The Identification of Cucumber TDC Genes and Analyses of Their Expression and Functions under Abiotic Stress Conditions
Abstract
1. Introduction
2. Materials and Methods
2.1. Identification and Analysis of TDC Genes in Cucumber
2.2. Analysis of Cis-Elements in the Promoter Regions
2.3. Plant Materials and Treatments
2.4. RNA Extraction and qRT-PCR Analysis
2.5. Subcellular Localization Analysis
2.6. Transient Overexpression of CsTDC Genes in Tobacco Leaves
2.7. Determination of TDC Activity and Melatonin Content
2.8. Transient Overexpression of CsTDC Genes and Abiotic Stress Treatments
2.9. Statistical Analyses
3. Results
3.1. Identification and Characterization of Cucumber TDC Genes
3.2. Identification of Cis-Elements in CsTDC Promoters
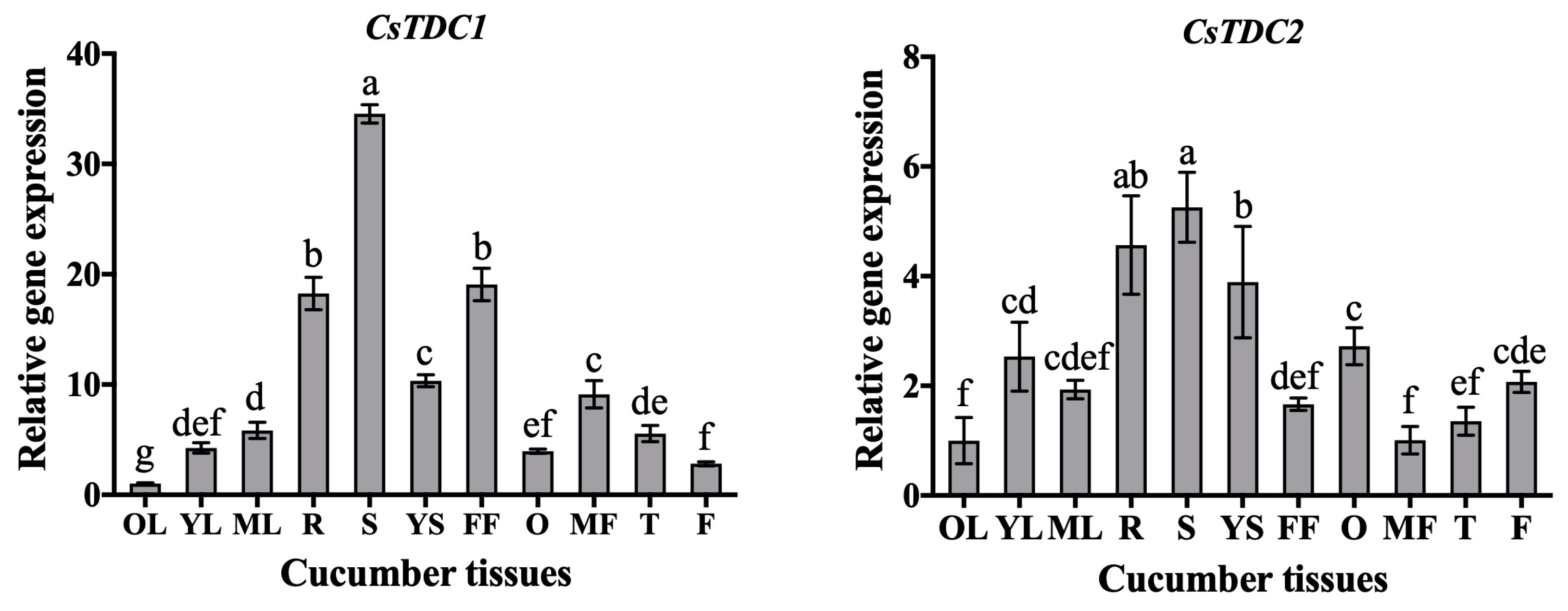
3.3. CsTDC Expression Profiles in Different Tissues
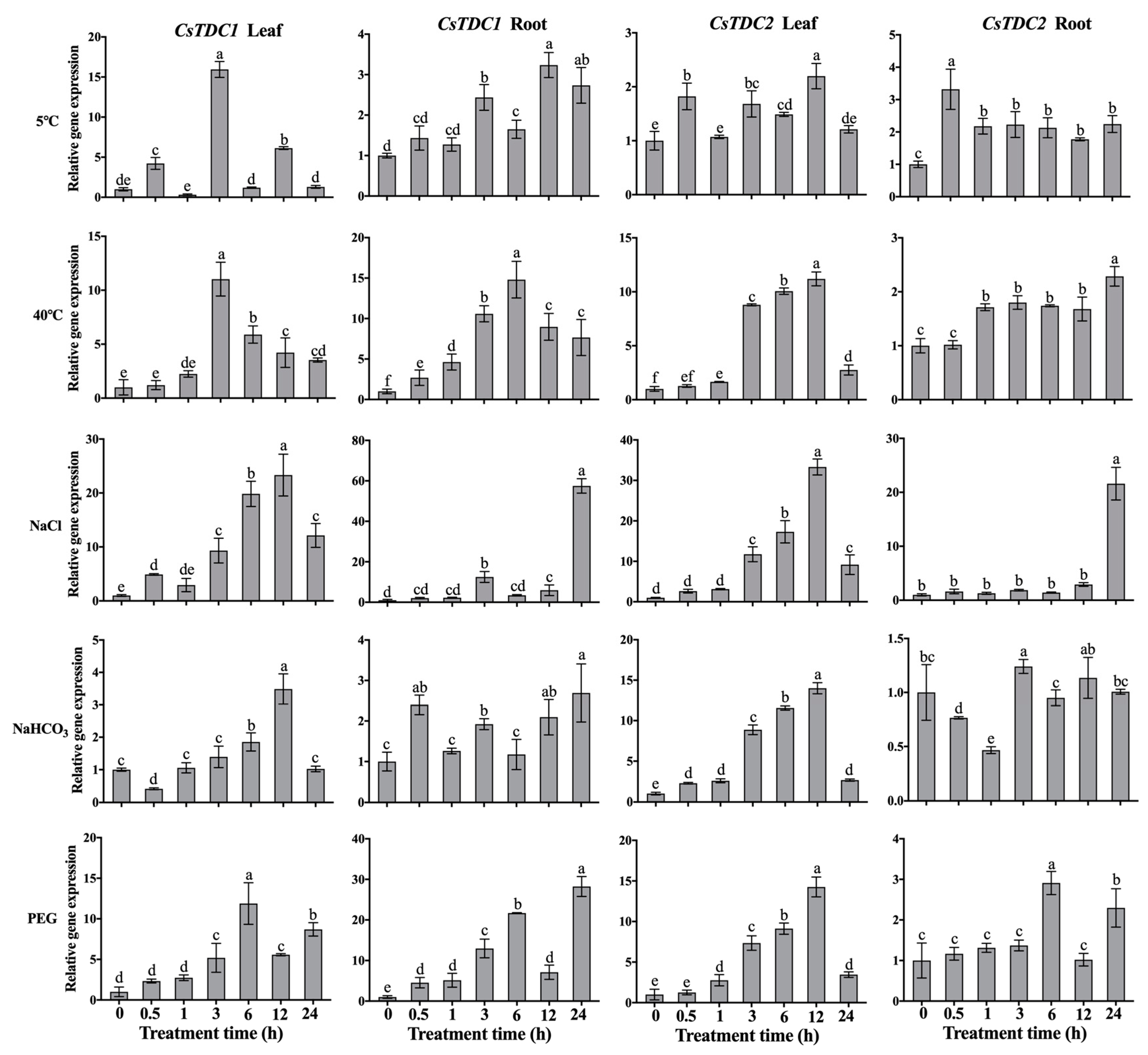
3.4. CsTDC Expression Profiles in Response to Various Abiotic Stresses and Exogenous Phytohormones
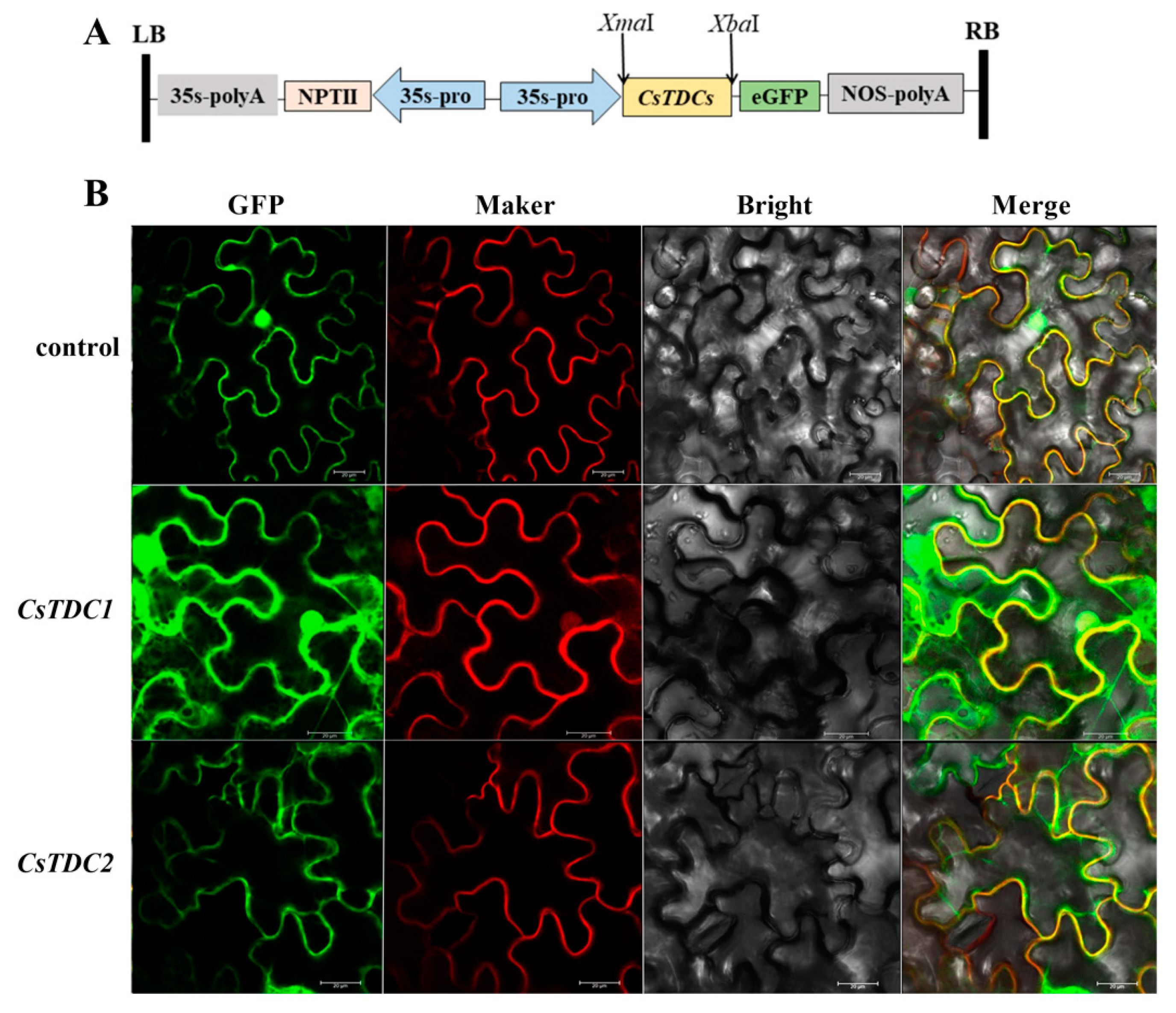
3.5. Subcellular Localization of CsTDC1 and CsTDC2
3.6. Transient Overexpression of CsTDC Genes in Tobacco Leaves Promoted Melatonin Biosynthesis
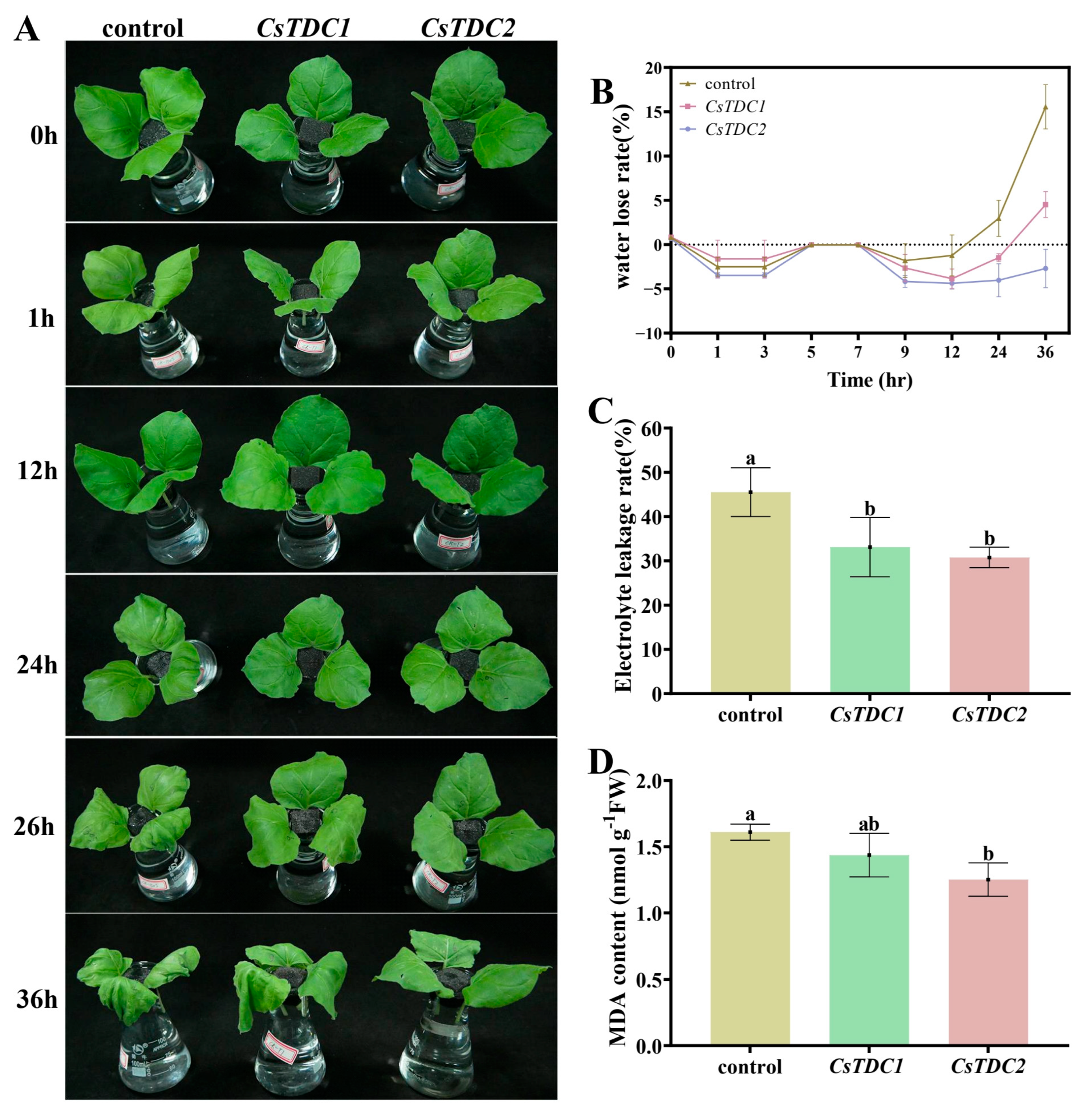
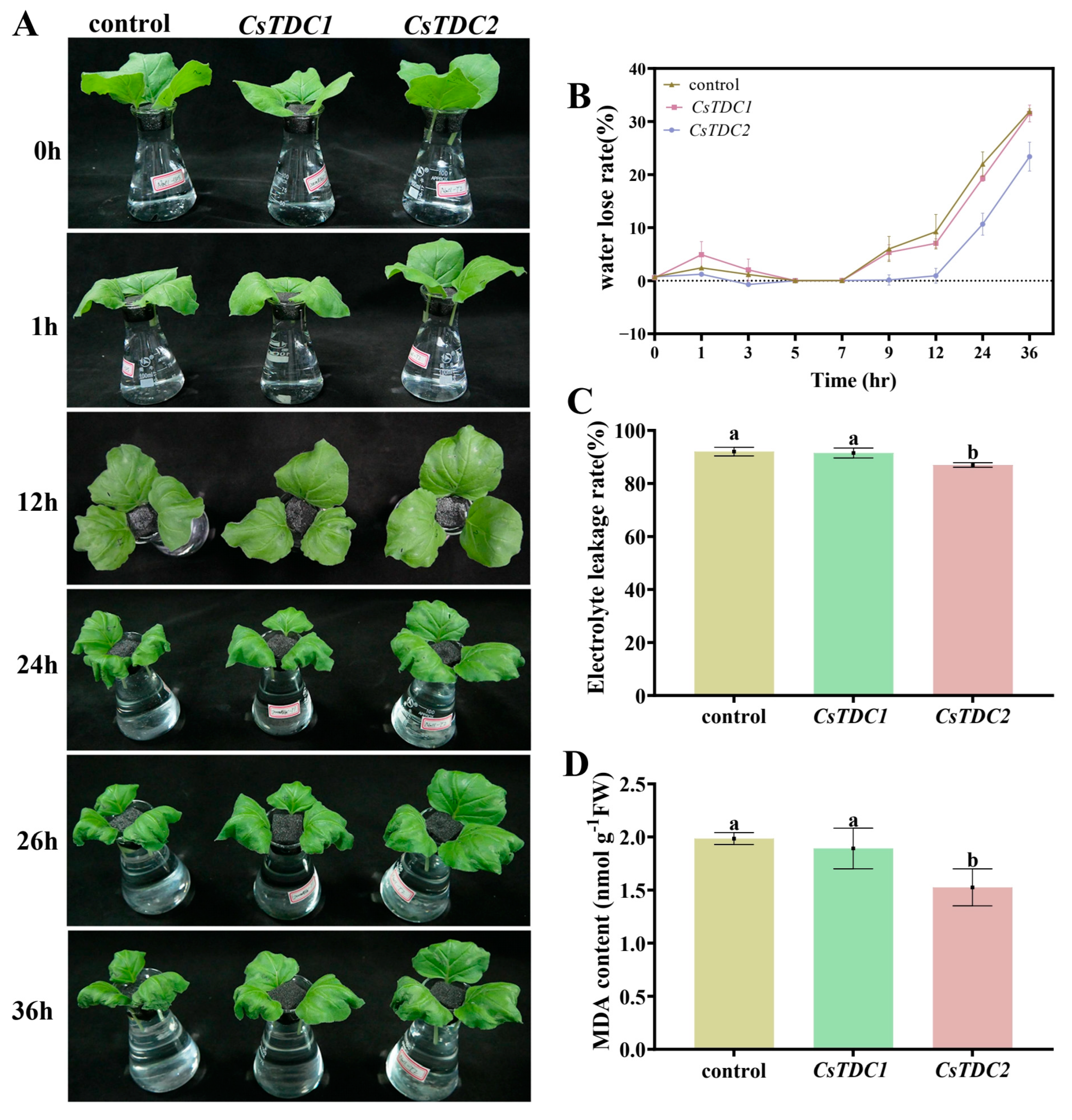
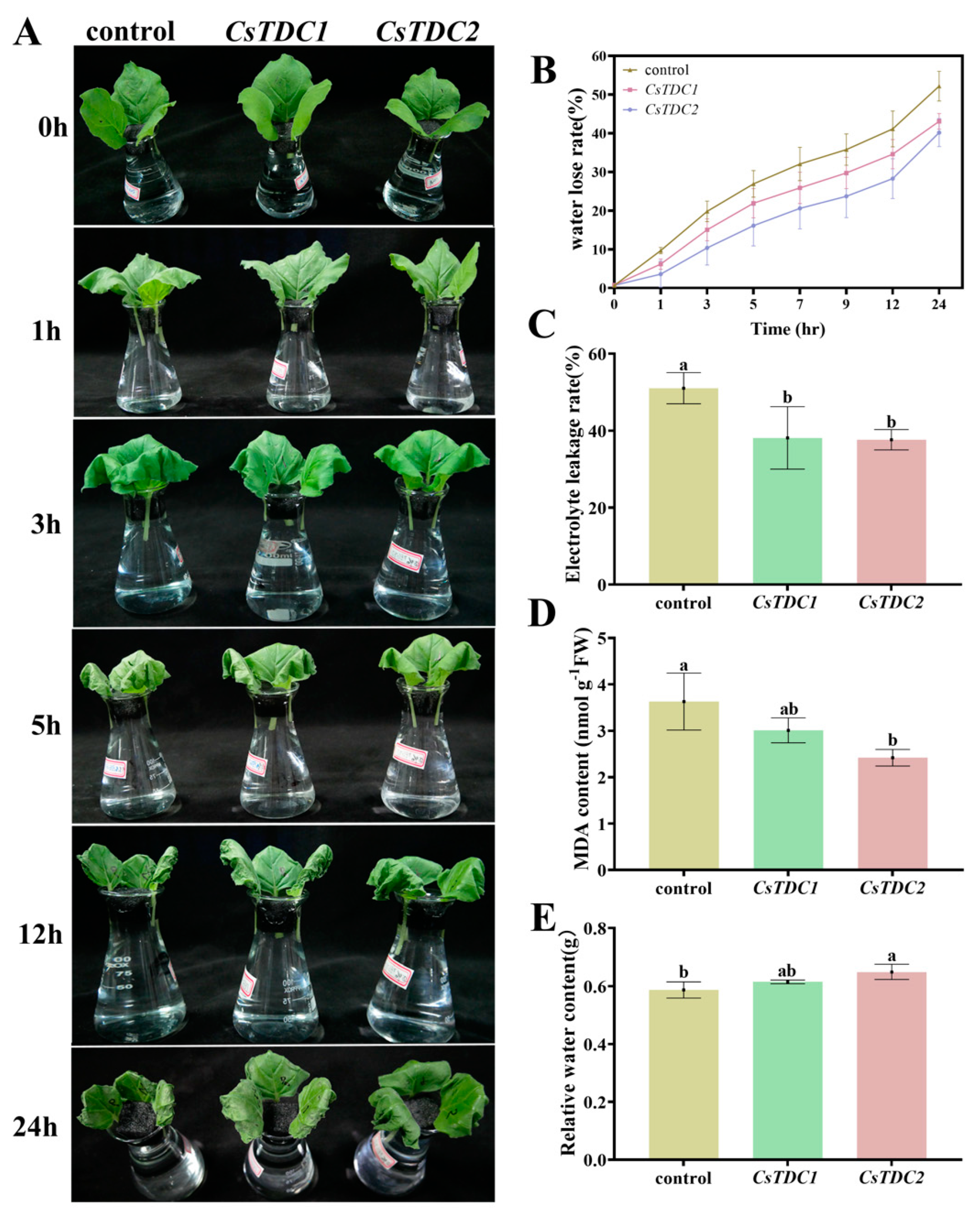
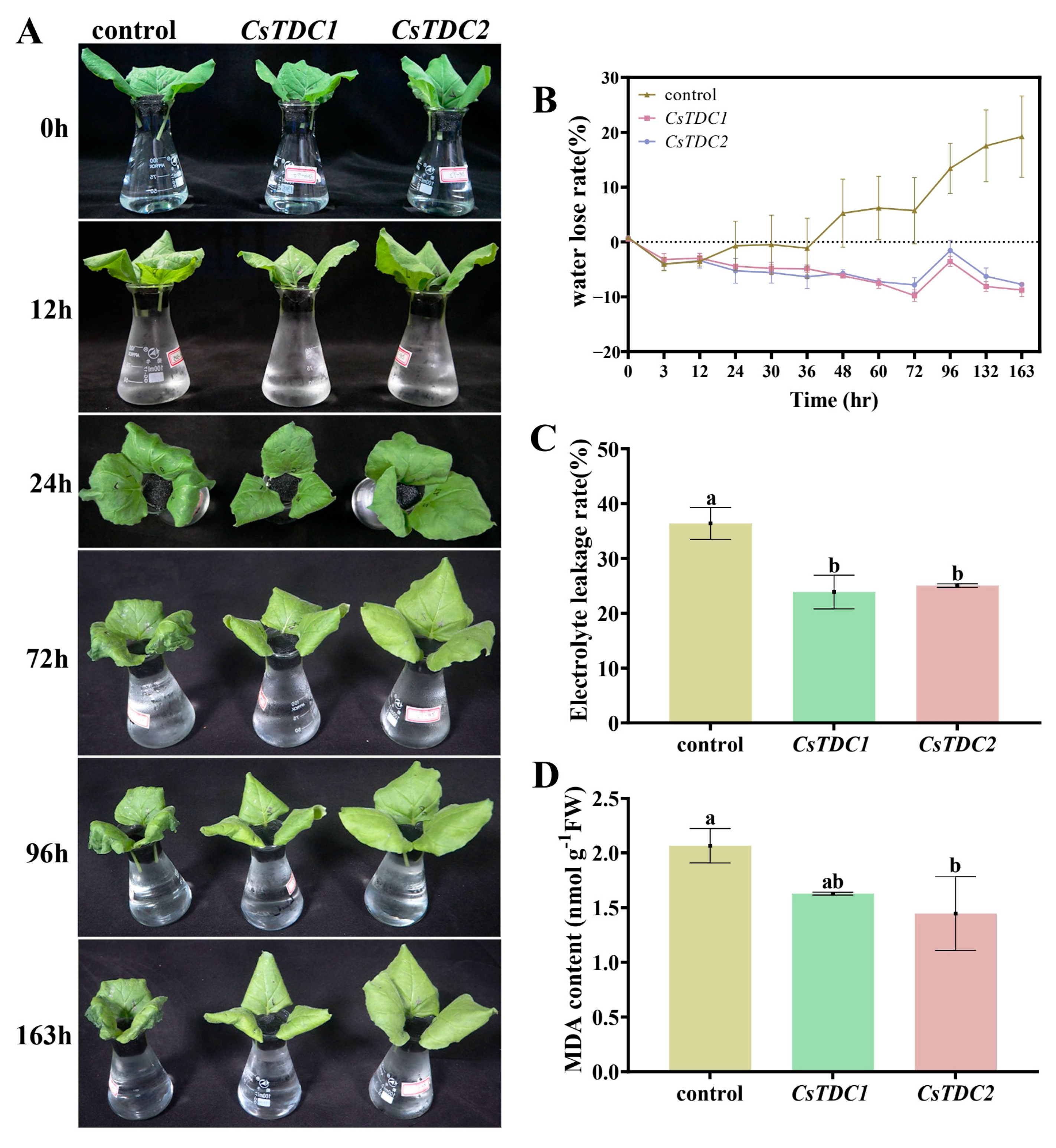
3.7. Transient Overexpression of CsTDC Genes Enhanced Abiotic Stress Tolerance in Tobacco
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Dubbels, R.; Reiter, R.; Klenke, E.; Goebel, A.; Schnakenberg, E.; Ehlers, C.; Schiwara, H.; Schloot, W. Melatonin in edible plants identified by radioimmunoassay and by high performance liquid chromatography-mass spectrometry. J. Pineal Res. 1995, 18, 28–31. [Google Scholar] [CrossRef]
- Hattori, A.; Migitaka, H.; Iigo, M.; Itoh, M.; Yamamoto, K.; Ohtani-Kaneko, R.; Hara, M.; Suzuki, T.; Reiter, R.J. Identification of melatonin in plants and its effects on plasma melatonin levels and binding to melatonin receptors in vertebrates. Biochem. Mol. Biol. Int. 1995, 35, 627–634. [Google Scholar]
- Wang, K.; Xing, Q.; Ahammed, G.J.; Zhou, J. Functions and prospects of melatonin in plant growth, yield, and quality. J. Exp. Bot. 2022, 73, 5928–5946. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.; Xu, X.; Li, L.; Sun, Q.; Wang, Q.; Huang, H.; Tong, Z.; Zhang, J. Melatonin-mediated development and abiotic stress tolerance in plants. Front. Plant Sci. 2023, 14, 1100827. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Tanveer, M.; Min, Y.; Shabala, S. Melatonin as a regulator of plant ionic homeostasis: Implications for abiotic stress tolerance. J. Exp. Bot. 2022, 73, 5886–5902. [Google Scholar] [CrossRef]
- Liu, N.; Gong, B.; Jin, Z.; Wang, X.; Wei, M.; Yang, F.; Li, Y.; Shi, Q. Sodic alkaline stress mitigation by exogenous melatonin in tomato needs nitric oxide as a downstream signal. J. Plant Physiol. 2015, 186, 68–77. [Google Scholar] [CrossRef] [PubMed]
- Zhao, D.; Zhang, X.; Wang, R.; Liu, D.; Sun, J.; Tao, J. Herbaceous peony tryptophan decarboxylase confers drought and salt stresses tolerance. Environ. Exp. Bot. 2019, 162, 345–356. [Google Scholar] [CrossRef]
- Sun, C.; Liu, L.; Wang, L.; Li, B.; Jin, C.; Lin, X. Melatonin: A master regulator of plant development and stress responses. J. Integr. Plant Biol. 2021, 63, 126–145. [Google Scholar] [CrossRef] [PubMed]
- Back, K.; Tan, D.X.; Reiter, R.J. Melatonin biosynthesis in plants: Multiple pathways catalyze tryptophan to melatonin in the cytoplasm or chloroplasts. J. Pineal Res. 2016, 61, 426–437. [Google Scholar] [CrossRef] [PubMed]
- Han, S.-W.; Shin, J.-S. Aromatic L-amino acid decarboxylases: Mechanistic features and microbial applications. Appl. Microbiol. Biot. 2022, 106, 4445–4458. [Google Scholar] [CrossRef]
- Facchini, P.J.; Huber-Allanach, K.L.; Tari, L.W. Plant aromatic L-amino acid decarboxylases: Evolution, biochemistry, regulation, and metabolic engineering applications. Phytochemistry 2000, 54, 121–138. [Google Scholar] [CrossRef]
- Torrens-Spence, M.P.; Chiang, Y.-C.; Smith, T.; Vicent, M.A.; Wang, Y.; Weng, J.-K. Structural basis for divergent and convergent evolution of catalytic machineries in plant aromatic amino acid decarboxylase proteins. Proc. Natl. Acad. Sci. USA 2020, 117, 10806–10817. [Google Scholar] [CrossRef]
- Back, K. Melatonin metabolism, signaling and possible roles in plants. Plant J. 2021, 105, 376–391. [Google Scholar] [CrossRef]
- De Luca, V.; Marineau, C.; Brisson, N. Molecular cloning and analysis of cDNA encoding a plant tryptophan decarboxylase: Comparison with animal dopa decarboxylases. Proc. Natl. Acad. Sci. USA 1989, 86, 2582–2586. [Google Scholar] [CrossRef]
- Lee, K.; Back, K. Melatonin-deficient rice plants show a common semidwarf phenotype either dependent or independent of brassinosteroid biosynthesis. J. Pineal Res. 2019, 66, e12537. [Google Scholar] [CrossRef] [PubMed]
- You, D.; Feng, Y.; Wang, C.; Sun, C.; Wang, Y.; Zhao, D.; Kai, G. Cloning, characterization, and enzymatic identification of a new tryptophan decarboxylase from Ophiorrhiza pumila. Biotechnol. Appl. Bioc. 2021, 68, 381–389. [Google Scholar] [CrossRef]
- De Masi, L.; Castaldo, D.; Pignone, D.; Servillo, L.; Facchiano, A. Experimental evidence and in silico identification of tryptophan decarboxylase in Citrus genus. Molecules 2017, 22, 272. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Zheng, M.; Long, H.; Deng, G.; Ishihara, A.; Liu, F.; Liang, J.; Pan, Z.; Yu, M. Molecular cloning and characterization of two genes encoding tryptophan decarboxylase from Aegilops variabilis with resistance to the cereal cyst nematode (Heterodera avenae) and root-knot nematode (Meloidogyne naasi). Plant Mol. Biol. Rep. 2016, 34, 273–282. [Google Scholar] [CrossRef]
- Qiao, C.; Chen, F.; Liu, Z.; Huang, T.; Li, W.; Zhang, G.; Luo, Y. Functional characterization of a catalytically promiscuous tryptophan decarboxylase from camptothecin-producing Camptotheca acuminata. Front. Plant Sci. 2022, 13, 987348. [Google Scholar] [CrossRef] [PubMed]
- Tsunoda, Y.; Hano, S.; Imoto, N.; Shibuya, T.; Ikeda, H.; Amagaya, K.; Kato, K.; Shirakawa, H.; Aso, H.; Kanayama, Y. Physiological roles of tryptophan decarboxylase revealed by overexpression of SlTDC1 in tomato. Sci. Hortic. 2021, 275, 109672. [Google Scholar] [CrossRef]
- Byeon, Y.; Park, S.; Lee, H.Y.; Kim, Y.S.; Back, K. Elevated production of melatonin in transgenic rice seeds expressing rice tryptophan decarboxylase. J. Pineal Res. 2014, 56, 275–282. [Google Scholar] [CrossRef] [PubMed]
- Kanjanaphachoat, P.; Wei, B.-Y.; Lo, S.-F.; Wang, I.-W.; Wang, C.-S.; Yu, S.-M.; Yen, M.-L.; Chiu, S.-H.; Lai, C.-C.; Chen, L.-J. Serotonin accumulation in transgenic rice by over-expressing tryptophan decarboxlyase results in a dark brown phenotype and stunted growth. Plant Mol. Biol. 2012, 78, 525–543. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Miao, L.; Huang, B.; Gao, L.; He, C.; Yan, Y.; Wang, J.; Yu, X.; Li, Y. Genome-wide identification and characterization of cucumber BPC transcription factors and their responses to abiotic stresses and exogenous phytohormones. Int. J. Mol. Sci. 2019, 20, 5048. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. 20 years of the SMART protein domain annotation resource. NAR 2018, 46, D493–D496. [Google Scholar] [CrossRef]
- Bateman, A.; Coin, L.; Durbin, R.; Finn, R.D.; Hollich, V.; Griffiths-Jones, S.; Khanna, A.; Marshall, M.; Moxon, S.; Sonnhammer, E.L. The Pfam protein families database. NAR 2004, 32, D138–D141. [Google Scholar] [CrossRef]
- Yu, C.S.; Chen, Y.C.; Lu, C.H.; Hwang, J.K. Prediction of protein subcellular localization. Proteins 2006, 64, 643–651. [Google Scholar] [CrossRef]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. NAR 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Hong, Y.; Pan, X.; Welti, R.; Wang, X. Phospholipase Dα3 is involved in the hyperosmotic response in Arabidopsis. Plant Cell 2008, 20, 803–816. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Sun, M.; Miao, L.; Di, Q.; Lv, L.; Yu, X.; Yan, Y.; He, C.; Wang, J.; Shi, A. Multifaceted regulatory functions of CsBPC2 in cucumber under salt stress conditions. Hortic. Res. 2023, 10, uhad051. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; An, B.; Shi, H.; Luo, H.; He, C. High concentration of melatonin regulates leaf development by suppressing cell proliferation and endoreduplication in Arabidopsis. Int. J. Mol. Sci. 2017, 18, 991. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Song, C.; Fang, S.; Wang, Z.; Song, S.; Jiao, J.; Wang, M.; Zheng, X.; Bai, T. Genome-wide identification and expression analysis of the ASMT gene family reveals their role in abiotic stress tolerance in apple. Sci. Hortic. 2022, 293, 110683. [Google Scholar] [CrossRef]
- Chang, J.; Guo, Y.; Yan, J.; Zhang, Z.; Yuan, L.; Wei, C.; Zhang, Y.; Ma, J.; Yang, J.; Zhang, X. The role of watermelon caffeic acid O-methyltransferase (ClCOMT1) in melatonin biosynthesis and abiotic stress tolerance. Hortic. Res. 2021, 8, 210. [Google Scholar] [CrossRef]
- Commisso, M.; Negri, S.; Gecchele, E.; Fazion, E.; Pontoriero, C.; Avesani, L.; Guzzo, F. Indolamine accumulation and TDC/T5H expression profiles reveal the complex and dynamic regulation of serotonin biosynthesis in tomato (Solanum lycopersicum L.). Front. Plant Sci. 2022, 13, 975434. [Google Scholar] [CrossRef]
- Qi, C.; Zhang, H.; Liu, Y.; Wang, X.; Dong, D.; Yuan, X.; Li, X.; Zhang, X.; Li, X.; Zhang, N. CsSNAT positively regulates salt tolerance and growth of cucumber by promoting melatonin biosynthesis. Environ. Exp. Bot. 2020, 175, 104036. [Google Scholar] [CrossRef]
- Zhao, D.; Luan, Y.; Shi, W.; Tang, Y.; Huang, X.; Tao, J. Melatonin enhances stem strength by increasing lignin content and secondary cell wall thickness in herbaceous peony. J. Exp. Bot. 2022, 73, 5974–5991. [Google Scholar] [CrossRef] [PubMed]
- Lv, Y.; Pan, J.; Wang, H.; Reiter, R.J.; Li, X.; Mou, Z.; Zhang, J.; Yao, Z.; Zhao, D.; Yu, D. Melatonin inhibits seed germination by crosstalk with abscisic acid, gibberellin, and auxin in Arabidopsis. J. Pineal Res. 2021, 70, e12736. [Google Scholar] [CrossRef]
- Zhang, N.; Zhao, B.; Zhang, H.J.; Weeda, S.; Yang, C.; Yang, Z.C.; Ren, S.; Guo, Y.D. Melatonin promotes water-stress tolerance, lateral root formation, and seed germination in cucumber (Cucumis sativus L.). J. Pineal Res. 2013, 54, 15–23. [Google Scholar] [CrossRef]
- Yang, W.-Y.; Zheng, Y.; Bahn, S.C.; Pan, X.-Q.; Li, M.-Y.; Vu, H.S.; Roth, M.R.; Scheu, B.; Welti, R.; Hong, Y.-Y. The patatin-containing phospholipase A pPLAIIα modulates oxylipin formation and water loss in Arabidopsis thaliana. Mol. Plant 2012, 5, 452–460. [Google Scholar] [CrossRef]
- Yang, N.; Sun, K.; Wang, X.; Wang, K.; Kong, X.; Gao, J.; Wen, D. Melatonin participates in selenium-enhanced cold tolerance of cucumber seedlings. Front. Plant Sci. 2021, 12, 786043. [Google Scholar] [CrossRef]
- Feng, Y.; Fu, X.; Han, L.; Xu, C.; Liu, C.; Bi, H.; Ai, X. Nitric oxide functions as a downstream signal for melatonin-induced cold tolerance in cucumber seedlings. Front. Plant Sci. 2021, 12, 686545. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Chen, Q.; Chen, W.; Guo, Q.; Xia, Y.; Wang, S.; Jing, D.; Liang, G. Physiological and transcription analyses reveal the regulatory mechanism of melatonin in inducing drought resistance in loquat (Eriobotrya japonica Lindl.) seedlings. Environ. Exp. Bot. 2021, 181, 104291. [Google Scholar] [CrossRef]
- Xu, L.; Xiang, G.; Sun, Q.; Ni, Y.; Jin, Z.; Gao, S.; Yao, Y. Melatonin enhances salt tolerance by promoting MYB108A-mediated ethylene biosynthesis in grapevines. Hortic. Res. 2019, 6, 114. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.; Jing, X.; Tang, H.; Li, X.; Gong, B.; Shi, Q. Using transcriptome to discover a novel melatonin-induced sodic alkaline stress resistant pathway in Solanum lycopersicum L. Plant Cell Physiol. 2019, 60, 2051–2064. [Google Scholar] [CrossRef]
- Fan, H.; Wang, S.; Wang, H.; Sun, M.; Wu, S.; Bao, W. Melatonin ameliorates the toxicity induced by deoxynivalenol in murine ovary granulosa cells by antioxidative and anti-inflammatory effects. Antioxidants 2021, 10, 1045. [Google Scholar] [CrossRef]


| Gene ID | Length (aa) | Molecular Weight (KD) | Chromosome | Location | pI | Strand Direction | Subcellular Location |
|---|---|---|---|---|---|---|---|
| CsaV3_1G036910 | 499 | 55.7 | 1 | 22,824,145–22,827,250 | 6.34 | – | Cytoplasm and plasma membrane |
| CsaV3_3G028450 | 486 | 54.6 | 3 | 24,862,649–24,886,753 | 5.79 | – | Cytoplasm and plasma membrane |
| Cis-Element | Function | CsTDC1 | CsTDC2 |
|---|---|---|---|
| Stress related | |||
| ARE | cis-element essential for the anaerobic induction | 2 | 5 |
| WRE3 | wound response elements | 1 | 1 |
| LTR | cis-element involved in low-temperature responsiveness | 0 | 1 |
| STRE | stress response elements | 4 | 2 |
| MYB | stress response elements | 1 | 3 |
| MYC | drought- and cold-responsive elements | 1 | 3 |
| Hormone related | |||
| ERE | ethylene-responsive element | 1 | 5 |
| ABRE | cis-element involved in abscisic acid responsiveness | 1 | 1 |
| CGTCA-motif | cis-element involved in MeJA-responsiveness | 0 | 2 |
| TCA-element | cis-element involved in salicylic acid responsiveness | 1 | 0 |
| TGA-element | auxin-responsive element | 0 | 1 |
| TGACG-motif | cis-element involved in MeJA-responsiveness | 0 | 2 |
| Development related | |||
| AACA_motif | involved in endosperm-specific negative expression | 1 | 0 |
| circadian | cis-acting regulatory element involved in circadian control | 1 | 0 |
| as-1 | cis-element involved in root-specific expression | 0 | 2 |
| CAT-box | cis-acting regulatory element related to meristem expression | 0 | 2 |
| Light related | |||
| AE-box | part of a module for light response | 0 | 1 |
| Box 4 | a conserved DNA module involved in light responsiveness | 4 | 7 |
| GATA-motif | part of a light-responsive element | 1 | 0 |
| GT1-motif | light responsive element | 0 | 3 |
| TCCC-motif | part of a light-responsive element | 1 | 0 |
| TCT-motif | part of a light-responsive element | 1 | 0 |
| G-Box | cis-element involved in light responsiveness | 1 | 2 |
| G-box | cis-element involved in light responsiveness | 0 | 1 |
| GA-motif | part of a light-responsive element | 0 | 1 |
| MRE | MYB binding site involved in light responsiveness | 4 | 1 |
| AAAC-motif | light responsive element | 1 | 0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Y.; Li, Q.; Liu, Y.; Wan, S.; Li, S. The Identification of Cucumber TDC Genes and Analyses of Their Expression and Functions under Abiotic Stress Conditions. Horticulturae 2024, 10, 307. https://doi.org/10.3390/horticulturae10040307
Zhang Y, Li Q, Liu Y, Wan S, Li S. The Identification of Cucumber TDC Genes and Analyses of Their Expression and Functions under Abiotic Stress Conditions. Horticulturae. 2024; 10(4):307. https://doi.org/10.3390/horticulturae10040307
Chicago/Turabian StyleZhang, Yiqiu, Qiuxia Li, Yu Liu, Shubei Wan, and Shuzhen Li. 2024. "The Identification of Cucumber TDC Genes and Analyses of Their Expression and Functions under Abiotic Stress Conditions" Horticulturae 10, no. 4: 307. https://doi.org/10.3390/horticulturae10040307
APA StyleZhang, Y., Li, Q., Liu, Y., Wan, S., & Li, S. (2024). The Identification of Cucumber TDC Genes and Analyses of Their Expression and Functions under Abiotic Stress Conditions. Horticulturae, 10(4), 307. https://doi.org/10.3390/horticulturae10040307






