492 |
| VvSWEET17a | Vitvi05g00013.t01 | GSVIVT01035138001 | XP_010649584.1 | 606 | 201 | Chr05: 123990—126791 (−) | 21,952.92 | 5.06 | 36.17 | 122.69 | 0.772 |
| VvSWEET17b | Vitvi14g00147.t01 | GSVIVT01031172001 | RVW97796.1 | 714 | 237 | Chr14: 1525182—1527230 (−) | 26,266.34 | 6.29 | 48.06 | 128.23 | 0.845 |
| VvSWEET17c | Vitvi14g00148.t01 | GSVIVT01031172001 | RVW97798.1 | 717 | 238 | Chr14: 1533798—1535492 (−) | 26,473.40 | 6.90 | 46.38 | 124.79 | 0.782 |
| VvSWEET17d | Vitvi14g00149.t01 | GSVIVT01031170001 | XP_002279031.1 | 897 | 298 | Chr17: 1541544—1545073 (−) | 32,764.71 | 9.68 | 37.96 | 107.58 | 0.424 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhong, L.; Xu, S.; Song, C.; Zhao, N.; Yang, Z.; Liu, Y.; Cui, X.; Zhang, J.; Li, Z.; Wang, X.; et al. Genome-Wide Identification, Characterization, and Expression Profile of SWEETs Gene Family in Grapevine (Vitis vinifera L.). Horticulturae 2024, 10, 428. https://doi.org/10.3390/horticulturae10050428
Zhong L, Xu S, Song C, Zhao N, Yang Z, Liu Y, Cui X, Zhang J, Li Z, Wang X, et al. Genome-Wide Identification, Characterization, and Expression Profile of SWEETs Gene Family in Grapevine (Vitis vinifera L.). Horticulturae. 2024; 10(5):428. https://doi.org/10.3390/horticulturae10050428
Chicago/Turabian StyleZhong, Linjing, Shuya Xu, Chenchen Song, Ning Zhao, Zhiqi Yang, Yanxiang Liu, Xiaoyue Cui, Jianxia Zhang, Zhi Li, Xiping Wang, and et al. 2024. "Genome-Wide Identification, Characterization, and Expression Profile of SWEETs Gene Family in Grapevine (Vitis vinifera L.)" Horticulturae 10, no. 5: 428. https://doi.org/10.3390/horticulturae10050428
APA StyleZhong, L., Xu, S., Song, C., Zhao, N., Yang, Z., Liu, Y., Cui, X., Zhang, J., Li, Z., Wang, X., & Gao, M. (2024). Genome-Wide Identification, Characterization, and Expression Profile of SWEETs Gene Family in Grapevine (Vitis vinifera L.). Horticulturae, 10(5), 428. https://doi.org/10.3390/horticulturae10050428






